Abstract
Each of the cyclohexenone rings in the title compound, C23H27NO6, adopts a half-chair (envelope) conformation with the C atom carrying the methyl groups lying out of the plane defined by the five remaining C atoms; the O atoms lie to the same side of the molecule as the respective >C(CH3)2 atoms. The hydroxy and carbonyl O atoms face each other and are orientated to allow for the formation of two intramolecular O—H⋯O hydrogen bonds. In the crystal, the presence of C—H⋯O contacts leads to the formation of supramolecular chains along the b axis. These aggregate into layers that stack along c.
Related literature
For the biological activity and uses of xanthenes, see: Jonathan et al. (1988 ▶); Pohlers & Scaiano (1997 ▶); Hilderbrand & Weissleder (2007 ▶). For background to xanthenedione derivatives, see: Hatakeyama et al. (1988 ▶); Shchekotikhin & Nikolaeva (2006 ▶).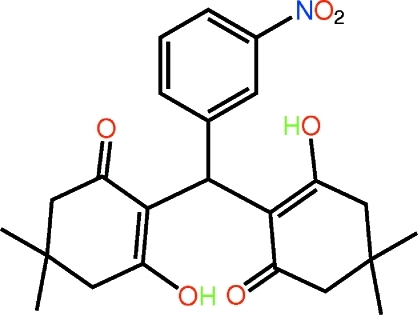
Experimental
Crystal data
C23H27NO6
M r = 413.46
Monoclinic,
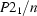
a = 14.2326 (10) Å
b = 8.6505 (6) Å
c = 16.8410 (12) Å
β = 97.796 (1)°
V = 2054.3 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.30 × 0.30 × 0.20 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.792, T max = 0.862
18883 measured reflections
4717 independent reflections
3928 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.112
S = 1.03
4717 reflections
279 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.37 e Å−3
Δρmin = −0.23 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810040079/hb5662sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810040079/hb5662Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1o⋯O4 | 0.85 (1) | 1.80 (1) | 2.6392 (13) | 167 (2) |
| O3—H3o⋯O2 | 0.86 (1) | 1.75 (1) | 2.5985 (13) | 170 (2) |
| C5—H5a⋯O4i | 0.99 | 2.34 | 3.2781 (16) | 158 |
| C9—H9⋯O6ii | 1.00 | 2.56 | 3.3431 (16) | 135 |
| C21—H21⋯O2iii | 0.95 | 2.44 | 3.3121 (16) | 152 |
Symmetry codes: (i) 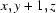 ; (ii)
; (ii) 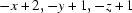 ; (iii)
; (iii) 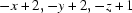 .
.
Acknowledgments
VV is grateful to the DST, India, for funding through the Young Scientist Scheme (Fast Track Proposal). The authors are also grateful to the University of Malaya for support of the crystallographic facility.
supplementary crystallographic information
Comment
Xanthenes are known for to possess various biological properties including anti-bacterial, anti-viral and anti-inflammatory activities (Jonathan et al., 1988). In particular, xanthenedione derivatives constitute a structural unit in several natural products (Hatakeyama et al., 1988), and they are valuable synthons because of the inherent reactivity of the in-built pyran ring (Shchekotikhin & Nikolaeva, 2006). Xanthene derivatives are also very useful and important organic compounds widely used as dyes (Hilderbrand & Weissleder, 2007), in laser technologies, and as fluorescent materials for visualization of biomolecules (Pohlers & Scaiano, 1997).
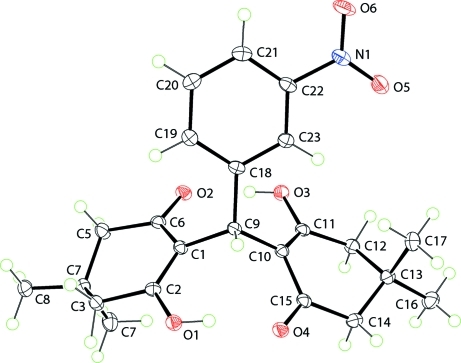
The molecular structure of the title compound, Fig. 1, features two cyclohexene rings, each with a half-chair conformation as, in each case, the C4 and C13 atoms, i.e. carrying two methyl groups, lie above the respective least-squares plane through the remaining five carbon atoms. For each ring, the O atoms lie to the same side of the molecule as the C4 or C13 atoms. The hydroxyl- and carbonyl-O atoms of one cyclohexene ring face the carbonyl- and hydroxyl-O atoms of the other to allow for the formation of intramolecular O—H···O hydrogen bonds, Table 1. The nitro group is co-planar with the benzene ring to which it is attached as seen in the O5—N1—C22—C21 torsion angle of -176.02 (11) °. The nitro-substituted benzene ring occupies a position almost side-on to both cyclohexene rings.
