Abstract
In the title compound, C5H8Cl2O6P2, the two six-membered rings display chair conformations. The P=O bond distances are 1.444 (2) and 1.446 (2) Å. Weak intermolecular C—H⋯O hydrogen bonds are present in the crystal structure.
Related literature
For applications of pentaerythritol diphosphonate compounds, see: Granzow (1981 ▶); Tanabe et al. (2005 ▶). For details of the preparation of the title compound, see: Li et al. (2002 ▶). For related compounds, see: Heinemann et al. (1994 ▶); Zhang et al. (2006 ▶). For bond-length, see: Allen et al. (1987 ▶); Elnagar et al. (2000 ▶).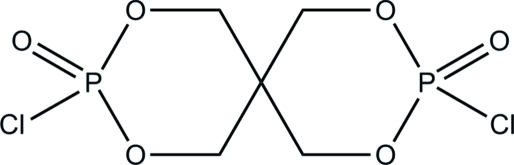
Experimental
Crystal data
C5H8Cl2O6P2
M r = 296.95
Orthorhombic,
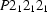
a = 6.0630 (5) Å
b = 12.7384 (10) Å
c = 13.4338 (10) Å
V = 1037.53 (14) Å3
Z = 4
Mo Kα radiation
μ = 0.94 mm−1
T = 185 K
0.12 × 0.10 × 0.08 mm
Data collection
Bruker SMART CCD 1000 area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.896, T max = 0.929
5317 measured reflections
1849 independent reflections
1662 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.031
wR(F 2) = 0.069
S = 1.00
1849 reflections
136 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.23 e Å−3
Absolute structure: Flack (1983 ▶), 747 Friedel pairs
Flack parameter: 0.18 (10)
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810043333/xu5060sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810043333/xu5060Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1A⋯O5i | 0.99 | 2.34 | 3.214 (4) | 147 |
| C1—H1B⋯O6ii | 0.99 | 2.31 | 3.252 (4) | 159 |
| C4—H4B⋯O5i | 0.99 | 2.36 | 3.260 (4) | 150 |
Symmetry codes: (i) 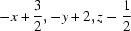 ; (ii)
; (ii) 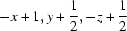 .
.
Acknowledgments
We thank Northeast Forestry University for financial support (graduate innovation funded projects GRAM09).
supplementary crystallographic information
Comment
Studies of pentaerythritol diphosphonate compounds have been significant interested. On one hand, the compounds have been reported to act as one of the most important reaction intermediates of fire retardant agents (Tanabe et al., 2005). On the other hand, it seems to be a good candidate in modifying the stability of polymers (Granzow, 1981). The findings have triggered the development of new flame retardant materials. As an extension of the work on the structural characterization of pentaerythritol diphosphonate compounds, the preparation and crystal structure of the title compound, (I), is proposed here.
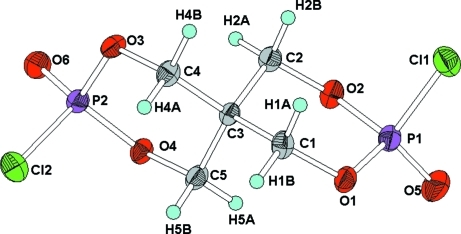
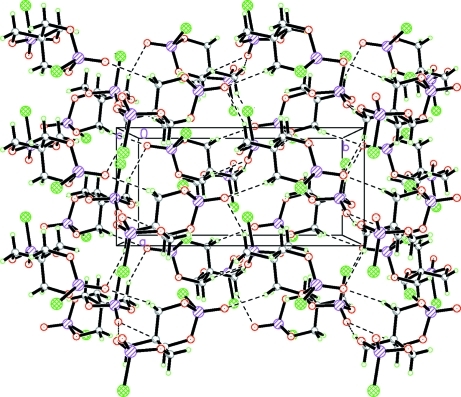
The asymmetric unit of (I) contain a spiro[5.5]undecane molecule (Fig. 1). Several compounds with similar structures have been reported previously (Heinemann et al., 1994; Zhang et al., 2006). The bond lengths and angles are within normal ranges (Allen et al., 1987; Elnagar et al., 2000). The six-membered rings of (I) have the chair conformation consistent with the steric difference in this conformation between opposite ends of the molecule. In addition, the C1—C3—C2 and C4—C3—C5 angles are in the range of 109.4 (3)–109.2 (3)°,the P—Cl bond lengths are 2.0050 (14) and 2.0047 (13) Å, respectively. In the crystal structure of (I), The non-classic C—H···O hydrogen bonds ranging from 3.099 (4) to 3.260 (4) Å contributed to the stability of the crystal packing.
