Abstract
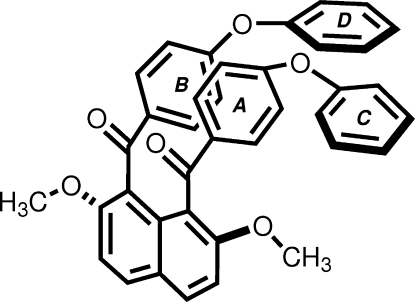
In the title molecule {systematic name: [2,7-dimethoxy-8-(4-phenoxybenzoyl)naphthalen-1-yl](4-phenoxyphenyl)methanone}, C38H28O6, the 4-phenoxybenzoyl units adopt a syn orientation with respect to the naphthalene ring system. The internal benzene rings, A and B, make dihedral angles of 86.72 (5) and 79.22 (5)° with the naphthalene ring system. The two terminal benzene rings, C and D, of the 4-phenoxybenzoyl groups are twisted with respect to benzene rings A and B, with dihedral angles of A/C = 62.72 (8) and B/D = 87.61 (6)°. In the crystal, H atoms in the naphthalene system make two types of intermolecular C—H⋯O interactions with the carbonyl O atom and the phenyl etheral O atom of neighbouring molecules. Molecules are further linked by C—H⋯π interactions involving a H atom of terminal benzene ring D and the π-system of the internal benzene ring A, forming dimers centered about an inversion center.
Related literature
For the syntheses of aroylated naphthalene compounds via electrophilic aromatic substitution of naphthalene derivatives, see: Okamoto & Yonezawa (2009 ▶). For the structures of closely related compounds, see: Nakaema et al. (2007 ▶, 2008 ▶); Mitsui et al. (2010 ▶); Muto et al. (2010 ▶); Watanabe et al. (2010a ▶,b
▶).
Experimental
Crystal data
C38H28O6
M r = 580.60
Monoclinic,

a = 12.0733 (4) Å
b = 12.4806 (4) Å
c = 19.8094 (6) Å
β = 91.115 (2)°
V = 2984.36 (15) Å3
Z = 4
Cu Kα radiation
μ = 0.71 mm−1
T = 193 K
0.50 × 0.30 × 0.30 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: numerical (NUMABS; Higashi, 1999 ▶) T min = 0.720, T max = 0.816
54106 measured reflections
5458 independent reflections
4862 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.099
S = 1.04
5458 reflections
400 parameters
H-atom parameters constrained
Δρmax = 0.16 e Å−3
Δρmin = −0.13 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 1998 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2004 ▶); program(s) used to solve structure: SIR2004 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPIII (Burnett & Johnson, 1996 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810042170/su2217sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810042170/su2217Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg3 is the centroid of ring A (C12–C17).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O2i | 0.95 | 2.44 | 3.1479 (17) | 131 |
| C6—H6⋯O6ii | 0.95 | 2.56 | 3.3293 (16) | 138 |
| C35—H35⋯Cg3iii | 0.95 | 2.78 | 3.6528 (17) | 153 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
The authors express their gratitude to Professor Keiichi Noguchi, Instrumentation Analysis Center, Tokyo University of Agriculture & Technology, for his technical advice. This work was partially supported by the Ogasawara Foundation for the Promotion of Science & Engineering, Tokyo, Japan.
supplementary crystallographic information
Comment
In the course of our studies on selective electrophilic aromatic aroylation of 2,7-dimethoxynaphthalene, peri-aroylnaphthalene compounds have proved to be formed regioselectively by the aid of a suitable acidic mediator (Okamoto & Yonezawa, 2009). Recently, we reported on the crystal structures of three such compounds, 1,8-dibenzoyl-2,7-dimethoxynaphthalene (Nakaema et al., 2008), bis(4-bromophenyl)(2,7-dimethoxynaphthalene-1,8-diyl)dimethanone (Watanabe et al., 2010a), and 1,8-bis(4-methylbenzoyl)-2,7-dimethoxynaphthalene (Muto et al., 2010). The aroyl groups at the 1,8-positions of the naphthalene rings in these compounds are twisted almost perpendicularly but a little tilted toward the exo sides against the naphthalene ring. In addition, 1,8-bis(4-chlorobenzoyl)-7-methoxynaphthalen-2-ol ethanol monosolvate (Mitsui et al., 2010), which was formed by selective demethylation at the 2-methoxy group of 1,8-bis(4-chlorobenzoyl)-2,7-dimethoxynaphthalene (Nakaema et al., 2007), solely has two syn-oriented chlorobenzoyl groups bonding to the naphthalene ring system, and in this molecule the 2-hydroxy group forms intramolecular hydrogen bonds with the carbonyl oxygen atom. As a part of our continuous studies on the molecular structures of this kind of homologous molecules, the crystal structure of the title compound, 1,8-bis(4-phenoxybenzoyl)-2,7-dimethoxynaphthalene, synthesized via nucleophilic aromatic substitution of (2,7-dimethoxynaphthalene-1,8-diyl)bis(4-fluorophenyl)dimethanone (Watanabe et al., 2010b), is reported herein.