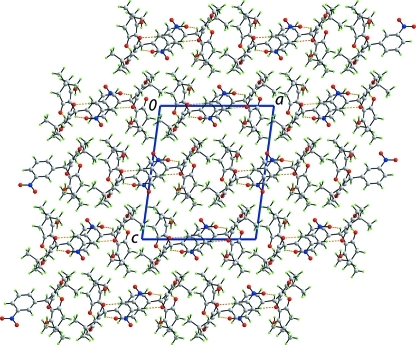
The most prominent intermolecular interactions in the crystal packing are of the type C—H···O, Table 1. These serve to link molecules into a supramolecular chain along the b axis, Fig. 2. The chains pack into layers in the ab plane which stack along the c axis, Fig. 3.
Experimental
A mixture of 3-nitrobenaldehyde (0.377 g, 0.0025 mol), dimedone (0.7 g, 0.005 mol) was refluxed in acetonitrile (20 ml) for 3 h. The progress of the reaction was monitored by TLC. After completion of the reaction, the solution was left for 2 days to precipitate the solid product. The product was recrystallized by slow evaporation of its acetonitrile solution which yielded colourless blocks of (I). Yield: 72%. M.pt. 443–445 K.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95 to 1.00 Å) and were included in the refinement in the riding model approximation, with Uiso(H) set to 1.2 to 1.5Uequiv(C). The O-bound H atoms were refined with the distance restraint O—H = 0.84±0.1 Å, and with Uiso(H) = 1.5Uequiv(O).
Figures
Fig. 1.
Molecular structure of (I) showing displacement ellipsoids at the 50% probability level.
Fig. 2.
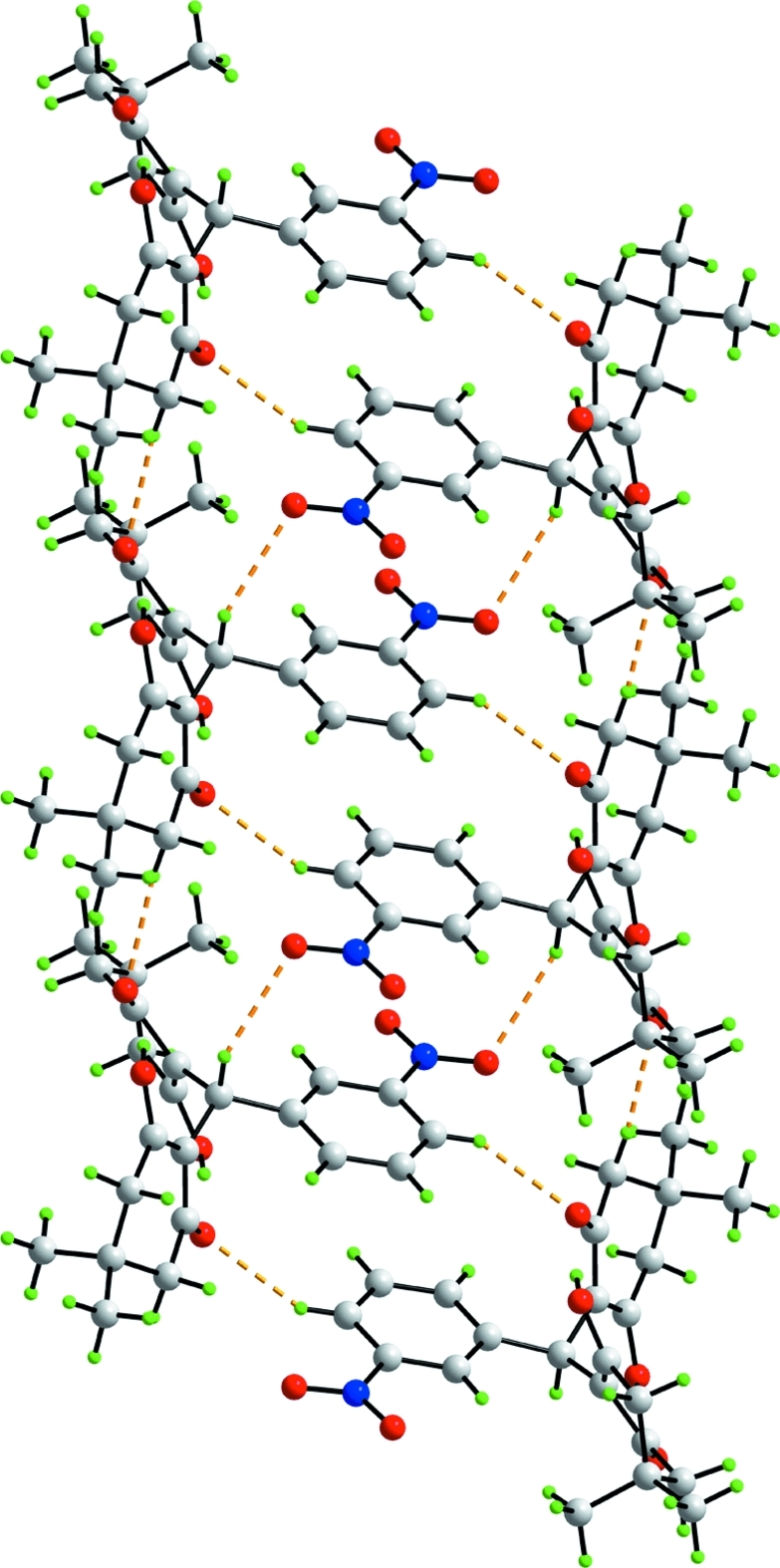
Supramolecular chain aligned along the b axis sustained by C—H···O contacts (shown as orange dashed lines).
Fig. 3.
Unit-cell contents for (I) viewed in projection along the b axis showing the stacking of layers along the c axis. The C—H···O contacts are shown as orange dashed lines.
Crystal data
| C23H27NO6 | F(000) = 880 |
| Mr = 413.46 | Dx = 1.337 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 5854 reflections |
| a = 14.2326 (10) Å | θ = 4.5–28.2° |
| b = 8.6505 (6) Å | µ = 0.10 mm−1 |
| c = 16.8410 (12) Å | T = 100 K |
| β = 97.796 (1)° | Block, colourless |
| V = 2054.3 (3) Å3 | 0.30 × 0.30 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX CCD diffractometer | 4717 independent reflections |
| Radiation source: fine-focus sealed tube | 3928 reflections with I > 2σ(I) |
| graphite | Rint = 0.036 |
| ω scans | θmax = 27.5°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −16→18 |
| Tmin = 0.792, Tmax = 0.862 | k = −11→11 |
