SUMMARY
Btn1p the yeast homolog of human CLN3, which is associated with juvenile Batten disease has been implicated in several cellular pathways. Yeast cells lacking BTN1 are unable to couple ATP hydrolysis and proton pumping activities by the vacuolar ATPase (V-ATPase). In this work, we demonstrate that changes in extracellular pH result in altered transcription of BTN1, as well as a change in the glycosylation state and localization of Btn1p. At high pH, Btn1p expression was increased and the protein was mainly located in vacuolar membranes. However, low pH decreased Btn1p expression and changed its location to undefined punctate membranes. Moreover, our results suggest that differential Btn1p localization may be regulated by its glycosylation state. Underlying pathogenic implications for Batten disease of altered cellular distribution of CLN3 are discussed.
INTRODUCTION
Juvenile neuronal lipofuscinosis (JNCL), or Batten disease, is a fatal pediatric neurodegenerative disease characterized pathologically by the accumulation of hydrophobic autofluorescent storage material within lysosomes of the patient’s cells. JNCL is an autosomal recessive disorder caused by mutations in the CLN3 gene (The International Batten Disease Consortium, 1995). Despite many studies, both the precise function of CLN3 or its location remains unclear (Jarvela et al., 1998; Kyttala et al., 2004; Storch et al., 2004).
Both Schizosaccharomyces pombe and Saccharomyces cerevisiae yeast models have been developed to study the primordial cellular function of CLN3. In both yeast models, human CLN3 complements deletion of the orthologous gene BTN1, indicating the protein function is conserved. Moreover, yeast Btn1p seems to reside in the vacuole, the analogous organelle to the lysosome (Pearce and Sherman, 1997; Croopnick et al., 1998; Pearce and Sherman, 1998; Kim et al., 2003; Gachet et al., 2005). Btn1p in Saccharomyces cerevisiae has been implicated in several different cellular pathways, including vacuolar pH homeostasis, small metabolite regulation and/or transport, and nitric oxide regulation (Pearce et al., 1999; Padilla-Lopez and Pearce, 2006; Vitiello et al., 2007; Cismondi et al., 2008). A previous work has shown that vacuolar pH disturbance found in yeast cells lacking Btn1p could be the result of an inability to regulate the coupling of ATP hydrolysis and proton pumping activities of the V-ATPase, which can be directly affected by the extracellular pH (Padilla-Lopez and Pearce, 2006).
In the present study we investigated the effect of extracellular pH on transcriptional and post-translational regulation of Btn1p. We report pH-dependent alterations in BTN1 transcription, and pH-dependent glycosylation of Btn1p. Furthermore, Btn1p localizes to different subcellular compartments in response to changes in extracellular pH.
As Btn1p is homologous to human CLN3, which is defective in the childhood neurodegenerative disorder, juvenile Batten disease, we propose that further studies may deduce a potential regulatory mechanism that controls CLN3 subcellular location by modifying the glycosylation state of the protein that will help us to understand the biochemical alterations leading to lysosomal storage and pathological consequences of this devastating disease.
RESULTS
BTN1 expression is pH-dependent
To measure the effect of pH on BTN1 mRNA levels, we utilized comparative real-time reverse transcriptase PCR (qRT-PCR). BTN1+ yeast were grown overnight in medium initially at pH 6.0 or 4.0, and then shifted to the test medium pH, of either 6.0 or 4.0, until the cells reached midlog phase. RNA levels after the pH shift were normalized to RNA levels before the shift. BTN1 expression in cells shifted from pH 6.0 to 4.0 was less than 5% of that in control cells (Fig. 1). Cells grown at pH 6.0 and then re-inoculated in medium at pH 6.0 showed no change in BTN1 mRNA levels. Conversely, yeast grown at pH 4.0 and then shifted to pH 6.0 had a corresponding eightfold increase in BTN1, whereas those re-inoculated in medium at pH 4.0 showed no change in transcript levels. Thus, BTN1 transcription is significantly altered by either an increase or a decrease in the pH of the extracellular medium.
Fig. 1.