In the title molecule, illustrated in Fig. 1, two intervenient benzene rings, A (C12–C17) and B (C25–C30), are in a syn orientation with respect to the naphthalene ring system (C1–C10), and make dihedral angles of 86.72 (5) and 79.22 (5)°, respectively, with the naphthalene ring system. Furthermore, the dihedral angles between benzene rings A and B and the terminal benzene rings C (C18–C23) and D (C31–C36)] are A/C = 62.72 (8), B/D = 87.61 (6)°. Benzene rings A and B are configurated almost parallel to one another, the dihedral angle A/B being only 12.20 (6)°. On the other hand, benzene rings C and D are far from being parallel to one another with a dihedral angle C/D of 64.10 (8)°.
In the crystal, hydrogen atoms in the naphthalene ring form two types of intermolecular C—H···O interactions with the carbonyl oxygen atom (C3—H3···O2i = 2.44 Å; see Fig. 2 and Table 1) and the phenyl ethereal oxygen atom (C6—H6···O6ii = 2.56 Å; see Fig. 2 and Table 1). Moreover, molecules are linked by C—H···π interactions forming dimeric pairs. The terminal benzene ring D acts as a hydrogen-bond donor and the π system of the intervenient benzene ring A (with centroid Cg3) of an adjacent molecule acts as an acceptor (C35—H35···Cg3iii = 2.78 Å; see Fig. 3 and Table 1).
Experimental
In a 10 ml one-necked flask equipped with a condenser, (2,7-dimethoxynaphthalene-1,8-diyl)bis(4-fluorophenyl)dimethanone (1.0 mmol, 432.4 mg), phenol (4.0 mmol, 376.4 mg), potassium carbonate (8.0 mmol, 1.10 g) and freshly distilled DMAc (2.0 ml) were stirred at 423 K for 6 h. This mixture was then added dropwise into methanol (20 ml) resulting in the formation of a pale yellow precipitate. The crude material was purified by column chromatography (silica gel, hexane: AcOEt= 2:1) to give the title compound (yield 104 mg, 18%). The isolated product was recrystallized from acetone to give block-like yellow single-crystals of the title compound. M.p. 441.6–444.4 K; 1HNMR δ (300 MHz, CDCl3): 3.72 (6H, s), 6.82–6.94 (4H, m), 7.09 (4H, d, J=7.5 Hz), 7.14–7.21 (4H, m), 7.36 (4H, t, J=8.4 Hz), 7.55–7.78 (4H,m), 7.92 (2H, d, J=7.5 Hz) p.p.m.; 13CNMR δ (75 MHz, CDCl3): 56.442, 111.128, 116.575, 120.274, 121.440, 124.345, 125.415, 129.467, 129.850, 131.360, 131.876, 133.424, 155.405, 155.998, 161.474, 195.057 p.p.m.; IR (KBr): 1673 (C=O), 1267 (Ar—O—Me) cm-1; HRMS (m/z): [M + H]+ calcd for C38H29O6, 581.1964 found, 581.82.
Refinement
All H atoms were found in a difference Fourier map and were subsequently refined as riding atoms: C—H = 0.95 (aromatic) and 0.98 (methyl) Å, with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The molecular structure of the title molecule, showing 50% probability displacement ellipsoids.
Fig. 2.
A partial crystal packing diagram of the title compound. The intermolecular C—H···O interactions are shown as double dashed lines (see Table 1 for details).
Fig. 3.
The dimeric pairs of the title molecule formed via C—H···π interactions, shown as double dashed lines (see Table 1 for details).
Crystal data
| C38H28O6 | F(000) = 1216 |
| Mr = 580.60 | Dx = 1.292 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54187 Å |
