Abstract
Antifreeze proteins protect several cold-blooded organisms from subzero environments by preventing death from freezing. The Type I antifreeze protein (AFP) isoform from Pseudopleuronectes americanus, named HPLC6, is a 37-residue protein that is a single α-helix. Mutational analysis of the protein showed that its alanine-rich face is important for binding to and inhibiting the growth of macromolecular ice. Almost all structural studies of HPLC6 involve the use of chemically synthesized protein as it requires a native N-terminal aspartate and an amidated C-terminus for full activity. Here, we examine the role of C-terminal amide and C-terminal arginine side chain in the activity, structure, and dynamics of nonamidated Arg37 HPLC6, nonamidated HPLC6 Ala37, amidated HPLC6 Ala37, and fully native HPLC6 using a recombinant bacterial system. The thermal hysteresis (TH) activities of the nonamidated mutants are 35% lower compared with amidated proteins, but analysis of the NMR data and circular dichroism spectra shows that they are all still α-helical. Relaxation data from the two nonamidated mutants indicate that the C-terminal residues are considerably more flexible than the rest of the protein because of the loss of the amide group, whereas the amidated Ala37 mutant has a C-terminus that is as rigid as the wild-type protein and has high TH activity. We propose that an increase in flexibility of the AFP causes it to lose activity because its dynamic nature prevents it from binding strongly to the ice surface.
Keywords: amidation, antifreeze protein, α-helicity, HPLC6, ice, dynamics, thermal hysteresis
Introduction
Cold-blooded organisms living in low-temperature environments face death from ice growth, either through mechanical damage or by dehydration because of the lack of liquid water. Some organisms produce large quantities of polyols to colligatively depress the freezing point, where the depression depends on the concentration of the molecules rather than on their chemical nature. Over 50 years ago, it was observed that the osmolarity of Arctic fish was insufficient to explain their ability to avoid freezing in −1.8°C sea water.1 This noncolligative protective strategy was subsequently shown to involve the production of antifreeze proteins (AFPs, also known as thermal hysteresis (TH) proteins, ice-binding proteins, or ice-structuring proteins). Discovered in fish,2 arthropods,3 plants,4 fungi,5 and bacteria,6 AFPs are thought to function by what is known as the Kelvin effect—the protein binds to the ice surface and forces the ice to grow as a curved front, at which point further ice growth becomes inhibited.7,8 The macroscopic, noncolligative nature of the freezing point depression results in a difference between the freezing and melting points of the solution. This difference is known as TH and is typically used to quantitate the ice-growth inhibition capabilities of an AFP.9 In addition, ice grown in the presence of a fish AFP forms a hexagonal bipyramid, compared with the round plate morphology seen with water alone.10
The Kelvin effect has been generally accepted as the mechanism by which ice growth is inhibited. How the AFP binds to the ice surface has been more difficult to determine as there are no consistently conserved motifs in AFP protein sequences or structures. The known and predicted structural folds include a single α-helical structure (Type I AFP),11 lectin folds (Type II AFP),12,13 mixed irregular α-helix and β-strand (Type III AFP),14,15 α-helical bundle (Type IV AFP),16 β-helical fold (some insect and plant AFPs),17–19 and polyproline Type II fold (snow flea AFP).20,21 The structures of Types II–IV AFPs and the insect AFPs are reviewed in Refs. 22–25.
The Type I AFPs are found in two flounders, several sculpins, and Liparis families and tend to be short (∼40 residues) or medium (∼100–200 residues) in length.10,26 The simplicity and relative abundance of the HPLC6 isoform from winter flounder (Pseudopleuronectes americanus) has made it a model system for the study of all AFPs. The protein is only 37 residues long, of which 23 are Ala. The X-ray crystallographic structure shows that nearly the entire protein is a single α-helix, with only the C-terminal Arg37 residue being a 310 helix.11 Solution NMR studies of HPLC6 in supercooled water27 and solid-state NMR studies of the protein in ice28 have shown that the protein maintains its α-helical secondary structure under conditions in which it would bind or be bound to an ice surface.
