Abstract
The asymmetric unit of the title compound, C7H5ClO3, contains two molecules; both feature an intramolecular O—H⋯O hydrogen bond, which generates an S(6) ring. In the crystal, both molecules form inversion dimers linked by pairs of O—H⋯O hydrogen bonds with R 2 2(8) ring motifs. The dimers are interlinked by C—H⋯O interactions.
Related literature
For biological background, see: Bright et al. (2010 ▶): Fattorusso et al. (2005 ▶); Miki et al. (2002 ▶). For a related structure, see: Raza et al. (2010 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶).
Experimental
Crystal data
C7H5ClO3
M r = 172.56
Monoclinic,
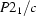
a = 23.526 (2) Å
b = 3.7972 (4) Å
c = 16.7321 (16) Å
β = 104.852 (5)°
V = 1444.8 (2) Å3
Z = 8
Mo Kα radiation
μ = 0.48 mm−1
T = 296 K
0.34 × 0.12 × 0.10 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.879, T max = 0.888
14048 measured reflections
3697 independent reflections
2444 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.138
S = 1.03
3697 reflections
211 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.38 e Å−3
Δρmin = −0.50 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810042042/hb5691sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810042042/hb5691Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2i | 0.83 (3) | 1.88 (3) | 2.710 (2) | 171 (3) |
| O3—H3⋯O2 | 0.80 (3) | 1.92 (3) | 2.620 (2) | 146 (3) |
| O4—H4A⋯O5ii | 0.93 (3) | 1.76 (3) | 2.694 (2) | 175 (2) |
| O6—H6⋯O5 | 0.87 (3) | 1.80 (3) | 2.606 (2) | 154 (3) |
| C5—H5⋯O6iii | 0.93 | 2.55 | 3.311 (3) | 139 |
Symmetry codes: (i) 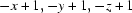 ; (ii)
; (ii) 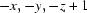 ; (iii)
; (iii) 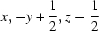 .
.
Acknowledgments
The authors acknowledge the provision of funds for the purchase of the diffractometer and encouragement by Dr Muhammad Akram Chaudhary, Vice Chancellor, University of Sargodha, Pakistan. ARR also acknowledges the Higher Education Commission, Government of Pakistan, for generous support of a research project (20–819).
supplementary crystallographic information
Comment
The benzoxazepines have a plethora of biological activities ranging from anti-inflammatory effect (Miki et al., 2002) to degenerative diseases like AIDS (Fattorusso et al., 2005) and cancer (Bright et al., 2010). Salicylic acid is an attractive substrate for the synthesis of 4,1-benzoxazepine. The objective of this work is to synthesize a variety of substituted salicylic acid derivatives as precursors for the asymmetric synthesis of 4,1-benzoxazepines by chiral-pool strategy.
We have reported the crystal structure of 2-methylamino-5-nitrobenzoic acid (Raza et al., 2010) and in continuation to synthesize substituted benzoic acid, the title compound (I, Fig. 1) is being reported.
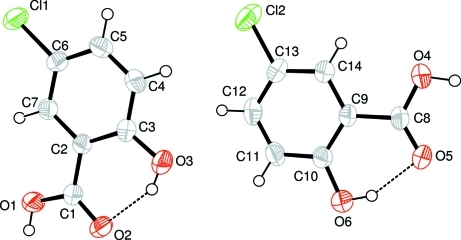
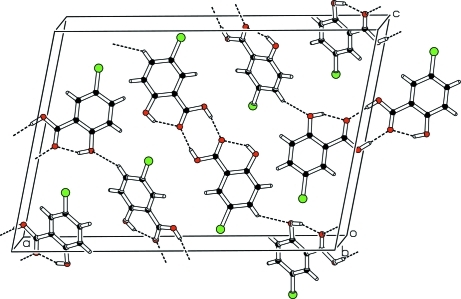
The title compound consists of two molecules in the crystallographic asymmetric unit which differ from each other geometrically. Both molecules, A (C1—C7/O1/O2/O3/CL1) and B (C8—C14/O4/O5/O6/CL2) are close to planar with r. m. s deviations of 0.023 and 0.007 Å, respectively. The dihedral angle between A/B is 1.77 (4)°. In each molecule, there exists an S(6) ring motif (Bernstein et al., 1995) due to intramolecular H-bonding of O—H···O type (Table 1, Fig. 1). The molecules form dimers with themselves due to intermolecular H-bondings of O—H···O type (Table 1, Fig. 2) with R22(8) ring motifs. These dimers are interlinked with each other due to H-bonding of C—H···O type (Fig. 2).