Experimental
The title compound was prepared by reaction of pentaerythritol with phosphorus oxychloride in acetonitrile according to the reported procedures (Li et al., 2002). Crystals were produced at the bottom of the vessel on slow evaporation of acetic acid solution.
Refinement
All H atoms were placed geometrically with C—H = 0.99 Å and refined using a riding atom model with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
A view of the title compound, showing the atomic numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
The crystal packing through C—H···O interactions along the c axis
Crystal data
| C5H8Cl2O6P2 | F(000) = 600 |
| Mr = 296.95 | Dx = 1.901 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2823 reflections |
| a = 6.0630 (5) Å | θ = 2.2–25.0° |
| b = 12.7384 (10) Å | µ = 0.94 mm−1 |
| c = 13.4338 (10) Å | T = 185 K |
| V = 1037.53 (14) Å3 | Block, colorless |
| Z = 4 | 0.12 × 0.10 × 0.08 mm |
Data collection
| Bruker SMART CCD 1000 area-detector diffractometer | 1849 independent reflections |
| Radiation source: fine-focus sealed tube | 1662 reflections with I > 2σ(I) |
| graphite | Rint = 0.036 |
| φ and ω scans | θmax = 25.1°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −5→7 |
| Tmin = 0.896, Tmax = 0.929 | k = −14→15 |
| 5317 measured reflections | l = −14→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.031 | H-atom parameters constrained |
| wR(F2) = 0.069 | w = 1/[σ2(Fo2) + (0.0283P)2 + 0.5846P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 1849 reflections | Δρmax = 0.27 e Å−3 |
| 136 parameters | Δρmin = −0.23 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 747 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.18 (10) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| P2 | 0.86704 (16) | 0.68037 (6) | 0.19061 (7) | 0.0195 (2) | |
| P1 | 0.61011 (17) | 0.94037 (7) | 0.51917 (6) | 0.0225 (2) | |
| Cl2 | 1.00827 (16) | 0.75305 (7) | 0.07460 (7) | 0.0322 (2) | |
| Cl1 | 0.28767 (15) | 0.96781 (7) | 0.53971 (7) | 0.0299 (2) | |
| O4 | 0.9665 (4) | 0.73565 (16) | 0.28413 (16) | 0.0197 (5) | |
| O2 | 0.6224 (4) | 0.82146 (16) | 0.48872 (15) | 0.0218 (5) | |
| O6 | 0.8993 (4) | 0.56810 (17) | 0.18897 (18) | 0.0274 (6) | |
| O3 | 0.6190 (4) | 0.71416 (16) | 0.18620 (16) | 0.0206 (5) | |
| O1 | 0.6715 (4) | 1.00446 (16) | 0.42462 (16) | 0.0227 (6) | |
| O5 | 0.7431 (5) | 0.96565 (19) | 0.60521 (16) | 0.0338 (6) | |