| Hall symbol: -P 2yn | Cell parameters from 37382 reflections |
| a = 12.0733 (4) Å | θ = 3.5–68.3° |
| b = 12.4806 (4) Å | µ = 0.71 mm−1 |
| c = 19.8094 (6) Å | T = 193 K |
| β = 91.115 (2)° | Block, yellow |
| V = 2984.36 (15) Å3 | 0.50 × 0.30 × 0.30 mm |
| Z = 4 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 5458 independent reflections |
| Radiation source: rotating anode | 4862 reflections with I > 2σ(I) |
| graphite | Rint = 0.038 |
| Detector resolution: 10.00 pixels mm-1 | θmax = 68.3°, θmin = 4.2° |
| ω scans | h = −14→14 |
| Absorption correction: numerical (NUMABS; Higashi, 1999) | k = −15→15 |
| Tmin = 0.720, Tmax = 0.816 | l = −23→23 |
| 54106 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H-atom parameters constrained |
| wR(F2) = 0.099 | w = 1/[σ2(Fo2) + (0.0496P)2 + 0.5589P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 5458 reflections | Δρmax = 0.16 e Å−3 |
| 400 parameters | Δρmin = −0.13 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00337 (19) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.96053 (8) | 0.06684 (7) | 0.81736 (5) | 0.0546 (2) | |
| O2 | 0.77701 (9) | −0.00287 (7) | 0.90988 (5) | 0.0586 (3) | |
| O3 | 0.97983 (8) | 0.27793 (8) | 0.73618 (5) | 0.0584 (3) | |
| O4 | 0.51924 (8) | 0.06935 (9) | 0.91941 (5) | 0.0615 (3) | |
| O5 | 1.08799 (8) | 0.36651 (9) | 1.06682 (5) | 0.0620 (3) | |
| O6 | 0.79851 (8) | 0.28865 (8) | 1.16825 (4) | 0.0556 (3) | |
| C1 | 0.83140 (10) | 0.20657 (9) | 0.79447 (6) | 0.0406 (3) | |
| C2 | 0.86712 (11) | 0.27048 (10) | 0.74219 (6) | 0.0471 (3) | |
| C3 | 0.79253 (12) | 0.32401 (11) | 0.69846 (7) | 0.0522 (3) | |
| H3 | 0.8189 | 0.3692 | 0.6637 | 0.063* | |
| C4 | 0.68212 (12) | 0.30989 (11) | 0.70687 (6) | 0.0510 (3) | |
| H4 | 0.6314 | 0.3447 | 0.6769 | 0.061* | |
| C5 | 0.64066 (10) | 0.24509 (10) | 0.75887 (6) | 0.0447 (3) | |
| C6 | 0.52610 (11) | 0.22967 (12) | 0.76518 (7) | 0.0542 (3) | |
| H6 | 0.4772 | 0.2613 | 0.7328 | 0.065* | |
| C7 | 0.48293 (11) | 0.17099 (13) | 0.81612 (7) | 0.0579 (4) | |
| H7 | 0.4053 | 0.1600 | 0.8186 | 0.069* | |
| C8 | 0.55556 (11) | 0.12678 (11) | 0.86504 (7) | 0.0493 (3) | |
| C9 | 0.66870 (10) | 0.13650 (9) | 0.86044 (6) | 0.0408 (3) | |
| C10 | 0.71611 (10) | 0.19418 (9) | 0.80557 (6) | 0.0394 (3) | |
| C11 | 0.92353 (10) | 0.15289 (9) | 0.83493 (6) | 0.0405 (3) | |
| C12 | 0.96973 (9) | 0.21139 (9) | 0.89451 (6) | 0.0388 (3) | |
| C13 | 0.92476 (9) | 0.30783 (9) | 0.91570 (6) | 0.0407 (3) | |
| H13 | 0.8647 | 0.3388 | 0.8909 | 0.049* | |
| C14 | 0.96670 (10) | 0.35918 (10) | 0.97258 (6) | 0.0446 (3) | |
| H14 | 0.9354 | 0.4250 | 0.9870 | 0.054* | |
| C15 | 1.05418 (10) | 0.31414 (11) | 1.00819 (6) | 0.0478 (3) | |
| C16 | 1.10038 (11) | 0.21766 (11) | 0.98832 (7) | 0.0535 (3) | |
| H16 | 1.1600 | 0.1867 | 1.0135 | 0.064* | |
| C17 | 1.05836 (11) | 0.16749 (10) | 0.93132 (7) | 0.0477 (3) | |
| H17 | 1.0903 | 0.1020 | 0.9169 | 0.057* | |
| C18 | 1.19700 (12) | 0.39961 (11) | 1.07515 (7) | 0.0522 (3) | |
| C19 | 1.27352 (14) | 0.39668 (15) | 1.02490 (8) | 0.0715 (4) | |
| H19 | 1.2535 | 0.3709 | 0.9812 | 0.086* | |