| 18883 measured reflections | l = −21→21 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.112 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0605P)2 + 0.67P] where P = (Fo2 + 2Fc2)/3 |
| 4717 reflections | (Δ/σ)max = 0.001 |
| 279 parameters | Δρmax = 0.37 e Å−3 |
| 2 restraints | Δρmin = −0.23 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.65593 (7) | 0.65589 (10) | 0.67564 (5) | 0.0159 (2) | |
| H1o | 0.6569 (15) | 0.594 (2) | 0.6365 (9) | 0.048 (6)* | |
| O2 | 0.73862 (7) | 1.02234 (10) | 0.49100 (5) | 0.0170 (2) | |
| O3 | 0.73479 (7) | 0.82683 (11) | 0.37346 (5) | 0.0170 (2) | |
| H3o | 0.7407 (13) | 0.8846 (19) | 0.4154 (8) | 0.036 (5)* | |
| O4 | 0.63116 (6) | 0.46796 (10) | 0.55110 (5) | 0.0163 (2) | |
| O5 | 0.99653 (7) | 0.53999 (12) | 0.35663 (6) | 0.0237 (2) | |
| O6 | 1.13360 (7) | 0.63202 (12) | 0.40630 (6) | 0.0259 (2) | |
| N1 | 1.04746 (8) | 0.61823 (13) | 0.40582 (7) | 0.0176 (2) | |
| C1 | 0.71426 (9) | 0.83167 (14) | 0.58531 (7) | 0.0129 (2) | |
| C2 | 0.67059 (9) | 0.79995 (14) | 0.65085 (7) | 0.0134 (2) | |
| C3 | 0.63844 (9) | 0.92120 (14) | 0.70461 (7) | 0.0153 (3) | |
| H3A | 0.6898 | 0.9405 | 0.7493 | 0.018* | |
| H3B | 0.5829 | 0.8817 | 0.7279 | 0.018* | |
| C4 | 0.61160 (9) | 1.07463 (14) | 0.66175 (7) | 0.0149 (3) | |
| C5 | 0.69030 (10) | 1.11682 (15) | 0.61177 (8) | 0.0184 (3) | |
| H5A | 0.6701 | 1.2088 | 0.5788 | 0.022* | |
| H5B | 0.7480 | 1.1453 | 0.6486 | 0.022* | |
| C6 | 0.71497 (9) | 0.98965 (15) | 0.55741 (8) | 0.0145 (3) | |
| C7 | 0.51615 (10) | 1.05773 (17) | 0.60799 (8) | 0.0217 (3) | |
| H7A | 0.4669 | 1.0306 | 0.6409 | 0.032* | |
| H7B | 0.5209 | 0.9761 | 0.5684 | 0.032* | |
| H7C | 0.4997 | 1.1557 | 0.5803 | 0.032* | |
| C8 | 0.60339 (10) | 1.20199 (15) | 0.72332 (8) | 0.0187 (3) | |
| H8A | 0.5531 | 1.1754 | 0.7554 | 0.028* | |
| H8B | 0.5880 | 1.3002 | 0.6955 | 0.028* | |
| H8C | 0.6638 | 1.2121 | 0.7585 | 0.028* | |
| C9 | 0.76179 (8) | 0.70610 (14) | 0.54208 (7) | 0.0124 (2) | |
| H9 | 0.7692 | 0.6185 | 0.5812 | 0.015* | |
| C10 | 0.70261 (9) | 0.63781 (14) | 0.46833 (7) | 0.0129 (2) | |
| C11 | 0.69851 (9) | 0.69205 (14) | 0.39210 (8) | 0.0139 (3) | |
| C12 | 0.65447 (9) | 0.60340 (15) | 0.31987 (8) | 0.0162 (3) | |
| H12A | 0.6947 | 0.6153 | 0.2767 | 0.019* | |