Several residues have been proposed to be important in maintaining the helical structure of HPLC6. The helix is stabilized by a salt bridge in the middle and cap structures at the two termini. The removal of the salt bridge between Asp18 and Glu22 by mutating residue 22 to Ala lowers the α-helical content and TH activity by 10%.29 In vivo, a C-terminal amide is added to neutralize the C-terminal negative dipole of the helix, and the Arg37 side chain has been proposed to form a hydrogen bond with a backbone carbonyl atom.11 The N-terminal residue, Asp1, forms a cap structure by hydrogen bonding to several nearby backbone amide groups. Both caps stabilize the unsatisfied backbone hydrogen bonds of the first and last four residues to prevent the helix from unwinding and neutralize the destabilizing helix dipoles.30
Many mutations have been made to Type I AFP HPLC6 to determine which residues are involved in inhibiting ice growth (see, for example, Refs. 29 and 31–36). The methyl groups of the Ala-rich face (Ala17 and Ala21, and equivalent positions) and the methyl groups of the four Thr residues (Thr2, Thr13, Thr24, and Thr35) were shown to be essential for full TH activity. Removal of the central two Thr methyl groups (by substitution with Ser)31,32,37 or changing Ala residues on the Ala17 face to Leu resulted in a >50% loss in activity.35 In contrast, removal of the Thr hydroxyl groups by mutation of the central two Thr residues to Val resulted in a moderate loss of activity (∼15% loss).31,37 These results suggest that van der Waals interactions between the methyl groups of these residues and the ice surface keep the protein bound to ice. Why hydrogen bonds are not involved and how van der Waals interactions promote ice binding are still unclear.
We examined the NMR structure of HPLC6 using a new expression system in bacteria that makes a recombinant AFP that has been named rHPLC6. Three mutants were also created, one lacking the C-terminal Arg37 side chain (rHPLC6-Ala37-NH2), one lacking the C-terminal amide (rHPLC6-Arg37), and one lacking both the C-terminal amide and the Arg37 side chain (rHPLC6-Ala37) to examine their importance in maintaining helical stability and full TH activity. Our experiments show that the loss of the amide causes an increase in the flexibility of the C-terminal region and a loss in TH activity, and that the Arg37 side chain has a minimal role in keeping the helix rigid so that it can interact effectively with ice.
Results
Expression and purification of Type I AFP and mutants
A number of Type I AFP expression systems have been used, but none have produced fully native HPLC6. To make rHPLC6, the recombinant protein rHPLC6-Ala37 was amidated through the transamidase activity of carboxypeptidase-Y (CPD-Y).38,39 For a description of the cloning and purification of rHPLC6-Ala37-NH2, rHPLC6-Arg37, and rHPLC6-Ala37 please see the Supporting Information. The C-terminal peptidase activity of CPD-Y can remove the terminal residue of a protein, and, when the reaction is performed in an excess of an amino acid (such as argininamide), perform the reverse reaction to attach the amidated amino acid. The correctly amidated-form rHPLC6 was separated from other products by reversed-phase HPLC. Despite the optimization of incubation time, temperature, protein concentration, and amidated amino acid concentration, the conversion rate was 25–30% for rHPLC6 and <4% for rHPLC6-Ala37-NH2. No substrate could be detected after the conversion reaction, but several other uncharacterized products were formed. The identity of the correct rHPLC6 product was shown by mass spectrometry (3242 ± 3 Da compared with the predicted value of 3242 Da) and by the observation that the amidated product eluted from the C18 column with a retention time identical to that of chemically synthesized Type I AFP.