| C20 | 1.38027 (17) | 0.4318 (2) | 1.03878 (11) | 0.0967 (7) | |
| H20 | 1.4343 | 0.4283 | 1.0046 | 0.116* | |
| C21 | 1.40910 (18) | 0.47185 (18) | 1.10142 (11) | 0.0946 (6) | |
| H21 | 1.4825 | 0.4962 | 1.1105 | 0.113* | |
| C22 | 1.33065 (17) | 0.47633 (14) | 1.15093 (9) | 0.0758 (5) | |
| H22 | 1.3501 | 0.5042 | 1.1942 | 0.091* | |
| C23 | 1.22396 (14) | 0.44059 (11) | 1.13820 (7) | 0.0602 (4) | |
| H23 | 1.1699 | 0.4441 | 1.1723 | 0.072* | |
| C24 | 0.73586 (10) | 0.08501 (9) | 0.91703 (6) | 0.0412 (3) | |
| C25 | 0.74864 (9) | 0.14370 (9) | 0.98181 (6) | 0.0388 (3) | |
| C26 | 0.69332 (10) | 0.23842 (10) | 0.99588 (6) | 0.0423 (3) | |
| H26 | 0.6442 | 0.2685 | 0.9629 | 0.051* | |
| C27 | 0.70890 (10) | 0.28958 (11) | 1.05731 (6) | 0.0458 (3) | |
| H27 | 0.6706 | 0.3542 | 1.0667 | 0.055* | |
| C28 | 0.78094 (10) | 0.24554 (10) | 1.10496 (6) | 0.0441 (3) | |
| C29 | 0.83819 (11) | 0.15178 (10) | 1.09148 (6) | 0.0487 (3) | |
| H29 | 0.8880 | 0.1224 | 1.1242 | 0.058* | |
| C30 | 0.82200 (11) | 0.10197 (10) | 1.03023 (6) | 0.0461 (3) | |
| H30 | 0.8614 | 0.0380 | 1.0207 | 0.055* | |
| C31 | 0.77444 (11) | 0.39731 (11) | 1.17747 (6) | 0.0477 (3) | |
| C32 | 0.68244 (12) | 0.42435 (12) | 1.21339 (7) | 0.0569 (4) | |
| H32 | 0.6344 | 0.3705 | 1.2300 | 0.068* | |
| C33 | 0.66113 (13) | 0.53178 (13) | 1.22497 (8) | 0.0651 (4) | |
| H33 | 0.5974 | 0.5521 | 1.2493 | 0.078* | |
| C34 | 0.73172 (13) | 0.60918 (13) | 1.20139 (8) | 0.0637 (4) | |
| H34 | 0.7169 | 0.6827 | 1.2097 | 0.076* | |
| C35 | 0.82377 (13) | 0.58022 (13) | 1.16573 (8) | 0.0625 (4) | |
| H35 | 0.8722 | 0.6339 | 1.1494 | 0.075* | |
| C36 | 0.84602 (12) | 0.47327 (12) | 1.15348 (7) | 0.0561 (3) | |
| H36 | 0.9095 | 0.4528 | 1.1290 | 0.067* | |
| C37 | 1.02198 (14) | 0.34475 (15) | 0.68454 (9) | 0.0741 (5) | |
| H37A | 1.1031 | 0.3439 | 0.6867 | 0.089* | |
| H37B | 0.9955 | 0.4182 | 0.6909 | 0.089* | |
| H37C | 0.9964 | 0.3182 | 0.6404 | 0.089* | |
| C38 | 0.40630 (15) | 0.0786 (2) | 0.93694 (12) | 0.1008 (7) | |
| H38A | 0.3948 | 0.0437 | 0.9806 | 0.121* | |
| H38B | 0.3598 | 0.0439 | 0.9023 | 0.121* | |
| H38C | 0.3863 | 0.1545 | 0.9401 | 0.121* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0598 (6) | 0.0431 (5) | 0.0604 (6) | 0.0113 (4) | −0.0108 (4) | −0.0065 (4) |
| O2 | 0.0817 (7) | 0.0420 (5) | 0.0514 (5) | 0.0130 (5) | −0.0147 (5) | −0.0058 (4) |
| O3 | 0.0499 (5) | 0.0617 (6) | 0.0636 (6) | 0.0012 (4) | 0.0039 (4) | 0.0206 (5) |
| O4 | 0.0488 (5) | 0.0698 (7) | 0.0658 (6) | −0.0101 (5) | 0.0012 (4) | 0.0149 (5) |
| O5 | 0.0549 (6) | 0.0761 (7) | 0.0546 (6) | −0.0006 (5) | −0.0087 (4) | −0.0167 (5) |
| O6 | 0.0719 (6) | 0.0533 (6) | 0.0411 (5) | 0.0076 (5) | −0.0123 (4) | −0.0064 (4) |
| C1 | 0.0459 (6) | 0.0357 (6) | 0.0399 (6) | 0.0017 (5) | −0.0052 (5) | 0.0007 (5) |
| C2 | 0.0492 (7) | 0.0438 (7) | 0.0481 (7) | 0.0032 (5) | −0.0007 (5) | 0.0045 (5) |
| C3 | 0.0610 (8) | 0.0491 (8) | 0.0462 (7) | 0.0042 (6) | −0.0033 (6) | 0.0116 (6) |
| C4 | 0.0593 (8) | 0.0491 (7) | 0.0440 (7) | 0.0091 (6) | −0.0120 (6) | 0.0050 (6) |
| C5 | 0.0491 (7) | 0.0430 (7) | 0.0414 (6) | 0.0049 (5) | −0.0106 (5) | −0.0018 (5) |