| H12B | 0.5916 | 0.6487 | 0.3006 | 0.019* | |
| C13 | 0.64204 (9) | 0.43071 (15) | 0.33584 (8) | 0.0162 (3) | |
| C14 | 0.59406 (10) | 0.41744 (16) | 0.41182 (8) | 0.0189 (3) | |
| H14A | 0.5281 | 0.4556 | 0.3999 | 0.023* | |
| H14B | 0.5913 | 0.3070 | 0.4268 | 0.023* | |
| C15 | 0.64394 (9) | 0.50637 (14) | 0.48217 (7) | 0.0140 (2) | |
| C16 | 0.57961 (10) | 0.35702 (17) | 0.26472 (8) | 0.0228 (3) | |
| H16A | 0.5718 | 0.2467 | 0.2752 | 0.034* | |
| H16B | 0.6097 | 0.3698 | 0.2161 | 0.034* | |
| H16C | 0.5174 | 0.4074 | 0.2573 | 0.034* | |
| C17 | 0.73790 (10) | 0.34775 (15) | 0.34742 (8) | 0.0201 (3) | |
| H17A | 0.7282 | 0.2378 | 0.3577 | 0.030* | |
| H17B | 0.7786 | 0.3932 | 0.3931 | 0.030* | |
| H17C | 0.7683 | 0.3592 | 0.2989 | 0.030* | |
| C18 | 0.86411 (8) | 0.74648 (14) | 0.53038 (7) | 0.0130 (2) | |
| C19 | 0.91956 (9) | 0.84390 (15) | 0.58406 (8) | 0.0156 (3) | |
| H19 | 0.8912 | 0.8949 | 0.6248 | 0.019* | |
| C20 | 1.01514 (9) | 0.86788 (15) | 0.57927 (8) | 0.0175 (3) | |
| H20 | 1.0509 | 0.9350 | 0.6165 | 0.021* | |
| C21 | 1.05918 (9) | 0.79508 (15) | 0.52075 (8) | 0.0171 (3) | |
| H21 | 1.1245 | 0.8105 | 0.5170 | 0.021* | |
| C22 | 1.00352 (9) | 0.69910 (15) | 0.46819 (7) | 0.0150 (3) | |
| C23 | 0.90774 (9) | 0.67322 (14) | 0.47154 (7) | 0.0144 (3) | |
| H23 | 0.8724 | 0.6062 | 0.4340 | 0.017* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0196 (5) | 0.0135 (4) | 0.0154 (4) | −0.0009 (4) | 0.0058 (4) | 0.0015 (3) |
| O2 | 0.0193 (5) | 0.0155 (5) | 0.0174 (4) | −0.0004 (4) | 0.0067 (4) | 0.0025 (3) |
| O3 | 0.0208 (5) | 0.0146 (4) | 0.0161 (5) | −0.0015 (4) | 0.0048 (4) | 0.0019 (4) |
| O4 | 0.0186 (5) | 0.0138 (4) | 0.0176 (4) | −0.0009 (3) | 0.0067 (4) | 0.0010 (3) |
| O5 | 0.0225 (5) | 0.0281 (5) | 0.0216 (5) | 0.0016 (4) | 0.0070 (4) | −0.0041 (4) |
| O6 | 0.0152 (5) | 0.0308 (6) | 0.0342 (6) | 0.0022 (4) | 0.0126 (4) | 0.0012 (5) |
| N1 | 0.0169 (6) | 0.0171 (5) | 0.0204 (6) | 0.0040 (4) | 0.0078 (4) | 0.0052 (4) |
| C1 | 0.0115 (6) | 0.0124 (6) | 0.0151 (6) | 0.0015 (4) | 0.0027 (5) | 0.0000 (4) |
| C2 | 0.0113 (6) | 0.0141 (6) | 0.0146 (6) | 0.0008 (5) | 0.0012 (4) | 0.0017 (5) |
| C3 | 0.0167 (6) | 0.0159 (6) | 0.0140 (6) | 0.0006 (5) | 0.0048 (5) | −0.0002 (5) |