Activity of Type I AFP and C-terminal mutants
To demonstrate that the recombinant Type I AFP rHPLC6 behaved the same way as the synthetic form, we examined its effect on ice crystal morphology and measured its TH. An ice crystal grown in the presence of synthetic AFP forms hexagonal bipyramids40; the identical morphology was seen for an ice crystal grown in the presence of the recombinant protein [Fig. 1(A)]. The TH measurements of recombinant and synthetic AFPs are also essentially identical [Fig. 1(B)]. All of these data (mass spectrometry, ice crystal morphology, and TH activity) prove that rHPLC6 is completely indistinguishable from HPLC6 produced by peptide synthesis.
Figure 1.
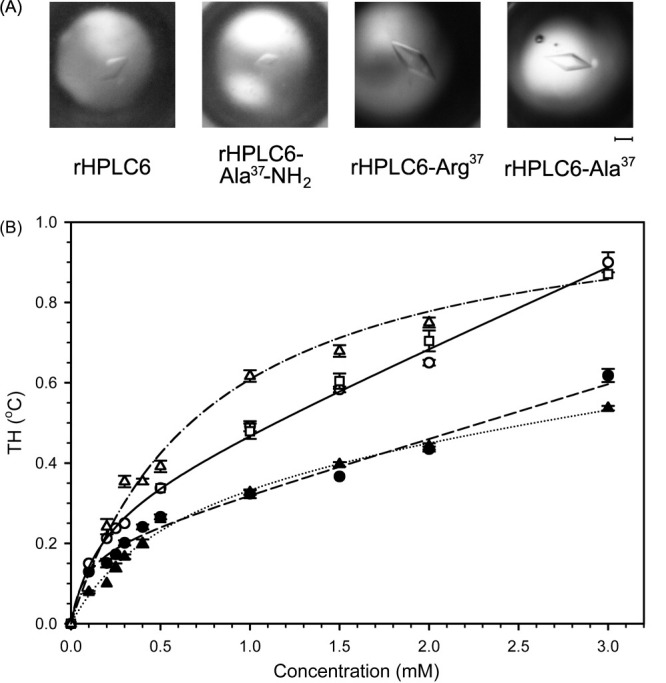
Activity of rHPLC6 and mutant constructs. (A) Ice crystal morphology of the four proteins in the thermal hysteretic gap. The identity of the protein is indicated below each photograph. The scale bar is the equivalent of 25 μm in length for all photographs. (B) Thermal hysteresis measurements. The activities of the constructs were measured using a nanoliter osmometer as described in the Material and Methods section. Synthetic HPLC6, open square; rHPLC6, open circle; rHPLC6-Ala37-NH2, open triangle; rHPLC6-Arg37, solid circle; rHPLC6-Ala37, solid triangle. The lines indicate the average trend for each of the proteins. Synthetic HPLC6 and rHPLC6, solid line; rHPLC6-Ala37-NH2, dotted and dashed line; rHPLC6-Arg37, dashed line; rHPLC6-Ala37, dotted line. The error bars represent the standard deviation of three independent measurements.
The effects of the rHPLC6-Ala37-NH2, rHPLC6-Arg37, and rHPLC6-Ala37 substitutions on the AFP activity were also examined. As can be seen in Figure 1, there is no difference with respect to ice crystal morphology, but the same is not true for the TH measurements [Fig. 1(B)]. Both rHPLC6-Ala37 and rHPLC6-Arg37 had very similar TH activities (∼65% activity compared with the wild-type protein), which is close to the 70% activity reported by Tong et al. for their recombinant HPLC6 isoform that also lacked a C-terminal amide.41 At 3 mM protein concentration, there is an apparent difference in TH between rHPLC6-Arg37 and rHPLC6-Ala37 (14% less activity for the Ala mutant). Higher concentrations of the Ala37 form undergo nonspecific aggregation as seen by increased linewidths in the 15N-HSQC spectra (data not shown), such that the loss of activity arises from aggregated protein not being able to inhibit ice growth. The rHPLC6-Ala37-NH2 protein had greater than wild-type activity at lower concentrations (∼105%) and similar activities at higher concentrations, which differs from the previously reported 100% activity for this mutant.29
Structural characterization
The structural effects of the C-terminal substitutions were examined using CD spectroscopy and NMR to determine why the nonamidated mutants had lost their activity. Previous studies of Type I AFP have shown that the loss of TH activity can correlate with a loss of helicity.10,29 To see whether this is the case here, the CD spectra of the four proteins at 3°C were compared (Fig. 2). The molar elliptical minima at 208 and 222 nm indicate that the wild-type protein rHPLC6 is α-helical, as has been shown in previous CD studies and from the X-ray crystallographic structure.10,11 The spectra of the two nonamidated mutants are identical to that of rHPLC6, showing that the alterations had no effect on the overall conformation, and that the loss of activity is not due to a loss of secondary structure. Likewise, the CD spectrum of rHPLC6-Ala37-NH2 is identical to all three other AFPs.