Experimental
A solution of Cu2Cl2 (3.46 g, 0.0375 mol) in HCl (10 ml) was added as drops to the diazonium salt of 5-amino-2-hydroxybenzioc acid (3.825 g, 0.025 mol), which was prepared by adding ice chilled aqueous solution of NaNO2 (2.58 g, 0.0375 mol) to the solution of 5-amino-2-hydroxybenzoic acid in EtOAc and H2SO4 (2.8 ml, 4.9 g, 0.05 mol). The temperature of the reaction mixture was controlled below 268 K. After the complete addition of Cu2Cl2, the reaction mixture was refluxed for one hour, cooled to room temperature, neutralized with aqueous NaHCO3 (10%) and extracted with EtOAc (3 × 25 ml). The organic layer was combined, dried over anhydrous Na2SO4, filtered, concentrated under reduced pressure and left for 48 h to afford light yellow needles of (I).
Refinement
The coordinates of hydroxy H-atoms are refined. The aryl H-atoms were positioned geometrically with (C—H = 0.93 Å) and refined as riding with Uiso(H) = xUeq(C, O), where x = 1.2 for all H atoms.
Figures
Fig. 1.
View of the title compound with displacement ellipsoids drawn at the 50% probability level. H-atoms are shown by small circles of arbitrary radius. The dotted lines indicate the intramolecular H-bonds.
Fig. 2.
The partial packing (PLATON; Spek, 2009) which shows that molecules form dimers and are interlinked.
Crystal data
| C7H5ClO3 | F(000) = 704 |
| Mr = 172.56 | Dx = 1.587 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 931 reflections |
| a = 23.526 (2) Å | θ = 2.8–26.0° |
| b = 3.7972 (4) Å | µ = 0.48 mm−1 |
| c = 16.7321 (16) Å | T = 296 K |
| β = 104.852 (5)° | Needle, light yellow |
| V = 1444.8 (2) Å3 | 0.34 × 0.12 × 0.10 mm |
| Z = 8 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 3697 independent reflections |
| Radiation source: fine-focus sealed tube | 2444 reflections with I > 2σ(I) |
| graphite | Rint = 0.047 |
| Detector resolution: 7.40 pixels mm-1 | θmax = 28.7°, θmin = 3.6° |
| ω scans | h = −31→31 |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | k = −4→5 |
| Tmin = 0.879, Tmax = 0.888 | l = −22→22 |
| 14048 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.138 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.073P)2] where P = (Fo2 + 2Fc2)/3 |
| 3697 reflections | (Δ/σ)max < 0.001 |
| 211 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.50 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.37512 (3) | 0.69966 (16) | 0.07736 (3) | 0.0501 (2) | |
| O1 | 0.49060 (7) | 0.6286 (5) | 0.38781 (10) | 0.0531 (6) | |
| O2 | 0.43512 (7) | 0.3536 (5) | 0.45885 (9) | 0.0497 (5) | |
| O3 | 0.32656 (7) | 0.1864 (5) | 0.38353 (10) | 0.0504 (6) | |
| C1 | 0.44188 (9) | 0.4711 (6) | 0.39317 (12) | 0.0372 (6) | |