| C4 | 0.0151 (6) | 0.0155 (6) | 0.0142 (6) | 0.0017 (5) | 0.0029 (5) | −0.0015 (5) |
| C5 | 0.0225 (7) | 0.0127 (6) | 0.0216 (6) | 0.0015 (5) | 0.0091 (5) | 0.0001 (5) |
| C6 | 0.0113 (6) | 0.0149 (6) | 0.0176 (6) | 0.0000 (5) | 0.0035 (5) | 0.0001 (5) |
| C7 | 0.0183 (7) | 0.0257 (7) | 0.0201 (7) | 0.0054 (5) | −0.0003 (5) | −0.0049 (5) |
| C8 | 0.0190 (6) | 0.0175 (6) | 0.0198 (6) | 0.0019 (5) | 0.0042 (5) | −0.0034 (5) |
| C9 | 0.0123 (6) | 0.0120 (6) | 0.0134 (6) | 0.0007 (4) | 0.0036 (4) | 0.0010 (4) |
| C10 | 0.0106 (5) | 0.0126 (6) | 0.0158 (6) | 0.0013 (5) | 0.0029 (5) | −0.0004 (5) |
| C11 | 0.0110 (6) | 0.0130 (6) | 0.0183 (6) | 0.0019 (4) | 0.0045 (5) | 0.0010 (5) |
| C12 | 0.0159 (6) | 0.0179 (6) | 0.0149 (6) | 0.0005 (5) | 0.0021 (5) | 0.0010 (5) |
| C13 | 0.0161 (6) | 0.0173 (6) | 0.0153 (6) | −0.0027 (5) | 0.0028 (5) | −0.0012 (5) |
| C14 | 0.0181 (6) | 0.0199 (7) | 0.0194 (6) | −0.0062 (5) | 0.0051 (5) | −0.0017 (5) |
| C15 | 0.0120 (6) | 0.0133 (6) | 0.0173 (6) | 0.0029 (5) | 0.0042 (5) | 0.0002 (5) |
| C16 | 0.0231 (7) | 0.0258 (7) | 0.0191 (7) | −0.0059 (6) | 0.0012 (5) | −0.0037 (6) |
| C17 | 0.0226 (7) | 0.0164 (6) | 0.0214 (7) | 0.0025 (5) | 0.0034 (5) | −0.0026 (5) |
| C18 | 0.0118 (6) | 0.0134 (6) | 0.0145 (6) | 0.0019 (5) | 0.0036 (5) | 0.0047 (4) |
| C19 | 0.0167 (6) | 0.0157 (6) | 0.0145 (6) | 0.0018 (5) | 0.0028 (5) | 0.0015 (5) |
| C20 | 0.0162 (6) | 0.0170 (6) | 0.0185 (6) | −0.0020 (5) | −0.0005 (5) | 0.0020 (5) |
| C21 | 0.0131 (6) | 0.0175 (6) | 0.0209 (6) | 0.0007 (5) | 0.0030 (5) | 0.0059 (5) |
| C22 | 0.0142 (6) | 0.0145 (6) | 0.0173 (6) | 0.0039 (5) | 0.0058 (5) | 0.0049 (5) |
| C23 | 0.0145 (6) | 0.0138 (6) | 0.0152 (6) | 0.0005 (5) | 0.0030 (5) | 0.0023 (5) |
Geometric parameters (Å, °)
| O1—C2 | 1.3395 (15) | C9—H9 | 1.0000 |
| O1—H1o | 0.851 (9) | C10—C11 | 1.3607 (17) |
| O2—C6 | 1.2436 (15) | C10—C15 | 1.4481 (17) |
| O3—C11 | 1.3299 (15) | C11—C12 | 1.5014 (18) |
| O3—H3o | 0.860 (9) | C12—C13 | 1.5323 (18) |
| O4—C15 | 1.2443 (15) | C12—H12A | 0.9900 |
| O5—N1 | 1.2275 (15) | C12—H12B | 0.9900 |
| O6—N1 | 1.2308 (14) | C13—C17 | 1.5304 (18) |
| N1—C22 | 1.4704 (16) | C13—C16 | 1.5303 (18) |