Figure 2.
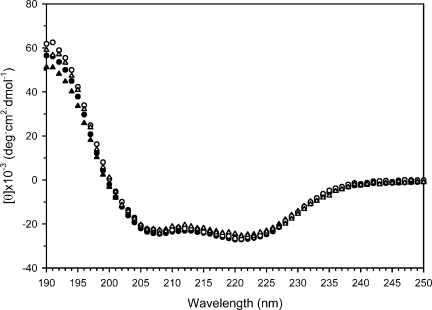
Global structure of rHPLC6 and mutants. The CD spectra of the proteins, collected at 3°C. rHPLC6, open circle; rHPLC6-Ala37-NH2, open triangle; rHPLC6-Arg37, solid circle; rHPLC6-Ala37, solid triangle.
Residue-specific changes in structure were examined by using 15N-labeled proteins and by collecting 15N-HSQC spectra [Fig. 3(A)]. The majority of the rHPLC6-Ala37-NH2, rHPLC6-Arg,37 and rHPLC6-Ala37 resonances are overlapped or nearly overlapped with those of rHPLC6. To show which regions may have been altered, a plot of the weighted chemical shift change of the backbone amide 1H and 15N atoms of the three mutant proteins is shown in Figure 3(B). Residues 1–25 in the mutant proteins have chemical shifts differences of <0.05 ppm, demonstrating that this part of the protein is completely unaffected by the mutations. Residues 26 and 27 show larger changes (0.05–0.10 ppm), whereas residues 28–34 show smaller changes (∼0.05 ppm). The largest chemical shift change occurs with residues 35 and 36 because of the loss of the amide and/or mutation of Arg37 to Ala37.
Figure 3.
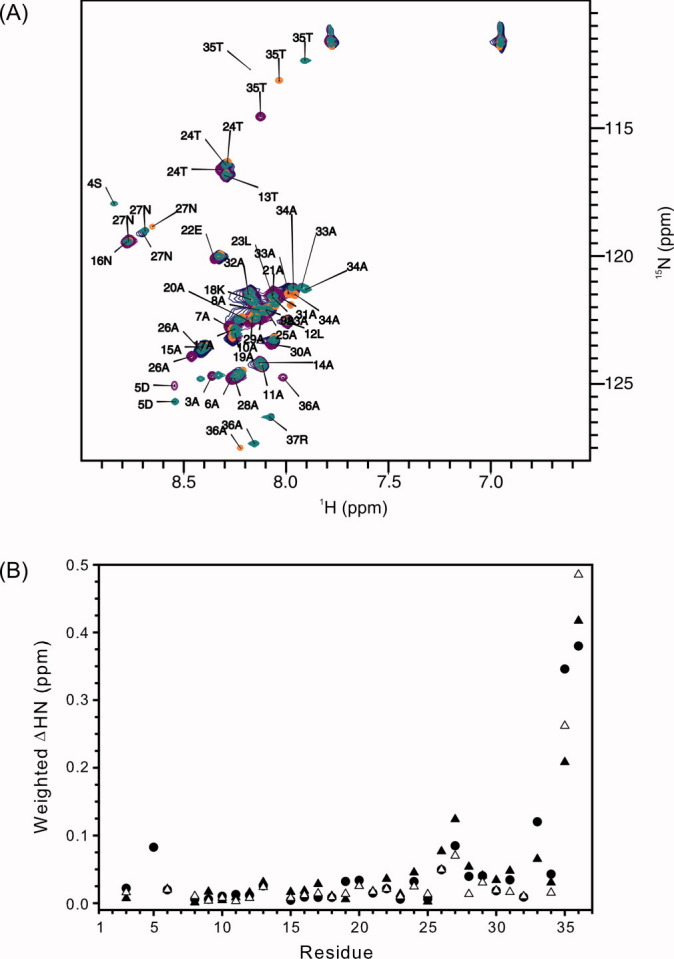
Local structure of rHPLC6, rHPLC6-Ala37-NH2, rHPLC6-Arg37, and rHPLC6-Ala37. (A) Overlapped 15N-HSQC spectra of rHPLC6 and mutants at 3°C. The residues of all of the assigned peaks of the wild-type protein are labeled with the residue number and single-letter amino acid code. Peaks of the mutant proteins that have shifted significantly [>0.05 ppm as shown in (B)] are also labeled. rHPLC6, purple; rHPLC6-Ala37-NH2, blue; rHPLC6-Arg37, green; rHPLC6-Ala37, orange. (B) Chemical shift changes of the mutants compared with the wild-type protein. Weighted changes in backbone 1H and 15N chemical shifts of rHPLC6-Ala37-NH2 (open triangles), rHPCL6-Arg37 (solid circles), and rHPLC6-Ala37 (solid triangles) are plotted on a per residue basis.
Effect of mutations on dynamics
Because the three C-terminal mutants did not have any significant structural differences compared with wild-type rHPLC6, relaxation experiments (15N-R1, 15N-R2, and 15N-NOE) of all four proteins were performed at 3°C to see whether an increase in dynamics due to the loss of the C-terminal functional groups would correlate with the changes in