| C2 | 0.39581 (8) | 0.4495 (5) | 0.31555 (11) | 0.0325 (6) | |
| C3 | 0.34043 (9) | 0.3106 (5) | 0.31526 (13) | 0.0363 (6) | |
| C4 | 0.29691 (10) | 0.3000 (6) | 0.24180 (14) | 0.0427 (7) | |
| C5 | 0.30768 (9) | 0.4181 (6) | 0.16973 (13) | 0.0425 (7) | |
| C6 | 0.36245 (9) | 0.5504 (5) | 0.16965 (12) | 0.0359 (6) | |
| C7 | 0.40631 (9) | 0.5677 (5) | 0.24125 (12) | 0.0355 (6) | |
| Cl2 | 0.13798 (3) | 0.73267 (17) | 0.21674 (4) | 0.0624 (3) | |
| O4 | 0.01191 (7) | 0.1719 (5) | 0.39980 (11) | 0.0604 (6) | |
| O5 | 0.06701 (7) | 0.1273 (5) | 0.52956 (10) | 0.0553 (6) | |
| O6 | 0.17545 (7) | 0.3485 (5) | 0.56545 (10) | 0.0538 (6) | |
| C8 | 0.06128 (9) | 0.2160 (6) | 0.45705 (14) | 0.0402 (7) | |
| C9 | 0.10935 (9) | 0.3749 (5) | 0.42825 (12) | 0.0349 (6) | |
| C10 | 0.16403 (9) | 0.4322 (6) | 0.48436 (13) | 0.0383 (7) | |
| C11 | 0.20911 (9) | 0.5788 (6) | 0.45616 (14) | 0.0447 (7) | |
| C12 | 0.20146 (10) | 0.6683 (6) | 0.37478 (15) | 0.0453 (8) | |
| C13 | 0.14736 (10) | 0.6137 (6) | 0.31970 (13) | 0.0413 (7) | |
| C14 | 0.10148 (9) | 0.4679 (6) | 0.34525 (13) | 0.0397 (7) | |
| H1 | 0.5163 (12) | 0.635 (7) | 0.4326 (18) | 0.0636* | |
| H3 | 0.3557 (12) | 0.189 (7) | 0.4210 (18) | 0.0604* | |
| H4 | 0.25996 | 0.21164 | 0.24137 | 0.0512* | |
| H5 | 0.27810 | 0.40924 | 0.12074 | 0.0510* | |
| H7 | 0.44297 | 0.65761 | 0.24065 | 0.0425* | |
| H4A | −0.0156 (12) | 0.058 (8) | 0.4222 (16) | 0.0725* | |
| H6 | 0.1424 (13) | 0.258 (7) | 0.5696 (19) | 0.0646* | |
| H11 | 0.24542 | 0.61752 | 0.49328 | 0.0536* | |
| H12 | 0.23233 | 0.76478 | 0.35677 | 0.0543* | |
| H14 | 0.06536 | 0.43138 | 0.30750 | 0.0476* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0588 (4) | 0.0597 (4) | 0.0332 (3) | −0.0012 (3) | 0.0142 (3) | 0.0039 (2) |
| O1 | 0.0349 (9) | 0.0858 (13) | 0.0354 (9) | −0.0154 (8) | 0.0034 (7) | 0.0057 (8) |
| O2 | 0.0443 (9) | 0.0749 (11) | 0.0300 (8) | −0.0096 (8) | 0.0096 (7) | 0.0028 (7) |
| O3 | 0.0427 (10) | 0.0719 (11) | 0.0399 (9) | −0.0131 (8) | 0.0167 (7) | 0.0048 (8) |
| C1 | 0.0340 (11) | 0.0433 (12) | 0.0359 (10) | −0.0004 (9) | 0.0119 (9) | −0.0002 (9) |
| C2 | 0.0314 (10) | 0.0349 (10) | 0.0318 (10) | 0.0006 (8) | 0.0092 (8) | −0.0018 (8) |
| C3 | 0.0356 (11) | 0.0370 (11) | 0.0395 (11) | −0.0004 (9) | 0.0153 (9) | −0.0010 (8) |
| C4 | 0.0324 (11) | 0.0477 (13) | 0.0473 (13) | −0.0058 (9) | 0.0091 (10) | −0.0004 (9) |
| C5 | 0.0358 (12) | 0.0474 (13) | 0.0405 (12) | −0.0017 (10) | 0.0028 (9) | −0.0011 (9) |
| C6 | 0.0408 (11) | 0.0353 (10) | 0.0319 (10) | 0.0017 (9) | 0.0098 (9) | −0.0008 (8) |
| C7 | 0.0331 (11) | 0.0386 (11) | 0.0358 (10) | −0.0009 (9) | 0.0109 (8) | −0.0028 (8) |