BTN1 mRNA level in response to extracellular pH determined by comparative RT2PCR. Cells were grown in medium at the initial pH value, then shifted to medium at the test pH and allowed to grow to OD600=0.5 prior to RNA isolation. Transcript levels were measured for five independent replicates and data analyzed by a Student’s t-test (P<0.0001). White bar: cells were grown at pH 4 and shifted to pH 6. Black bar: cells were grown at pH 6 and shifted to pH 4. Values are arbitrary units relative to mRNA expression of yeast cells grown and shifted to the same pH (horizontal line; attributed as 1). Cells grown at pH 6 then shifted to pH 4 showed BTN1 expression that was less than 5% of that in control cells, whereas shifting from pH 4 to pH 6 resulted in increased expression of 7.63-fold compared with controls.
Differential pH-dependent posttranslational modification of Btn1p
BTN1 mRNA levels are altered by changes in the pH of the extracellular environment (Fig. 1). To investigate whether there was a corresponding change in the amount of Btn1p protein, we immunoblotted lysates from cells expressing a functional C-terminally V5-tagged Btn1p. This plasmid construct allowed for expression of the protein at endogenous levels since it contained one kilobase of DNA sequence upstream of BTN1, which contains the promoter for BTN1, followed by the BTN1 open reading frame and V5 epitope tag. Yeast grown at pH 4.0 and pH 6.0 exhibited a significant difference in Btn1p levels (Fig. 2A). As determined by densitometry followed by normalization to an Actin loading control, cells grown at pH 6.0 had an approximate 1.7-fold increase in Btn1p compared with cells grown at pH 4.0 (Fig. 2B). However, Btn1p from cells grown at pH 6.0, but not pH 4.0 appeared as two bands, with one band at approximately 44 kDa and another at 37 kDa (Fig. 2A). It should be noted that, since no pH-dependent changes were found in the 37 kDa band expression, the increased level of Btn1p at pH 6.0 is a function of an increase of the 44 kDa band exclusively.
Fig. 2.
Btn1p is regulated by the extracellular pH. (A) Expression of Btn1p measured in whole cell extracts from cells expressing Btn1p-V5. 50 μg of protein was run on SDS-PAGE and immunoblotted using an anti-V5 antibody (1:5000). (B) Btn1p levels in cells grown at pH 6.0 were approximately 1.7-fold higher than in those grown at pH 4.0 as determined by densitometry of Btn1p bands. (C) 50 μg of whole cell lysate from cells expressing Btn1p-V5 was harvested and treated for 2 hours with 1000 units PNGase F, followed by SDS-PAGE and immunoblotting. (D) Immunoblotting for Btn1p glycosylation mutants demonstrates that loss of Btn1p glycosylation occurs when N206, N175, N206 and all three putative asparagines are mutated. Actin was used as a loading control.
Computer-based analysis of the Btn1p sequence predicts the presence of three glycosylation sites on Btn1p; N175, N178 and N206, and the human ortholog of Btn1p, CLN3 has been shown to be glycosylated (Storch et al., 2007), suggesting that Btn1p is glycosylated. To test the glycosylation state of Btn1p, cell extracts were treated with PNGase F prior to immunoblotting for Btn1p-V5. The PNGase F treatment resulted in the reduction of the 44 kDa band, indicating that Btn1p is indeed glycosylated. Furthermore, since the 44 kDa band is significantly enriched in cells grown at pH 6.0 versus pH 4.0, we predict that Btn1p glycosylation is regulated by pH (Fig. 2C).
To confirm the PNGase F treatment results and identify which residue(s) of Btn1p are glycosylated, site-directed mutagenesis was performed on the three putative glycosylation sites of our V5-tagged Btn1p construct. Each of the predicted glycosylated residues was mutated individually, and in combination, to glutamine. Whole cell extracts from cells grown at pH 4 and 6 were immunoblotted for Btn1p-V5 to determine which residues are glycosylated. Expression of the unglycosylated triple mutant was limited to cells grown at pH 4.0, with virtually no detectable Btn1p-V5 in the cells that were grown at pH 6.0, indicating that glycosylation is probably necessary for stabilization of the protein at pH 6.0 (Fig. 2D). Individually, mutations N175Q and N178Q did not noticeably alter the size of the protein, indicating that those residues by themselves are not sufficient for Btn1p glycosylation. However, N206Q alone resulted in a decrease in the 44 kDa band and a subsequent enrichment of the 37 kDa Btn1p-V5 band when cells were grown at pH 6.0, suggesting that this residue is required for glycosylation of Btn1p. Curiously, a combination of the N175Q and N178Q mutations resulted in expression of glycosylated Btn1p only in cells grown at pH 4.0 (Fig. 2D). Furthermore, combination of N178Q and N206Q resulted in an increase in the level of glycosylation of Btn1p compared with the alleles containing the N206Q residue alone, indicating that N178Q may also be glycosylated (Fig. 2D). This result is comparable with what has been observed in the human homolog to Btn1p, CLN3, which is also glycosylated (Storch et al., 2007).