TH activity. In all four proteins, several residues are missing in the data because of resonance overlap (Ala7 and Ala14) or a lack of assignments (N-terminal residues Asp1, Thr2, and Ser4). The C-terminal residue of rHPLC6-Ala37-NH2 and rHPLC6 is missing because of a lack of labeled alaninamide and argininamide. For the resonances that were assigned, the relaxation measurement results are shown in Figure 4. For the R1 values of rHPLC6, the average was 1.28 ± 0.05 s−1. For rHPLC6-Arg37, rHPLC6-Ala37, and rHPLC6-Ala37-NH2, the R1 averages were 1.37 ± 0.06, 1.33 ± 0.05, and 1.3 ± 0.1 s−1, respectively. The R1 measurements are essentially the same for all four proteins, and no region of the protein shows any large variation from this average value [Fig. 4(A)]. For the wild-type protein, the average R2 value was 12 ± 2 s−1, with only residues 34–36 showing a slight decrease [Fig. 4(B)]. Similarly, the average R2 value was 11 ± 1 s−1 for rHPLC6-Ala37-NH2, with residues 34 and 36 also showing a slight decrease. For the nonamidated mutant proteins, the average R2 value over residues 1–30 was essentially the same as the wild-type protein, with rHPLC6-Arg37 being 13 ± 1 s−1 and rHPLC6-Ala37 being 11.6 ± 0.8 s−1. Over the same region, the average NOE ratio for rHPLC6 was 0.8 ± 0.2 and for rHPLC6-Ala37-NH2 was 0.8 ± 0.1, with the ratio being 0.84 ± 0.07 for rHPLC6-Arg37 and 0.82 ± 0.08 for rHPLC6-Ala37 [Fig. 4(C)]. One exception in this region is Asp5, which showed an increased R2 value compared with other N-terminal residues. Interestingly, residues located before and after this residue (Ala3 and Ala6) do not show this increase, indicating that the N-terminal cap may stabilize residues 1–4 and helical contacts stabilize residue 6 onward, but residue 5 is not stabilized, making it more flexible. Although only one residue is affected, the increase in dynamics near the N-terminus is somewhat similar to the “hinge” seen in the shorthorn sculpin ss3 AFP structure.42 For the wild-type protein rHPLC6 and rHPLC6-Ala37-NH2, the relatively constant R1, R2, and NOE values of residues 1–30 show that this part of the helix is quite rigid. This also applies to the other two mutants, but for residues 31 onward the four proteins display differences in their relaxation data.