| Cl2 | 0.0825 (5) | 0.0671 (5) | 0.0442 (3) | −0.0108 (3) | 0.0284 (3) | 0.0069 (3) |
| O4 | 0.0345 (9) | 0.0962 (14) | 0.0511 (10) | −0.0143 (9) | 0.0121 (8) | 0.0148 (9) |
| O5 | 0.0414 (9) | 0.0852 (13) | 0.0429 (9) | −0.0100 (8) | 0.0173 (7) | 0.0136 (8) |
| O6 | 0.0402 (9) | 0.0821 (12) | 0.0387 (9) | −0.0076 (9) | 0.0093 (7) | 0.0030 (8) |
| C8 | 0.0313 (11) | 0.0481 (13) | 0.0447 (12) | 0.0001 (9) | 0.0159 (10) | 0.0023 (9) |
| C9 | 0.0326 (11) | 0.0374 (11) | 0.0386 (11) | 0.0008 (9) | 0.0163 (9) | 0.0002 (8) |
| C10 | 0.0357 (11) | 0.0429 (12) | 0.0378 (11) | 0.0017 (9) | 0.0123 (9) | −0.0023 (9) |
| C11 | 0.0317 (11) | 0.0528 (14) | 0.0499 (13) | −0.0065 (10) | 0.0113 (10) | −0.0034 (10) |
| C12 | 0.0413 (13) | 0.0420 (12) | 0.0597 (15) | −0.0090 (10) | 0.0261 (11) | −0.0056 (10) |
| C13 | 0.0517 (14) | 0.0385 (11) | 0.0400 (11) | −0.0019 (10) | 0.0230 (10) | −0.0004 (9) |
| C14 | 0.0374 (11) | 0.0450 (12) | 0.0386 (11) | −0.0018 (9) | 0.0130 (9) | 0.0005 (9) |
Geometric parameters (Å, °)
| Cl1—C6 | 1.741 (2) | C4—C5 | 1.370 (3) |
| Cl2—C13 | 1.740 (2) | C5—C6 | 1.383 (3) |
| O1—C1 | 1.316 (3) | C6—C7 | 1.368 (3) |
| O2—C1 | 1.234 (3) | C4—H4 | 0.9300 |
| O3—C3 | 1.351 (3) | C5—H5 | 0.9300 |
| O1—H1 | 0.83 (3) | C7—H7 | 0.9300 |
| O3—H3 | 0.80 (3) | C8—C9 | 1.468 (3) |
| O4—C8 | 1.313 (3) | C9—C14 | 1.399 (3) |
| O5—C8 | 1.233 (3) | C9—C10 | 1.402 (3) |
| O6—C10 | 1.352 (3) | C10—C11 | 1.383 (3) |
| O4—H4A | 0.93 (3) | C11—C12 | 1.370 (3) |
| O6—H6 | 0.87 (3) | C12—C13 | 1.382 (3) |
| C1—C2 | 1.465 (3) | C13—C14 | 1.375 (3) |
| C2—C7 | 1.402 (3) | C11—H11 | 0.9300 |
| C2—C3 | 1.404 (3) | C12—H12 | 0.9300 |
| C3—C4 | 1.384 (3) | C14—H14 | 0.9300 |
| C1—O1—H1 | 113.3 (19) | C6—C7—H7 | 120.00 |
| C3—O3—H3 | 108 (2) | C2—C7—H7 | 120.00 |
| C8—O4—H4A | 109.9 (16) | O5—C8—C9 | 122.4 (2) |
| C10—O6—H6 | 103 (2) | O4—C8—C9 | 115.14 (19) |
| O1—C1—C2 | 115.11 (17) | O4—C8—O5 | 122.4 (2) |
| O2—C1—C2 | 122.3 (2) | C8—C9—C14 | 120.85 (19) |
| O1—C1—O2 | 122.58 (19) | C8—C9—C10 | 119.74 (18) |
| C1—C2—C7 | 120.64 (18) | C10—C9—C14 | 119.4 (2) |
| C3—C2—C7 | 119.43 (18) | O6—C10—C11 | 117.7 (2) |
| C1—C2—C3 | 119.93 (17) | O6—C10—C9 | 123.2 (2) |
| C2—C3—C4 | 119.24 (19) | C9—C10—C11 | 119.09 (19) |
| O3—C3—C4 | 117.2 (2) | C10—C11—C12 | 121.5 (2) |
| O3—C3—C2 | 123.58 (19) | C11—C12—C13 | 119.3 (2) |
| C3—C4—C5 | 120.7 (2) | Cl2—C13—C12 | 118.86 (18) |
| C4—C5—C6 | 120.2 (2) | Cl2—C13—C14 | 120.14 (17) |
| Cl1—C6—C7 | 119.94 (17) | C12—C13—C14 | 121.0 (2) |
| C5—C6—C7 | 120.69 (19) | C9—C14—C13 | 119.7 (2) |
| Cl1—C6—C5 | 119.37 (16) | C10—C11—H11 | 119.00 |
| C2—C7—C6 | 119.77 (19) | C12—C11—H11 | 119.00 |