It should be noted that that mutation of one or two putative sites alters the accessibility of the remaining glycosylation sites by variable site occupancy, a condition where the N-linked glycosylation of select asparagine residues occurs in a manner dependent upon the location of the target sequence, conformation of the protein and the identity of specific residues that flank the target sequence (Jones et al., 2005). Therefore, it may be that N206 is the sole residue glycosylated in Btn1p, however, when N178 is mutated in concert, N175 becomes accessible for glycosylation. This may also explain the finding that levels of glycosylation in response to extracellular pH varied from construct to construct, with several constructs appearing fully glycosylated at pH 4.0 whereas others were either not expressed or not glycosylated at each pH with no discernable pattern. We conclude from these experiments that normal Btn1p is glycosylated at residue N206 and this glycosylation is pH dependent.
Differential pH-dependent localization of Btn1p
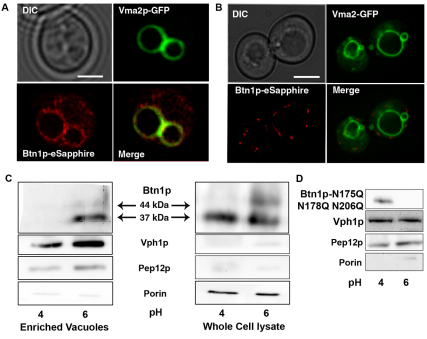
Glycosylation often shields lysosomal and vacuolar proteins from degradation by blocking the access of hydrolases to otherwise exposed residues. Given the potential role that N-linked glycosylation plays in protecting Btn1p at the vacuolar membrane, we surmised that altering the glycosylation of the protein would alter the localization of Btn1p relative to the extracellular pH. To test this, we investigated Btn1p localization at different extracellular pHs. Btn1p was C-terminally tagged with eSapphire, a UV inducible variant of GFP (Sheff and Thorn, 2004). Like the Btn1p-V5 construct, this plasmid is expressed at endogenous Btn1p levels. GFP-tagged Vma2p, a V-ATPase V1 subunit protein (kindly provided by Patricia Kane, SUNY Upstate) was used to identify the vacuole. Btn1p-GFP was previously shown to be functional and when overexpressed localized to the vacuole (Croopnick et al., 1998; Pearce and Sherman, 1998). Here, we confirm that endogenously expressed Btn1p-eSapphire also localizes to the vacuole at pH 6, as determined by Vma2p-GFP colocalization (Fig. 3A). However, Btn1p in cells grown in medium at pH 4.0 localized to puncta (Fig. 3B). The location of Vma2p remained unchanged, indicating the integrity of the vacuolar membrane was intact, and thus the alteration in Btn1p localization was not merely a by-product of altered vacuolar morphology. Attempts to show colocalization of Btn1p at pH 4.0 with standard organellar and typical vesicular trafficking intermediates failed to identify these puncta.
Fig. 3.
Localization of Btn1p is dependent upon the pH of the growth media. (A) Yeast co-expressing Btn1p-eSapphire and Vma2p-GFP grown for 6 hours in SC-URA at pH 6.0 clearly showing that Btn1p predominantly co-localizes with Vma2p in the vacuolar membrane. (B) Same strain grown in SC-URA at pH 4.0 for 6 hours showing that Btn1p is not in the vacuolar membrane but predominantly in unidentified puncta. (C) Vacuolar enrichment from strains expressing Btn1p-V5 grown at pH 4.0 and 6.0 showing that Btn1p co-fractionates with the vacuolar membrane protein Vph1p after growth at pH 6.0 but not at pH 4.0. (D) Vacuoles from btn1-Δ cells expressing Btn1p-N175Q, N178Q, N206Q-V5 (all three putative N-glycosylation sites mutated).