| C6 | 0.0494 (7) | 0.0581 (8) | 0.0545 (8) | 0.0052 (6) | −0.0163 (6) | 0.0021 (6) |
| C7 | 0.0407 (7) | 0.0666 (9) | 0.0660 (9) | −0.0031 (6) | −0.0106 (6) | 0.0031 (7) |
| C8 | 0.0481 (7) | 0.0477 (7) | 0.0519 (7) | −0.0055 (6) | −0.0038 (6) | 0.0015 (6) |
| C9 | 0.0445 (6) | 0.0374 (6) | 0.0400 (6) | −0.0013 (5) | −0.0073 (5) | −0.0025 (5) |
| C10 | 0.0452 (6) | 0.0351 (6) | 0.0377 (6) | 0.0013 (5) | −0.0081 (5) | −0.0038 (5) |
| C11 | 0.0411 (6) | 0.0356 (6) | 0.0448 (6) | 0.0009 (5) | −0.0015 (5) | 0.0042 (5) |
| C12 | 0.0381 (6) | 0.0349 (6) | 0.0433 (6) | −0.0012 (5) | −0.0016 (5) | 0.0054 (5) |
| C13 | 0.0369 (6) | 0.0359 (6) | 0.0492 (7) | −0.0001 (5) | −0.0035 (5) | 0.0058 (5) |
| C14 | 0.0423 (6) | 0.0381 (6) | 0.0535 (7) | −0.0013 (5) | 0.0008 (5) | −0.0017 (5) |
| C15 | 0.0455 (7) | 0.0513 (7) | 0.0465 (7) | −0.0035 (6) | −0.0041 (5) | −0.0029 (6) |
| C16 | 0.0509 (7) | 0.0552 (8) | 0.0537 (8) | 0.0085 (6) | −0.0150 (6) | 0.0020 (6) |
| C17 | 0.0497 (7) | 0.0411 (7) | 0.0521 (7) | 0.0088 (5) | −0.0073 (6) | 0.0027 (5) |
| C18 | 0.0560 (8) | 0.0464 (7) | 0.0534 (7) | 0.0011 (6) | −0.0158 (6) | 0.0014 (6) |
| C19 | 0.0657 (10) | 0.0865 (12) | 0.0619 (9) | −0.0152 (8) | −0.0117 (7) | −0.0112 (8) |
| C20 | 0.0701 (11) | 0.1271 (18) | 0.0927 (14) | −0.0304 (12) | −0.0038 (10) | −0.0231 (13) |
| C21 | 0.0769 (12) | 0.1078 (16) | 0.0979 (15) | −0.0283 (11) | −0.0281 (11) | −0.0112 (12) |
| C22 | 0.0966 (13) | 0.0629 (10) | 0.0665 (10) | −0.0146 (9) | −0.0363 (10) | −0.0002 (8) |
| C23 | 0.0814 (10) | 0.0475 (8) | 0.0508 (8) | −0.0021 (7) | −0.0179 (7) | 0.0023 (6) |
| C24 | 0.0459 (6) | 0.0358 (6) | 0.0418 (6) | −0.0033 (5) | −0.0029 (5) | 0.0021 (5) |
| C25 | 0.0422 (6) | 0.0348 (6) | 0.0392 (6) | −0.0049 (5) | −0.0026 (5) | 0.0039 (5) |
| C26 | 0.0403 (6) | 0.0453 (7) | 0.0411 (6) | 0.0026 (5) | −0.0060 (5) | 0.0010 (5) |
| C27 | 0.0447 (6) | 0.0462 (7) | 0.0464 (7) | 0.0065 (5) | −0.0040 (5) | −0.0048 (5) |
| C28 | 0.0478 (7) | 0.0461 (7) | 0.0382 (6) | −0.0042 (5) | −0.0034 (5) | −0.0021 (5) |
| C29 | 0.0580 (8) | 0.0434 (7) | 0.0441 (7) | 0.0031 (6) | −0.0138 (6) | 0.0043 (5) |
| C30 | 0.0567 (7) | 0.0348 (6) | 0.0463 (7) | 0.0020 (5) | −0.0076 (5) | 0.0034 (5) |
| C31 | 0.0519 (7) | 0.0522 (8) | 0.0387 (6) | −0.0001 (6) | −0.0077 (5) | −0.0075 (5) |
| C32 | 0.0554 (8) | 0.0596 (9) | 0.0558 (8) | −0.0123 (7) | 0.0045 (6) | −0.0102 (7) |
| C33 | 0.0582 (8) | 0.0671 (10) | 0.0706 (10) | −0.0053 (7) | 0.0151 (7) | −0.0195 (8) |
| C34 | 0.0663 (9) | 0.0533 (8) | 0.0719 (10) | −0.0056 (7) | 0.0077 (7) | −0.0161 (7) |
| C35 | 0.0629 (9) | 0.0602 (9) | 0.0646 (9) | −0.0141 (7) | 0.0077 (7) | −0.0052 (7) |
| C36 | 0.0517 (8) | 0.0661 (9) | 0.0506 (7) | −0.0023 (7) | 0.0063 (6) | −0.0075 (7) |
| C37 | 0.0649 (9) | 0.0804 (11) | 0.0774 (11) | 0.0013 (8) | 0.0147 (8) | 0.0301 (9) |
| C38 | 0.0584 (10) | 0.1314 (19) | 0.1134 (16) | −0.0008 (11) | 0.0215 (10) | 0.0495 (14) |
Geometric parameters (Å, °)
| O1—C11 | 1.2167 (14) | C18—C23 | 1.3826 (19) |
| O2—C24 | 1.2135 (15) | C19—C20 | 1.384 (2) |
| O3—C2 | 1.3714 (16) | C19—H19 | 0.9500 |