| C5—C4—H4 | 120.00 | C11—C12—H12 | 120.00 |
| C3—C4—H4 | 120.00 | C13—C12—H12 | 120.00 |
| C4—C5—H5 | 120.00 | C9—C14—H14 | 120.00 |
| C6—C5—H5 | 120.00 | C13—C14—H14 | 120.00 |
| O1—C1—C2—C3 | −175.24 (19) | O4—C8—C9—C10 | −179.8 (2) |
| O1—C1—C2—C7 | 4.5 (3) | O4—C8—C9—C14 | −0.2 (3) |
| O2—C1—C2—C3 | 4.0 (3) | O5—C8—C9—C10 | −0.4 (3) |
| O2—C1—C2—C7 | −176.3 (2) | O5—C8—C9—C14 | 179.1 (2) |
| C1—C2—C3—O3 | −1.1 (3) | C8—C9—C10—O6 | −0.1 (3) |
| C1—C2—C3—C4 | 178.4 (2) | C8—C9—C10—C11 | 179.3 (2) |
| C7—C2—C3—O3 | 179.12 (19) | C14—C9—C10—O6 | −179.7 (2) |
| C7—C2—C3—C4 | −1.3 (3) | C14—C9—C10—C11 | −0.2 (3) |
| C1—C2—C7—C6 | −178.98 (19) | C8—C9—C14—C13 | −179.5 (2) |
| C3—C2—C7—C6 | 0.8 (3) | C10—C9—C14—C13 | 0.1 (3) |
| O3—C3—C4—C5 | −179.4 (2) | O6—C10—C11—C12 | 179.4 (2) |
| C2—C3—C4—C5 | 1.0 (3) | C9—C10—C11—C12 | −0.1 (3) |
| C3—C4—C5—C6 | −0.1 (3) | C10—C11—C12—C13 | 0.5 (3) |
| C4—C5—C6—Cl1 | −179.91 (18) | C11—C12—C13—Cl2 | 179.34 (18) |
| C4—C5—C6—C7 | −0.5 (3) | C11—C12—C13—C14 | −0.7 (3) |
| Cl1—C6—C7—C2 | 179.56 (15) | Cl2—C13—C14—C9 | −179.65 (17) |
| C5—C6—C7—C2 | 0.2 (3) | C12—C13—C14—C9 | 0.4 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.83 (3) | 1.88 (3) | 2.710 (2) | 171 (3) |
| O3—H3···O2 | 0.80 (3) | 1.92 (3) | 2.620 (2) | 146 (3) |
| O4—H4A···O5ii | 0.93 (3) | 1.76 (3) | 2.694 (2) | 175 (2) |
| O6—H6···O5 | 0.87 (3) | 1.80 (3) | 2.606 (2) | 154 (3) |
| C5—H5···O6iii | 0.93 | 2.55 | 3.311 (3) | 139 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y, −z+1; (iii) x, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5691).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bright, S. A., McElligott, A. M., O’Connell, J. W., O’Connor, L., Carroll, P., Campiani, G., Deininger, M. W., Conneally, E., Lawler, M., Williams, D. C. & Zisterer, D. M. (2010). Br. J. Cancer, 102, 1474–1482. [DOI] [PMC free article] [PubMed]
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Fattorusso, C., Gemma, S., Butini, S., Huleatt, P., Catalanotti, B., Persico, M., Angelis, M. D., Fiorini, I., Nacci, V., Ramunno, A., Rodriquez, M., Greco, G., Novellino, E., Bergamini, A., Marini, S., Coletta, M., Maga, G., Spadari, S. & Campiani, G. (2005). J. Med. Chem.48, 7153–7165. [DOI] [PubMed]
- Miki, T., Kori, M., Mabuchi, H., Tozawa, R. I., Nishimoto, T., Sugiyama, Y., Teshima, K. & Yukimasa, H. (2002). J. Med. Chem.45, 4571–4580. [DOI] [PubMed]
- Raza, A. R., Rubab, S. L. & Tahir, M. N. (2010). Acta Cryst. E66, o1484. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810042042/hb5691sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810042042/hb5691Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report