To confirm pH-dependent Btn1p localization, we fractionated cells expressing a functional BTN1-V5 using differential centrifugation (Fig. 3C). Vacuoles from cells that were grown at pH 4 had considerably less Btn1p compared with vacuoles from cells grown at pH 6.0. Vph1p, a component of the V0 V-ATPase subunit, was used as a vacuolar marker and loading control since V-ATPase levels do not change in response to pH (Padilla-Lopez and Pearce, 2006). Additionally, antibodies against Pep12p (endosome) and Porin (mitochondria) were used to assess the vacuolar purity.
When all putative Btn1p glycosylation sites were mutated, the level of detectable Btn1p-N175Q, N178Q, N206Q-V5 was decreased in vacuoles derived from cells grown at pH 6.0 (Fig. 3D). Interestingly, this triple-mutant Btn1p was present in the vacuolar membrane fraction of cells grown at pH 4.0 which might suggest a requirement for vacuolar localization to be trafficked to the unidentified puncta. However, we were unable to identify Btn1p-N175Q, N178Q, N206Q-eSapphire using epifluorescence microscopy (data not shown). Although not confirmed experimentally, we expect that unglycosylated Btn1p levels are too low to be seen and that it could be degraded at the vacuole. To further test whether glycosylation is required for our observed pH-dependent localization of Btn1p, Btn1p-N206Q-eSapphire was examined by microscopy at both pH 4.0 and 6.0. Again, despite numerous attempts and despite detecting the tagged protein in cell extracts we were again unable to identify any N206Q-Btn1p in the cells (data not shown). We conclude that Btn1p-N206Q probably has too short a half life in the cells. Thus, while the different glycosylation states of Btn1p at different pHs may be involved in the differential localization of this protein, we cannot definitively show that unglycosylated forms of Btn1p are unable to respond as normal Btn1p in response to pH.
DISCUSSION
Btn1p is the yeast homolog to human CLN3 (Pearce and Sherman, 1997) with mutations in CLN3 resulting in Batten disease. Previous studies have demonstrated that expression of human CLN3 in this yeast model functionally complement an absence of Btn1p (Pearce and Sherman, 1998; Kim et al., 2003) indicating that the primordial function of CLN3 is conserved in yeast and that findings from this model will certainly provides clues about the cellular consequences of not having this protein present. Studies of both yeast and mammalian systems have revealed many anomalies in vacuolar or lysosomal function. Curiously though, although both yeast Btn1p and human CLN3 have been localized to this compartment, some studies reveal that CLN3 may in fact localize to other membranes (Phillips et al., 2005).
The V-ATPase acts in concert with a variety of additional ion channels to maintain the pH homeostasis of the vacuole (Forgac, 1999; Graham et al., 2000; Kane and Parra, 2000). It establishes an electrochemical gradient across the vacuolar membrane via the coupling of two specific activities, ATP hydrolysis and pumping of protons into the vacuolar lumen (Ohsumi and Anraku, 1981; Russnak et al., 2001; Shimazu et al., 2005). Regulation of the V-ATPase complex in S. cerevisiae occurs by the reversible disassembly of its two subunits, the integral membrane pore forming V0, and the peripherally associated V1, which is responsible for the hydrolysis of ATP. ATP-driven proton transport only occurs efficiently when these two subunits are associated (Kane and Parra, 2000). At elevated growth pH (6.0) we report increased transcription of BTN1, presence of a glycosylated Btn1p and a vacuolar localization of Btn1p, which corresponds to our previous observation that coupling of V-ATPase and proton pumping is disrupted in btn1-Δ during growth at pH 6.0, but not pH 4.0 suggesting that the function of Btn1p at the vacuole predominately occurs at elevated pH.