| O3—C37 | 1.4217 (17) | C20—C21 | 1.376 (3) |
| O4—C8 | 1.3727 (16) | C20—H20 | 0.9500 |
| O4—C38 | 1.418 (2) | C21—C22 | 1.378 (3) |
| O5—C18 | 1.3863 (17) | C21—H21 | 0.9500 |
| O5—C15 | 1.3871 (15) | C22—C23 | 1.382 (2) |
| O6—C28 | 1.3771 (14) | C22—H22 | 0.9500 |
| O6—C31 | 1.3996 (16) | C23—H23 | 0.9500 |
| C1—C2 | 1.3829 (17) | C24—C25 | 1.4832 (16) |
| C1—C10 | 1.4219 (17) | C25—C26 | 1.3887 (17) |
| C1—C11 | 1.5145 (16) | C25—C30 | 1.3938 (16) |
| C2—C3 | 1.4061 (18) | C26—C27 | 1.3839 (17) |
| C3—C4 | 1.358 (2) | C26—H26 | 0.9500 |
| C3—H3 | 0.9500 | C27—C28 | 1.3847 (17) |
| C4—C5 | 1.4092 (19) | C27—H27 | 0.9500 |
| C4—H4 | 0.9500 | C28—C29 | 1.3877 (18) |
| C5—C6 | 1.4045 (19) | C29—C30 | 1.3738 (18) |
| C5—C10 | 1.4339 (16) | C29—H29 | 0.9500 |
| C6—C7 | 1.359 (2) | C30—H30 | 0.9500 |
| C6—H6 | 0.9500 | C31—C32 | 1.3729 (19) |
| C7—C8 | 1.4070 (19) | C31—C36 | 1.374 (2) |
| C7—H7 | 0.9500 | C32—C33 | 1.385 (2) |
| C8—C9 | 1.3761 (17) | C32—H32 | 0.9500 |
| C9—C10 | 1.4323 (17) | C33—C34 | 1.376 (2) |
| C9—C24 | 1.5139 (16) | C33—H33 | 0.9500 |
| C11—C12 | 1.4871 (16) | C34—C35 | 1.377 (2) |
| C12—C13 | 1.3887 (17) | C34—H34 | 0.9500 |
| C12—C17 | 1.3952 (16) | C35—C36 | 1.384 (2) |
| C13—C14 | 1.3838 (17) | C35—H35 | 0.9500 |
| C13—H13 | 0.9500 | C36—H36 | 0.9500 |
| C14—C15 | 1.3783 (18) | C37—H37A | 0.9800 |
| C14—H14 | 0.9500 | C37—H37B | 0.9800 |
| C15—C16 | 1.3873 (19) | C37—H37C | 0.9800 |
| C16—C17 | 1.3792 (18) | C38—H38A | 0.9800 |
| C16—H16 | 0.9500 | C38—H38B | 0.9800 |
| C17—H17 | 0.9500 | C38—H38C | 0.9800 |
| C18—C19 | 1.372 (2) | ||
| C2—O3—C37 | 118.15 (11) | C21—C20—H20 | 119.6 |
| C8—O4—C38 | 118.24 (12) | C19—C20—H20 | 119.6 |
| C18—O5—C15 | 120.22 (11) | C20—C21—C22 | 119.42 (18) |
| C28—O6—C31 | 117.95 (10) | C20—C21—H21 | 120.3 |
| C2—C1—C10 | 119.93 (11) | C22—C21—H21 | 120.3 |
| C2—C1—C11 | 114.52 (11) | C21—C22—C23 | 120.55 (16) |
| C10—C1—C11 | 125.54 (10) | C21—C22—H22 | 119.7 |
| O3—C2—C1 | 115.37 (11) | C23—C22—H22 | 119.7 |
| O3—C2—C3 | 122.61 (12) | C22—C23—C18 | 119.11 (17) |
| C1—C2—C3 | 122.01 (12) | C22—C23—H23 | 120.4 |
| C4—C3—C2 | 118.77 (12) | C18—C23—H23 | 120.4 |
| C4—C3—H3 | 120.6 | O2—C24—C25 | 120.76 (11) |
| C2—C3—H3 | 120.6 | O2—C24—C9 | 120.79 (11) |
| C3—C4—C5 | 121.84 (12) | C25—C24—C9 | 118.44 (10) |
| C3—C4—H4 | 119.1 | C26—C25—C30 | 118.80 (11) |
| C5—C4—H4 | 119.1 | C26—C25—C24 | 123.49 (10) |
| C6—C5—C4 | 120.53 (11) | C30—C25—C24 | 117.70 (11) |
| C6—C5—C10 | 119.75 (12) | C27—C26—C25 | 120.77 (11) |
| C4—C5—C10 | 119.71 (12) | C27—C26—H26 | 119.6 |
| C7—C6—C5 | 122.17 (12) | C25—C26—H26 | 119.6 |
| C7—C6—H6 | 118.9 | C26—C27—C28 | 119.28 (12) |
| C5—C6—H6 | 118.9 | C26—C27—H27 | 120.4 |
| C6—C7—C8 | 118.68 (13) | C28—C27—H27 | 120.4 |
| C6—C7—H7 | 120.7 | O6—C28—C27 | 123.30 (11) |
| C8—C7—H7 | 120.7 | O6—C28—C29 | 115.86 (11) |
| O4—C8—C9 | 115.52 (11) | C27—C28—C29 | 120.81 (11) |
| O4—C8—C7 | 122.74 (12) | C30—C29—C28 | 119.26 (11) |
| C9—C8—C7 | 121.73 (12) | C30—C29—H29 | 120.4 |
| C8—C9—C10 | 120.43 (11) | C28—C29—H29 | 120.4 |
| C8—C9—C24 | 115.57 (11) | C29—C30—C25 | 121.08 (12) |