Figure 4.
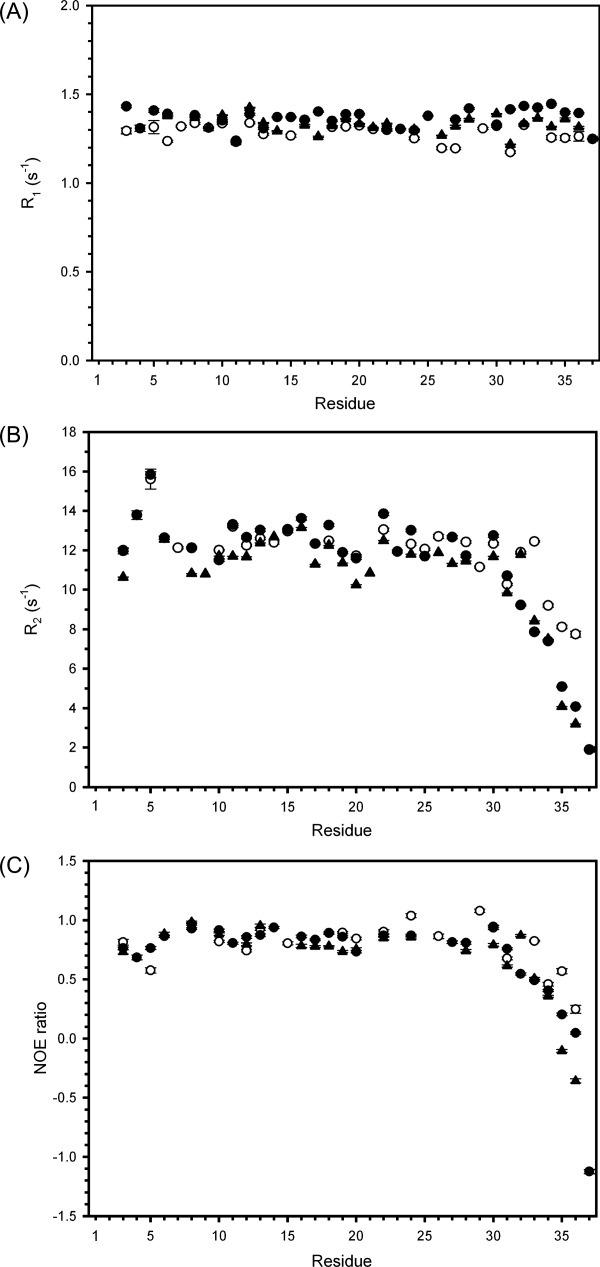
Relaxation data of rHPLC6 and mutants. The (A) R1, (B) R2, and (C) 15N-NOE data are plotted on a per residue basis. Error bars represent the error in fitting the relaxation decay curves as described in CCPNMR. rHPLC6, open circle; rHPLC6-Ala37-NH2, open triangle; rHPLC6-Arg37, solid circle; rHPLC6-Ala37, solid triangle.