It has been previously established that V-ATPase subunits assemble or disassemble in the presence or absence of glucose, respectively (Kane, 1995). Moreover, it has recently been shown that V-ATPase assembly can be regulated by other environmental factors (Arai et al., 1989; Oluwatosin and Kane, 1997; Qi and Forgac, 2007; Chung et al., 2003). We confirmed the previously reported vacuolar location of Btn1p, however, this only occurs when the cells are grown at an initial extracellular pH of at least 6.0. When the cells are grown at pH 4.0, Btn1p localization shifts to a series of unidentified punctate foci around the cell periphery. Interestingly, at a growth pH of 6.0 when Btn1p predominantly localizes to the vacuole, the predominant form of Btn1p is glycosylated, whereas, at pH 4.0 when Btn1p is not associated with the vacuole, Btn1p is not glycosylated, suggesting that glycosylation may be required for this vacuolar localization. Thus, it is possible that Btn1p imparts a regulatory function at the vacuole, and perhaps on coupling of V-ATPase activity with proton pumping at this elevated pH. Whether glycosylation of Btn1p and subsequent localization to the vacuole are simply the positioning of this protein in a compartment to perform its regulatory role, or whether the posttranslational modification is required for regulation of the activity of Btn1p itself will require further study.
In contrast to our results, a recent study using Schizosaccharomyces pombe reported that btn1, the S. pombe homolog of Btn1p, localizes predominately to the Golgi, trafficking to the vacuole solely to be degraded (Codlin and Mole, 2009). However, this study, unlike the previous reports on S. cerevisiae Btn1p, likely expressed btn1 above endogenous levels, which could result in altered localization patterns within the cell. Moreover, studies on Btn1 localization in S. pombe were not performed at different pHs so we cannot directly compare these studies. Nevertheless, in terms of the functional characterization of Btn1p, retention of Btn1p at the Golgi would leave it readily available for rapid transport to the vacuole when required in response to pH stress. In regard to the puncta that we see Btn1p localized to at pH 4.0, there are a large number of vesicular intermediates involved in protein transport that would be candidates for these foci and a major insufficiency in our study is that we have failed to colocalize Btn1p with any known organelle, subcellular marker or trafficking intermediate including the Golgi. Thus, we can only speculate that Btn1p is sequestered in this unidentified puncta until its activity or function is literally put to work at elevated pH (6.0) when it is mobilized to the vacuole. It is also possible that a function exists for Btn1p at lower pH (4.0) in these puncta.
Thus, our results indicate that the subcellular localization of Btn1p is pH dependent and that this localization is most likely regulated at the posttranslational level through glycosylation state. A recent study demonstrated that Btn1p functionally interacts with Sdo1p, a protein involved in ribosomal maturation (Menne et al., 2007; Vitiello et al., 2010), and that Sdo1p may regulate Btn1p function. Interestingly, interaction between Btn1p and Sdo1p only occurs in undefined punctuate spots, similar to the ones found in this work (Vitiello et al., 2010). This observation suggests a potential role of Sdo1p in regulating Btn1p by controlling its cellular localization. Whether Btn1p glycosylation state impacts or mediates a novel translational control mechanism needs to be investigated.
Most studies investigating CLN3 localization has not taken into account either pH or glycosylation as potential factors for regulating CLN3 distribution among cell membranes. Although it has been reported that changes on N-glycosylation sites of lysosomal proteins such as CLN5 could result either in altered localization or retention in the ER (Lebrun et al., 2009), previous studies with mammalian CLN3 suggest that glycosylation is not a necessary signal for delivery of the protein to the lysosomal membrane (Storch et al., 2007). Rather, a combination of posttranslational modifications, glycosylation and prenylation work together to target CLN3 to the lysosome. However, this study was performed by overexpressing CLN3, which potentially could alter the normal distribution of the protein within the cell. Nevertheless, several studies using a variety of cell types and expression systems have reported that CLN3 may localize to other non lysosomal membranes (reviewed by Phillips et al., 2005).
Finally, if CLN3 also has altered subcellular location, this may not just occur in response to environmental changes such as pH. Many different cells types exist in humans that could potentially have a need to regulate CLN3 through altered localization. One paradox in studying Batten disease that remains unsolved is the fact that autofluorescent storage materials accumulates in lysosomes. Although it is generally considered that most cell types ultimately accumulate this material, the rate of this accumulation is highly variable between cells. Our model implies that Btn1p localizes to the vacuole at elevated pH and when pH regulation is most required, and that perhaps the vacuole is the site of active Btn1p function, and that under conditions when pH regulation is not needed that Btn1p is not localized to the vacuole. As the yeast vacuole is the analogous organelle to the lysosome, it is possible that accumulation of autofluorescent storage material may occur more rapidly in the cells that have lost functional CLN3 when these cells typically have CLN3 localized to the lysosomal membrane. Thus, human cells that would ordinarily localize CLN3 to another subcompartment would be less prone to lysosomal dysfunction and thus accumulation of lysosomal storage material. If this is the case, therapeutically, cells most likely to accumulate storage material could be preferentially targeted with agents that might aid dissipation of this material.