| C1—C2 | 1.3662 (17) | C13—C14 | 1.5343 (18) |
| C1—C6 | 1.4456 (17) | C14—C15 | 1.5072 (18) |
| C1—C9 | 1.5169 (16) | C14—H14A | 0.9900 |
| C2—C3 | 1.4971 (17) | C14—H14B | 0.9900 |
| C3—C4 | 1.5341 (17) | C16—H16A | 0.9800 |
| C3—H3A | 0.9900 | C16—H16B | 0.9800 |
| C3—H3B | 0.9900 | C16—H16C | 0.9800 |
| C4—C8 | 1.5282 (17) | C17—H17A | 0.9800 |
| C4—C5 | 1.5337 (18) | C17—H17B | 0.9800 |
| C4—C7 | 1.5340 (18) | C17—H17C | 0.9800 |
| C5—C6 | 1.5029 (17) | C18—C23 | 1.3913 (17) |
| C5—H5A | 0.9900 | C18—C19 | 1.3988 (18) |
| C5—H5B | 0.9900 | C19—C20 | 1.3893 (18) |
| C7—H7A | 0.9800 | C19—H19 | 0.9500 |
| C7—H7B | 0.9800 | C20—C21 | 1.3886 (19) |
| C7—H7C | 0.9800 | C20—H20 | 0.9500 |
| C8—H8A | 0.9800 | C21—C22 | 1.3821 (19) |
| C8—H8B | 0.9800 | C21—H21 | 0.9500 |
| C8—H8C | 0.9800 | C22—C23 | 1.3902 (18) |
| C9—C10 | 1.5223 (17) | C23—H23 | 0.9500 |
| C9—C18 | 1.5360 (16) | ||
| C2—O1—H1o | 109.0 (14) | O3—C11—C12 | 112.91 (11) |
| C11—O3—H3o | 108.3 (13) | C10—C11—C12 | 123.24 (11) |
| O6—N1—O5 | 123.66 (11) | C11—C12—C13 | 113.65 (10) |
| O6—N1—C22 | 117.97 (11) | C11—C12—H12A | 108.8 |
| O5—N1—C22 | 118.36 (11) | C13—C12—H12A | 108.8 |
| C2—C1—C6 | 118.45 (11) | C11—C12—H12B | 108.8 |
| C2—C1—C9 | 121.69 (11) | C13—C12—H12B | 108.8 |
| C6—C1—C9 | 119.85 (11) | H12A—C12—H12B | 107.7 |
| O1—C2—C1 | 123.08 (11) | C17—C13—C16 | 108.50 (11) |
| O1—C2—C3 | 112.96 (10) | C17—C13—C12 | 110.94 (11) |
| C1—C2—C3 | 123.90 (11) | C16—C13—C12 | 109.71 (11) |
| C2—C3—C4 | 113.50 (10) | C17—C13—C14 | 110.40 (11) |
| C2—C3—H3A | 108.9 | C16—C13—C14 | 110.16 (11) |
| C4—C3—H3A | 108.9 | C12—C13—C14 | 107.12 (11) |
| C2—C3—H3B | 108.9 | C15—C14—C13 | 113.70 (11) |
| C4—C3—H3B | 108.9 | C15—C14—H14A | 108.8 |
| H3A—C3—H3B | 107.7 | C13—C14—H14A | 108.8 |
| C8—C4—C5 | 109.24 (11) | C15—C14—H14B | 108.8 |
| C8—C4—C7 | 109.03 (11) | C13—C14—H14B | 108.8 |
| C5—C4—C7 | 110.68 (11) | H14A—C14—H14B | 107.7 |
| C8—C4—C3 | 109.97 (10) | O4—C15—C10 | 121.35 (12) |
| C5—C4—C3 | 108.08 (10) | O4—C15—C14 | 118.96 (11) |
| C7—C4—C3 | 109.83 (11) | C10—C15—C14 | 119.65 (11) |
| C6—C5—C4 | 114.11 (11) | C13—C16—H16A | 109.5 |
| C6—C5—H5A | 108.7 | C13—C16—H16B | 109.5 |
| C4—C5—H5A | 108.7 | H16A—C16—H16B | 109.5 |