| C10—C9—C24 | 124.00 (10) | C29—C30—H30 | 119.5 |
| C1—C10—C9 | 125.36 (10) | C25—C30—H30 | 119.5 |
| C1—C10—C5 | 117.62 (11) | C32—C31—C36 | 122.03 (13) |
| C9—C10—C5 | 117.00 (11) | C32—C31—O6 | 118.55 (13) |
| O1—C11—C12 | 121.75 (11) | C36—C31—O6 | 119.34 (12) |
| O1—C11—C1 | 120.60 (11) | C31—C32—C33 | 118.59 (14) |
| C12—C11—C1 | 117.59 (10) | C31—C32—H32 | 120.7 |
| C13—C12—C17 | 118.87 (11) | C33—C32—H32 | 120.7 |
| C13—C12—C11 | 121.50 (10) | C34—C33—C32 | 120.34 (14) |
| C17—C12—C11 | 119.60 (11) | C34—C33—H33 | 119.8 |
| C14—C13—C12 | 120.57 (11) | C32—C33—H33 | 119.8 |
| C14—C13—H13 | 119.7 | C35—C34—C33 | 120.08 (15) |
| C12—C13—H13 | 119.7 | C35—C34—H34 | 120.0 |
| C15—C14—C13 | 119.50 (12) | C33—C34—H34 | 120.0 |
| C15—C14—H14 | 120.2 | C34—C35—C36 | 120.31 (14) |
| C13—C14—H14 | 120.2 | C34—C35—H35 | 119.8 |
| C14—C15—O5 | 116.51 (12) | C36—C35—H35 | 119.8 |
| C14—C15—C16 | 121.14 (12) | C31—C36—C35 | 118.64 (13) |
| O5—C15—C16 | 122.21 (12) | C31—C36—H36 | 120.7 |
| C17—C16—C15 | 118.87 (12) | C35—C36—H36 | 120.7 |
| C17—C16—H16 | 120.6 | O3—C37—H37A | 109.5 |
| C15—C16—H16 | 120.6 | O3—C37—H37B | 109.5 |
| C16—C17—C12 | 121.04 (12) | H37A—C37—H37B | 109.5 |
| C16—C17—H17 | 119.5 | O3—C37—H37C | 109.5 |
| C12—C17—H17 | 119.5 | H37A—C37—H37C | 109.5 |
| C19—C18—C23 | 121.03 (14) | H37B—C37—H37C | 109.5 |
| C19—C18—O5 | 123.84 (12) | O4—C38—H38A | 109.5 |
| C23—C18—O5 | 115.09 (13) | O4—C38—H38B | 109.5 |
| C18—C19—C20 | 119.02 (16) | H38A—C38—H38B | 109.5 |
| C18—C19—H19 | 120.5 | O4—C38—H38C | 109.5 |
| C20—C19—H19 | 120.5 | H38A—C38—H38C | 109.5 |
| C21—C20—C19 | 120.8 (2) | H38B—C38—H38C | 109.5 |
| C37—O3—C2—C1 | −178.33 (13) | C18—O5—C15—C14 | −123.87 (13) |
| C37—O3—C2—C3 | 1.6 (2) | C18—O5—C15—C16 | 60.33 (18) |
| C10—C1—C2—O3 | 179.42 (11) | C14—C15—C16—C17 | 0.9 (2) |
| C11—C1—C2—O3 | −1.79 (16) | O5—C15—C16—C17 | 176.54 (12) |
| C10—C1—C2—C3 | −0.50 (19) | C15—C16—C17—C12 | −1.0 (2) |
| C11—C1—C2—C3 | 178.30 (12) | C13—C12—C17—C16 | 0.78 (19) |
| O3—C2—C3—C4 | 178.22 (12) | C11—C12—C17—C16 | −177.51 (12) |
| C1—C2—C3—C4 | −1.9 (2) | C15—O5—C18—C19 | 8.8 (2) |
| C2—C3—C4—C5 | 1.3 (2) | C15—O5—C18—C23 | −173.45 (12) |
| C3—C4—C5—C6 | −178.26 (13) | C23—C18—C19—C20 | 2.4 (3) |
| C3—C4—C5—C10 | 1.5 (2) | O5—C18—C19—C20 | 179.97 (17) |
| C4—C5—C6—C7 | −177.53 (14) | C18—C19—C20—C21 | −1.7 (3) |
| C10—C5—C6—C7 | 2.7 (2) | C19—C20—C21—C22 | 0.3 (4) |
| C5—C6—C7—C8 | 1.7 (2) | C20—C21—C22—C23 | 0.3 (3) |
| C38—O4—C8—C9 | 164.34 (16) | C21—C22—C23—C18 | 0.4 (3) |
| C38—O4—C8—C7 | −17.1 (2) | C19—C18—C23—C22 | −1.7 (2) |
| C6—C7—C8—O4 | 177.86 (13) | O5—C18—C23—C22 | −179.52 (13) |
| C6—C7—C8—C9 | −3.7 (2) | C8—C9—C24—O2 | 98.63 (15) |
| O4—C8—C9—C10 | 179.63 (11) | C10—C9—C24—O2 | −82.66 (16) |
| C7—C8—C9—C10 | 1.0 (2) | C8—C9—C24—C25 | −80.37 (14) |
| O4—C8—C9—C24 | −1.60 (17) | C10—C9—C24—C25 | 98.34 (14) |
| C7—C8—C9—C24 | 179.81 (12) | O2—C24—C25—C26 | −172.30 (12) |
| C2—C1—C10—C9 | −175.50 (12) | C9—C24—C25—C26 | 6.70 (17) |
| C11—C1—C10—C9 | 5.84 (19) | O2—C24—C25—C30 | 9.04 (18) |
| C2—C1—C10—C5 | 3.26 (17) | C9—C24—C25—C30 | −171.96 (11) |