METHODS
Media
Unless otherwise indicated, all strains were maintained in YPD medium (1% yeast extract, 2% peptone, 2% dextrose). Strains harboring a plasmid were grown in synthetic complete medium (6.7 mg/ml yeast nitrogen base without amino acids, 5 mg/ml ammonium sulfate, 2% dextrose, and all amino acids except uracil, asparagine, glutamine, proline, alanine, cystine and glycine) minus the appropriate auxotrophic markers. Microscopy was performed on cells grown in YNB medium (0.67% yeast nitrogen base without amino acids, 2% dextrose and necessary auxotrophic amino acids) plus 50 mM MES to buffer at the indicated pHs.
Yeast strains
Wild-type yeast strain B11718 was purchased from ResGen. The btn1-Δ strain was created by S.P.V. by homologous recombination of a KAN cassette at the BTN1 loci followed by removal of the cassette by loxP-mediated excision (Guldener et al., 1996). The Vma2p-GFP strain was kindly donated by Patricia Kane (SUNY Upstate Medical University, NY).
Plasmids
BTN1 was amplified from plasmid pAA1793 using forward and reverse primers engineered with a SalI tag (italics) on both (forward: 5′-GTCGACATGAGTGACAAATCTCAT-3′; reverse: 5′-GTC-GACTTCCATCCTACACCAAAG-3′). The amplicon was then subcloned into pCR Blunt (Invitrogen) followed by digestion with SalI to excise BTN1. Full-length BTN1 was then ligated into the SalI site of pKT150 in frame with the eSapphire fluorophore. BTN1 orientation was confirmed by digestion with PvuI. The BTN1-eSapphire gene was then amplified [forward primer (Btn1nhe1F) 5′-GCTAGCTAGGTGACACTATAGAAC-3′, and reverse primer (Btn1Xho1R) 5′-CTCGAGCGCTTATTTAGAAGTGGC-3′) and cloned into NheI and XhoI sites in the MCS of pAA1793. Directionality was confirmed by restriction digest using HindIII. Constructs were then sequenced to ensure no random mutations were introduced during the PCR process.
Quantification of BTN1 mRNA using RT-PCR
Yeast cells were grown to midlog phase in media at either pH 4 or 6 and either inoculated into media at the same pH or inoculated into media at the other pH. RNA was extracted using standard methods. Random hexamer priming with or without reverse transcriptase was used to synthesize cDNA from 5 μg RNA using the First Strand Synthesis III kit according to the manufacturer’s protocol (Invitrogen). Reactions containing 2 μl cDNA, 0.25 μM primers specific to ACT1 (forward: 5′-ATGGTCGGTATGGGTCAAAA-3′; reverse: 5′-AACCAGCGTAAATTGGAACG-3′) or BTN1 (forward: 5′-CCT-GACTTACCAAAGTCTTC-3′; reverse: 5′-TCATTCTCATAA-GATGTCCA-3′) and iQ SYBR Green Supermix (Bio-Rad) were run on an icycler (Bio-Rad) using the following reaction parameters: 95°C for 2 minutes, 1 cycle, then 95°C for 20 seconds, 51°C for 20 seconds, 72°C for 45 seconds, 40 cycles, followed by 95°C for 1 minute and 55°C for 1 minute. Changes in transcript levels were analyzed using REST software (Pfaffl et al., 2002). PCR efficiencies were analyzed using DART-PCR software (Peirson et al., 2003), with both sets of primers having 100% efficiency.
Microscopy
Yeast expressing Vma2p-GFP and Btn1p-eSapphire were grown overnight in SC-URA. Cells were harvested the next day, washed twice in ddH2O and re-innoculated into YNB pH 4.0 or 6.0 and grown to an OD600 of 0.8. Microscopy was performed using an Olympus BX61 epifluorescence microscope with a 1.3 NA, 100× oil immersion lens, using a DAPI filter to visualize Btn1p-eSapphire and a 488 nm green filter for GFP. Image deconvolution was performed using Autoquant X2 (Media Cybernetics). Image overlays and additional processing was done using ImageJ.