| C6—C5—H5B | 108.7 | C13—C16—H16C | 109.5 |
| C4—C5—H5B | 108.7 | H16A—C16—H16C | 109.5 |
| H5A—C5—H5B | 107.6 | H16B—C16—H16C | 109.5 |
| O2—C6—C1 | 121.46 (12) | C13—C17—H17A | 109.5 |
| O2—C6—C5 | 119.71 (11) | C13—C17—H17B | 109.5 |
| C1—C6—C5 | 118.79 (11) | H17A—C17—H17B | 109.5 |
| C4—C7—H7A | 109.5 | C13—C17—H17C | 109.5 |
| C4—C7—H7B | 109.5 | H17A—C17—H17C | 109.5 |
| H7A—C7—H7B | 109.5 | H17B—C17—H17C | 109.5 |
| C4—C7—H7C | 109.5 | C23—C18—C19 | 117.87 (11) |
| H7A—C7—H7C | 109.5 | C23—C18—C9 | 120.68 (11) |
| H7B—C7—H7C | 109.5 | C19—C18—C9 | 121.04 (11) |
| C4—C8—H8A | 109.5 | C20—C19—C18 | 121.57 (12) |
| C4—C8—H8B | 109.5 | C20—C19—H19 | 119.2 |
| H8A—C8—H8B | 109.5 | C18—C19—H19 | 119.2 |
| C4—C8—H8C | 109.5 | C19—C20—C21 | 120.88 (12) |
| H8A—C8—H8C | 109.5 | C19—C20—H20 | 119.6 |
| H8B—C8—H8C | 109.5 | C21—C20—H20 | 119.6 |
| C1—C9—C10 | 115.85 (10) | C22—C21—C20 | 116.90 (12) |
| C1—C9—C18 | 113.02 (10) | C22—C21—H21 | 121.6 |
| C10—C9—C18 | 114.39 (10) | C20—C21—H21 | 121.6 |
| C1—C9—H9 | 103.9 | C21—C22—C23 | 123.42 (12) |
| C10—C9—H9 | 103.9 | C21—C22—N1 | 118.75 (11) |
| C18—C9—H9 | 103.9 | C23—C22—N1 | 117.83 (11) |
| C11—C10—C15 | 118.08 (11) | C22—C23—C18 | 119.37 (12) |
| C11—C10—C9 | 125.74 (11) | C22—C23—H23 | 120.3 |
| C15—C10—C9 | 116.15 (10) | C18—C23—H23 | 120.3 |
| O3—C11—C10 | 123.83 (12) | ||
| C6—C1—C2—O1 | 171.69 (11) | C10—C11—C12—C13 | −17.54 (17) |
| C9—C1—C2—O1 | −8.61 (19) | C11—C12—C13—C17 | −71.32 (14) |
| C6—C1—C2—C3 | −11.38 (18) | C11—C12—C13—C16 | 168.81 (11) |
| C9—C1—C2—C3 | 168.32 (11) | C11—C12—C13—C14 | 49.24 (14) |
| O1—C2—C3—C4 | −153.63 (11) | C17—C13—C14—C15 | 68.28 (14) |
| C1—C2—C3—C4 | 29.16 (17) | C16—C13—C14—C15 | −171.90 (11) |
| C2—C3—C4—C8 | −166.16 (11) | C12—C13—C14—C15 | −52.62 (14) |
| C2—C3—C4—C5 | −46.99 (14) | C11—C10—C15—O4 | −167.09 (11) |
| C2—C3—C4—C7 | 73.84 (14) | C9—C10—C15—O4 | 11.17 (17) |
| C8—C4—C5—C6 | 171.38 (11) | C11—C10—C15—C14 | 10.51 (18) |
| C7—C4—C5—C6 | −68.56 (14) | C9—C10—C15—C14 | −171.23 (11) |
| C3—C4—C5—C6 | 51.74 (14) | C13—C14—C15—O4 | −157.72 (12) |
| C2—C1—C6—O2 | −167.47 (12) | C13—C14—C15—C10 | 24.63 (17) |
| C9—C1—C6—O2 | 12.83 (18) | C1—C9—C18—C23 | −159.11 (11) |