| C3 | 0.6534 (6) | 0.8561 (2) | 0.3088 (2) | 0.0170 (7) | |
| C5 | 0.9032 (6) | 0.8438 (2) | 0.3065 (3) | 0.0208 (8) | |
| H5A | 0.9656 | 0.8912 | 0.2553 | 0.025* | |
| H5B | 0.9652 | 0.8644 | 0.3718 | 0.025* | |
| C4 | 0.5581 (6) | 0.8229 (2) | 0.2084 (2) | 0.0198 (8) | |
| H4A | 0.3955 | 0.8295 | 0.2098 | 0.024* | |
| H4B | 0.6152 | 0.8697 | 0.1555 | 0.024* | |
| C1 | 0.5966 (6) | 0.9720 (2) | 0.3254 (2) | 0.0211 (7) | |
| H1A | 0.6694 | 1.0155 | 0.2740 | 0.025* | |
| H1B | 0.4353 | 0.9822 | 0.3198 | 0.025* | |
| C2 | 0.5455 (6) | 0.7897 (3) | 0.3905 (2) | 0.0207 (8) | |
| H2A | 0.3833 | 0.7978 | 0.3870 | 0.025* | |
| H2B | 0.5810 | 0.7147 | 0.3796 | 0.025* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| P2 | 0.0192 (5) | 0.0181 (4) | 0.0211 (5) | −0.0002 (4) | 0.0003 (4) | −0.0033 (4) |
| P1 | 0.0263 (5) | 0.0223 (5) | 0.0189 (5) | −0.0018 (4) | 0.0020 (4) | −0.0028 (4) |
| Cl2 | 0.0331 (5) | 0.0376 (5) | 0.0259 (5) | −0.0039 (5) | 0.0073 (4) | 0.0002 (4) |
| Cl1 | 0.0282 (5) | 0.0299 (5) | 0.0317 (5) | 0.0023 (4) | 0.0074 (4) | −0.0002 (4) |
| O4 | 0.0187 (13) | 0.0183 (12) | 0.0221 (12) | 0.0044 (11) | −0.0028 (10) | −0.0046 (10) |
| O2 | 0.0295 (13) | 0.0182 (11) | 0.0178 (12) | 0.0025 (11) | −0.0017 (11) | 0.0003 (9) |
| O6 | 0.0283 (14) | 0.0192 (12) | 0.0346 (14) | 0.0004 (11) | 0.0046 (13) | −0.0042 (11) |
| O3 | 0.0205 (12) | 0.0169 (11) | 0.0245 (12) | −0.0013 (10) | −0.0035 (12) | −0.0037 (10) |
| O1 | 0.0307 (15) | 0.0169 (11) | 0.0206 (12) | −0.0025 (11) | 0.0042 (11) | −0.0037 (10) |
| O5 | 0.0393 (16) | 0.0398 (15) | 0.0223 (12) | −0.0063 (14) | −0.0051 (13) | −0.0065 (11) |
| C3 | 0.0198 (18) | 0.0146 (15) | 0.0166 (16) | 0.0006 (13) | −0.0007 (15) | −0.0015 (13) |
| C5 | 0.0248 (19) | 0.0164 (16) | 0.0212 (18) | −0.0009 (15) | −0.0002 (17) | −0.0046 (14) |
| C4 | 0.0219 (19) | 0.0151 (16) | 0.0223 (19) | 0.0042 (15) | −0.0024 (15) | −0.0015 (14) |
| C1 | 0.028 (2) | 0.0183 (17) | 0.0167 (17) | 0.0027 (16) | 0.0007 (16) | 0.0000 (14) |
| C2 | 0.024 (2) | 0.0189 (18) | 0.0196 (18) | 0.0002 (15) | 0.0002 (15) | −0.0014 (14) |
Geometric parameters (Å, °)
| P2—O6 | 1.444 (2) | C3—C5 | 1.523 (5) |
| P2—O4 | 1.561 (2) | C3—C4 | 1.528 (4) |
| P2—O3 | 1.565 (2) | C3—C2 | 1.532 (4) |
| P2—Cl2 | 2.0047 (13) | C3—C1 | 1.533 (4) |
| P1—O5 | 1.446 (2) | C5—H5A | 0.9900 |
| P1—O1 | 1.555 (2) | C5—H5B | 0.9900 |
| P1—O2 | 1.571 (2) | C4—H4A | 0.9900 |