Membrane preparation
600 ml of yeast cells were grown to OD600 1.0–1.4 at 30°C with orbital shaking. Yeast were collected, washed and resuspended in 10 ml buffer A (1.2 M sorbitol, 20 mM KPO4). 2 mg/ml 20T Zymolase was added and the samples were incubated at 30°C for 1 hour. After spheroplasting, cells were homogenized by 25 strokes in a Dounce homogenizer. Unlysed cells and other debris were pelleted by centrifugation at 500 g for 5 minutes, with the supernatant collected and used as the whole cell lysate. Vacuoles were isolated as previously described (Ohsumi and Anraku, 1981). Protein concentration was determined by Lowry assay (Lowry et al., 1951). Membrane fractions were analyzed by SDS-PAGE and western bloting using monoclonal antibodies against Vma2p (13D11) and Vph1p (10D7), alkaline phosphatase (1D3), carboxypeptidase Y (10A5), all from Molecular Probes, and the V5 epitope antibody from Invitrogen (all 1:5000).
PNGaseF treatment
PNGaseF treatment was performed according to the manufacturer’s instructions (New England Biolabs).
Site-directed mutagenesis of BTN1
Putative glycosylation sites were mutated by dpn1-mediated mutagenesis as previously described (Fisher and Pei, 1997).
Acknowledgments
This research was funded in part by NIH grant RO1-NS036610 and the Beat Batten Foundation.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
D.M.W. performed experiments, analyzed the data and prepared the manuscript; S.P.-L., S.P.V. and D.A.P. interpreted the data and prepared the manuscript.
REFERENCES
- Arai H., Pink S., Forgac M. (1989). Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry 28, 3075–3082 [DOI] [PubMed] [Google Scholar]
- Chung J.H., Lester R.L., Dickson R.C. (2003). Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J. Biol. Chem. 278, 28872–28881 [DOI] [PubMed] [Google Scholar]
- Cismondi I.A., Kohan R., Ghio A., Ramirez A.M., Halac I.N. (2008). Gene symbol: CLN6. Disease: Neuronal ceroid lipofuscinosis, late Infantile. Hum. Genet. 124, 324. [PubMed] [Google Scholar]
- Codlin S., Mole S.E. (2009). S. pombe btn1, the orthologue of the Batten disease gene CLN3, is required for vacuole protein sorting of Cpy1p and Golgi exit of Vps10p. J. Cell. Sci. 122, 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croopnick J.B., Choi H.C., Mueller D.M. (1998). The subcellular location of the yeast Saccharomyces cerevisiae homologue of the protein defective in the juvenile form of Batten disease. Biochem. Biophys. Res. Commun. 250, 335–341 [DOI] [PubMed] [Google Scholar]
- Fisher C.L., Pei G.K. (1997). Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23, 570–571, 574 [DOI] [PubMed] [Google Scholar]
- Forgac M. (1999). Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 274, 12951–12954 [DOI] [PubMed] [Google Scholar]
- Gachet Y., Codlin S., Hyams J.S., Mole S.E. (2005). btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell. Sci. 118, 5525–5536 [DOI] [PubMed] [Google Scholar]
- Graham L.A., Powell B., Stevens T.H. (2000). Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J. Exp. Biol. 203, 61–70 [DOI] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H. (1996). A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvela I., Sainio M., Rantamaki T., Olkkonen V.M., Carpen O., Peltonen L., Jalanko A. (1998). Biosynthesis and intracellular targeting of the CLN3 protein defective in Batten disease. Hum. Mol. Genet. 7, 85–90 [DOI] [PubMed] [Google Scholar]
- Jones D.C., Mehlert A., Guther M.L., Ferguson M.A. (2005). Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J. Biol. Chem. 280, 35929–35942 [DOI] [PubMed] [Google Scholar]
- Kane P.M. (1995). Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270, 17025–17032 [PubMed] [Google Scholar]
- Kane P.M., Parra K.J. (2000). Assembly and regulation of the yeast vacuolar H(+)-ATPase. J. Exp. Biol. 203, 81–87 [DOI] [PubMed] [Google Scholar]