| C2—C1—C6—C5 | 15.06 (18) | C10—C9—C18—C23 | −23.66 (16) |
| C9—C1—C6—C5 | −164.65 (11) | C1—C9—C18—C19 | 28.36 (16) |
| C4—C5—C6—O2 | 145.36 (12) | C10—C9—C18—C19 | 163.81 (11) |
| C4—C5—C6—C1 | −37.13 (16) | C23—C18—C19—C20 | 0.13 (18) |
| C2—C1—C9—C10 | 96.76 (14) | C9—C18—C19—C20 | 172.86 (11) |
| C6—C1—C9—C10 | −83.54 (14) | C18—C19—C20—C21 | −0.2 (2) |
| C2—C1—C9—C18 | −128.46 (12) | C19—C20—C21—C22 | 0.12 (19) |
| C6—C1—C9—C18 | 51.23 (15) | C20—C21—C22—C23 | −0.03 (19) |
| C1—C9—C10—C11 | 88.93 (15) | C20—C21—C22—N1 | −179.31 (11) |
| C18—C9—C10—C11 | −45.23 (16) | O6—N1—C22—C21 | 4.50 (17) |
| C1—C9—C10—C15 | −89.18 (13) | O5—N1—C22—C21 | −176.02 (11) |
| C18—C9—C10—C15 | 136.66 (11) | O6—N1—C22—C23 | −174.82 (11) |
| C15—C10—C11—O3 | 167.35 (11) | O5—N1—C22—C23 | 4.65 (17) |
| C9—C10—C11—O3 | −10.7 (2) | C21—C22—C23—C18 | −0.02 (19) |
| C15—C10—C11—C12 | −14.28 (18) | N1—C22—C23—C18 | 179.27 (11) |
| C9—C10—C11—C12 | 167.64 (11) | C19—C18—C23—C22 | −0.03 (18) |
| O3—C11—C12—C13 | 160.99 (11) | C9—C18—C23—C22 | −172.79 (11) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1o···O4 | 0.85 (1) | 1.80 (1) | 2.6392 (13) | 167 (2) |
| O3—H3o···O2 | 0.86 (1) | 1.75 (1) | 2.5985 (13) | 170 (2) |
| C5—H5a···O4i | 0.99 | 2.34 | 3.2781 (16) | 158 |
| C9—H9···O6ii | 1.00 | 2.56 | 3.3431 (16) | 135 |
| C21—H21···O2iii | 0.95 | 2.44 | 3.3121 (16) | 152 |
Symmetry codes: (i) x, y+1, z; (ii) −x+2, −y+1, −z+1; (iii) −x+2, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5662).
References
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hatakeyama, S., Ochi, N., Numata, H. & Takano, S. (1988). Chem. Commun. pp. 1202–1204.
- Hilderbrand, S. A. & Weissleder, R. (2007). Tetrahedron Lett.48, 4383–4385. [DOI] [PMC free article] [PubMed]
- Jonathan, R. D., Srinivas, K. R. & Glen, E. B. (1988). Eur. J. Med. Chem.23, 111–117.
- Pohlers, G. & Scaiano, J. C. (1997). Chem. Mater.9, 3222–3230.
- Shchekotikhin, Y. M. & Nikolaeva, T. G. (2006). Chem. Heterocycl. Compd, 42, 28–33.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810040079/hb5662sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810040079/hb5662Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report