| C11—C1—C10—C5 | −175.39 (11) | C30—C25—C26—C27 | −1.23 (18) |
| C8—C9—C10—C1 | −177.92 (12) | C24—C25—C26—C27 | −179.88 (11) |
| C24—C9—C10—C1 | 3.42 (19) | C25—C26—C27—C28 | 0.25 (19) |
| C8—C9—C10—C5 | 3.31 (17) | C31—O6—C28—C27 | −22.47 (18) |
| C24—C9—C10—C5 | −175.35 (11) | C31—O6—C28—C29 | 159.22 (12) |
| C6—C5—C10—C1 | 176.01 (11) | C26—C27—C28—O6 | −177.55 (11) |
| C4—C5—C10—C1 | −3.76 (17) | C26—C27—C28—C29 | 0.68 (19) |
| C6—C5—C10—C9 | −5.12 (17) | O6—C28—C29—C30 | 177.76 (12) |
| C4—C5—C10—C9 | 175.10 (11) | C27—C28—C29—C30 | −0.6 (2) |
| C2—C1—C11—O1 | −87.22 (15) | C28—C29—C30—C25 | −0.4 (2) |
| C10—C1—C11—O1 | 91.50 (16) | C26—C25—C30—C29 | 1.33 (19) |
| C2—C1—C11—C12 | 90.10 (13) | C24—C25—C30—C29 | −179.95 (12) |
| C10—C1—C11—C12 | −91.18 (14) | C28—O6—C31—C32 | 107.42 (14) |
| O1—C11—C12—C13 | −177.79 (11) | C28—O6—C31—C36 | −75.86 (15) |
| C1—C11—C12—C13 | 4.92 (16) | C36—C31—C32—C33 | 0.7 (2) |
| O1—C11—C12—C17 | 0.44 (18) | O6—C31—C32—C33 | 177.33 (13) |
| C1—C11—C12—C17 | −176.85 (11) | C31—C32—C33—C34 | −0.7 (2) |
| C17—C12—C13—C14 | −0.39 (18) | C32—C33—C34—C35 | 0.4 (3) |
| C11—C12—C13—C14 | 177.86 (11) | C33—C34—C35—C36 | −0.1 (2) |
| C12—C13—C14—C15 | 0.28 (18) | C32—C31—C36—C35 | −0.4 (2) |
| C13—C14—C15—O5 | −176.41 (11) | O6—C31—C36—C35 | −177.04 (12) |
| C13—C14—C15—C16 | −0.6 (2) | C34—C35—C36—C31 | 0.1 (2) |
Hydrogen-bond geometry (Å, °)
| Cg3 is the centroid of ring A (C12–C17). |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O2i | 0.95 | 2.44 | 3.1479 (17) | 131 |
| C6—H6···O6ii | 0.95 | 2.56 | 3.3293 (16) | 138 |
| C35—H35···Cg3iii | 0.95 | 2.78 | 3.6528 (17) | 153 |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2; (ii) x−1/2, −y+1/2, z−1/2; (iii) −x+2, −y+1, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2217).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst.38, 381–388.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory. Tennessee, USA.
- Higashi, T. (1999). NUMABS Rigaku Corporation, Tokyo, Japan.
- Mitsui, R., Nagasawa, A., Noguchi, K., Okamoto, A. & Yonezawa, N. (2010). Acta Cryst. E66, o1790. [DOI] [PMC free article] [PubMed]
- Muto, T., Kato, Y., Nagasawa, A., Okamoto, A. & Yonezawa, N. (2010). Acta Cryst. E66, o2752. [DOI] [PMC free article] [PubMed]
- Nakaema, K., Okamoto, A., Noguchi, K. & Yonezawa, N. (2007). Acta Cryst. E63, o4120.
- Nakaema, K., Watanabe, S., Okamoto, A., Noguchi, K. & Yonezawa, N. (2008). Acta Cryst. E64, o807. [DOI] [PMC free article] [PubMed]
- Okamoto, A. & Yonezawa, N. (2009). Chem. Lett.38, 914–915.
- Rigaku (1998). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2004). CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Watanabe, S., Nagasawa, A., Okamoto, A., Noguchi, K. & Yonezawa, N. (2010b). Acta Cryst. E66, o329. [DOI] [PMC free article] [PubMed]
- Watanabe, S., Nakaema, K., Muto, T., Okamoto, A. & Yonezawa, N. (2010a). Acta Cryst. E66, o403. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810042170/su2217sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810042170/su2217Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