- Kim Y., Ramirez-Montealegre D., Pearce D.A. (2003). A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl. Acad. Sci. USA 100, 15458–15462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A., Ihrke G., Vesa J., Schell M.J., Luzio J.P. (2004). Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol. Biol. Cell 15, 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun A.H., Storch S., Ruschendorf F., Schmiedt M.L., Kyttala A., Mole S.E., Kitzmuller C., Saar K., Mewasingh L.D., Boda V., et al. (2009). Retention of lysosomal protein CLN5 in the endoplasmic reticulum causes neuronal ceroid lipofuscinosis in Asian sibship. Hum. Mutat. 30, E651–E661 [DOI] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- Menne T.F., Goyenechea B., Sanchez-Puig N., Wong C.C., Tonkin L.M., Ancliff P.J., Brost R.L., Costanzo M., Boone C., Warren A.J. (2007). The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 39, 486–495 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y., Anraku Y. (1981). Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 256, 2079–2082 [PubMed] [Google Scholar]
- Oluwatosin Y.E., Kane P.M. (1997). Mutations in the CYS4 gene provide evidence for regulation of the yeast vacuolar H+-ATPase by oxidation and reduction in vivo. J. Biol. Chem. 272, 28149–28157 [DOI] [PubMed] [Google Scholar]
- Padilla-Lopez S., Pearce D.A. (2006). Saccharomyces cerevisiae lacking Btn1p modulate vacuolar ATPase activity to regulate pH imbalance in the vacuole. J. Biol. Chem. 281, 10273–10280 [DOI] [PubMed] [Google Scholar]
- Pearce D.A., Sherman F. (1997). BTN1, a yeast gene corresponding to the human gene responsible for Batten’s disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast 13, 691–697 [DOI] [PubMed] [Google Scholar]
- Pearce D.A., Sherman F. (1998). A yeast model for the study of Batten disease. Proc. Natl. Acad. Sci. USA 95, 6915–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D.A., Ferea T., Nosel S.A., Das B., Sherman F. (1999). Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 22, 55–58 [DOI] [PubMed] [Google Scholar]
- Peirson S.N., Butler J.N., Foster R.G. (2003). Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S.N., Benedict J.W., Weimer J.M., Pearce D.A. (2005). CLN3, the protein associated with batten disease: structure, function and localization. J. Neurosci. Res. 79, 573–583 [DOI] [PubMed] [Google Scholar]
- Qi J., Forgac M. (2007). Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J. Biol. Chem. 282, 24743–24751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R., Konczal D., McIntire S.L. (2001). A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 276, 23849–23857 [DOI] [PubMed] [Google Scholar]
- Sheff M.A., Thorn K.S. (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 [DOI] [PubMed] [Google Scholar]
- Shimazu M., Sekito T., Akiyama K., Ohsumi Y., Kakinuma Y. (2005). A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J. Biol. Chem. 280, 4851–4857 [DOI] [PubMed] [Google Scholar]
- Storch S., Pohl S., Braulke T. (2004). A dileucine motif and a cluster of acidic amino acids in the second cytoplasmic domain of the batten disease-related CLN3 protein are required for efficient lysosomal targeting. J. Biol. Chem. 279, 53625–53634 [DOI] [PubMed] [Google Scholar]
- Storch S., Pohl S., Quitsch A., Falley K., Braulke T. (2007). C-terminal prenylation of the CLN3 membrane glycoprotein is required for efficient endosomal sorting to lysosomes. Traffic 8, 431–444 [DOI] [PubMed] [Google Scholar]
- The International Batten Disease Consortium (1995). Isolation of a novel gene underlying Batten disease, CLN3. Cell 82, 949–957 [DOI] [PubMed] [Google Scholar]
- Vitiello S.P., Wolfe D.M., Pearce D.A. (2007). Absence of Btn1p in the yeast model for juvenile Batten disease may cause arginine to become toxic to yeast cells. Hum. Mol. Genet. 16, 1007–1016 [DOI] [PubMed] [Google Scholar]
- Vitiello S.P., Benedict J.W., Padilla-Lopez S., Pearce D.A. (2010). Interaction between Sdo1p and Btn1p in the Saccharomyces cerevisiae model for Batten disease. Hum. Mol. Genet. 19, 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]