| P1—Cl1 | 2.0050 (14) | C4—H4B | 0.9900 |
| O4—C5 | 1.461 (3) | C1—H1A | 0.9900 |
| O2—C2 | 1.457 (4) | C1—H1B | 0.9900 |
| O3—C4 | 1.464 (4) | C2—H2A | 0.9900 |
| O1—C1 | 1.467 (4) | C2—H2B | 0.9900 |
| O6—P2—O4 | 114.00 (13) | O4—C5—H5A | 109.4 |
| O6—P2—O3 | 113.70 (14) | C3—C5—H5A | 109.4 |
| O4—P2—O3 | 106.12 (13) | O4—C5—H5B | 109.4 |
| O6—P2—Cl2 | 112.82 (11) | C3—C5—H5B | 109.4 |
| O4—P2—Cl2 | 104.61 (10) | H5A—C5—H5B | 108.0 |
| O3—P2—Cl2 | 104.71 (10) | O3—C4—C3 | 110.2 (3) |
| O5—P1—O1 | 113.75 (14) | O3—C4—H4A | 109.6 |
| O5—P1—O2 | 113.36 (14) | C3—C4—H4A | 109.6 |
| O1—P1—O2 | 106.39 (12) | O3—C4—H4B | 109.6 |
| O5—P1—Cl1 | 113.26 (12) | C3—C4—H4B | 109.6 |
| O1—P1—Cl1 | 104.73 (10) | H4A—C4—H4B | 108.1 |
| O2—P1—Cl1 | 104.49 (11) | O1—C1—C3 | 109.5 (2) |
| C5—O4—P2 | 119.3 (2) | O1—C1—H1A | 109.8 |
| C2—O2—P1 | 119.24 (19) | C3—C1—H1A | 109.8 |
| C4—O3—P2 | 119.6 (2) | O1—C1—H1B | 109.8 |
| C1—O1—P1 | 121.3 (2) | C3—C1—H1B | 109.8 |
| C5—C3—C4 | 109.2 (3) | H1A—C1—H1B | 108.2 |
| C5—C3—C2 | 112.5 (3) | O2—C2—C3 | 111.0 (3) |
| C4—C3—C2 | 108.6 (3) | O2—C2—H2A | 109.4 |
| C5—C3—C1 | 109.0 (3) | C3—C2—H2A | 109.4 |
| C4—C3—C1 | 108.0 (3) | O2—C2—H2B | 109.4 |
| C2—C3—C1 | 109.4 (3) | C3—C2—H2B | 109.4 |
| O4—C5—C3 | 111.2 (3) | H2A—C2—H2B | 108.0 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1A···O5i | 0.99 | 2.34 | 3.214 (4) | 147 |
| C1—H1B···O6ii | 0.99 | 2.31 | 3.252 (4) | 159 |
| C4—H4B···O5i | 0.99 | 2.36 | 3.260 (4) | 150 |
Symmetry codes: (i) −x+3/2, −y+2, z−1/2; (ii) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5060).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Elnagar, H. Y., Ranken, P. F. & Fronczek, F. R. (2000). Acta Cryst. C56, 905–906. [DOI] [PubMed]
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Granzow, A. H. (1981). US Patent 4 257 931.
- Heinemann, F. W., Hartung, H., Kugler, S. & Kircheiss, A. (1994). Z. Kristallogr.209, 558–559.
- Li, B., Sun, C.-Y. & Zhang, X.-C. (2002). CN Patent 1 414 000A.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tanabe, S., Yanagida, T., Imamura, K., Tando, K. & Taketani, Y. (2005). EP Patent 1 586 576.
- Zhang, Y.-H., Wang, X.-H., Liu, S. & Yao, C. (2006). Acta Cryst. E62, o2620–o2621.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810043333/xu5060sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810043333/xu5060Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report