Decreasing R2 values and decreasing NOE ratios of all four proteins show that they have increased flexibility for the C-terminal residues compared with the rest of the protein. The R2 and NOE data for rHPLC6 and rHPLC6-Ala37-NH2 indicate that the C-terminal residues are only slightly more flexible than the central and N-terminal residues. In contrast, the R2 and NOE values for rHPLC6-Arg37 and rHPLC6-Ala37 at the C-terminus changed substantially, which shows that residues 31–37 are flexible compared with the rest of the protein. The change in the dynamics of these two proteins is somewhat different, with the Ala37 form showing a slightly larger increase in flexibility as assessed by these relaxation parameters compared with Arg37. Because this is the most substantial difference observed between the nonamidated mutants and the amidated proteins, it likely represents the cause for the loss of TH activity that was observed.
Discussion
Wild-type protein expression and structure
Several systems have been developed to express recombinant Type I AFP HPLC6 isoform in bacteria, including an artificially longer variant of the HPLC6 isoform,43 the pro-form of HPLC6 fused to galactosidase,44 and the HPLC6 mature isoform.41 All of these constructs, however, are not structurally identical to the native isoform, and as such they are cannot achieve the same level of TH activity or change ice crystal morphology in the same way. Here, we created a recombinant expression system for fully native, isotopically labeled, HPLC6 isoform to determine what effect the C-terminal amide and Arg37 side chain have on the protein's structure and function.
An earlier study using 13Cα relaxation experiments suggested that the C-terminal Thr35 and Arg37 might be more flexible than other parts of HPLC6, but the weak signal due to the use of natural abundance carbon prevented a definitive statement from being made.28 We show here that there is a moderate increase in the flexibility at the C-terminus of Type I AFP HPLC6, demonstrating that a correctly positioned Thr35 methyl group on the Ala-rich face is not critical for activity. A study on the Thr35 to Ser mutant supports our proposal, as the mutant was 90% active compared with TH measurements made with wild-type protein.37
Loss of amidation compared with loss of arginine side chain
The data show that the loss of both the C-terminal amide and side chain (rHPLC6-Ala37) compared with the loss of the C-terminal amide (rHPLC6-Arg37) did not have a significantly additive effect on the activity, structure, or relaxation measurements. With respect to the activity measurements, the difference between rHPLC6-Arg37 and rHPLC6-Ala37 was the same at a concentration of 2 mM or less (65% compared with the wild-type rHPLC6 at a concentration of 1 mM). The CD analysis showed no significant change in helical content among all three proteins, and the relaxation data showed that residues 31–37 of the mutants are more flexible than those in the wild-type protein, with rHPLC6-Ala37 being somewhat more flexible than the Arg37 protein. Similarly, the loss of the side chain but the retention of the amide in rHPLC6-Ala37-NH2 showed that the C-terminal residues are slightly more rigid than those of the wild-type protein, which may explain the incrementally greater activity of the mutant.
The structure of the C-terminal cap from the X-ray crystallographic study,11 shown in Figure 5, provides an explanation as to why amidation is more important for structural stability than the side chain. An inspection of Arg37 shows that the side chain could form a hydrogen bond with the carbonyl of Ala33. However, the position of Arg37 side chain in the crystal is held in place by two hydrogen bonds to the side chain Asp5 of a nearby symmetry mate. In comparison, the only crystal contacts of the amide cap of Arg37 are with two water molecules, showing that its position is not restrained in the crystal. The C-terminal amide could therefore make a hydrogen bond to the carbonyl of Ala34 or Thr35. Thus, the amide group is important in stabilizing these two residues, whereas the Arg37 side chain probably has only a moderate effect on C-terminal stabilization by possibly weakly hydrogen bonding to a nearby carbonyl. The greater than wild-type TH of the rHPLC6-Ala37-NH2 mutant indicates that any stabilization by the arginine side chain is not critical for its activity. The unimportance of the Arg side chain is also supported by the study of Doig and Baldwin using nonamidated, model α-helical peptides.45 In that work, the substitution of the C-terminal residue Arg with Ala only decreased the helicity by ∼2%.
Figure 5.
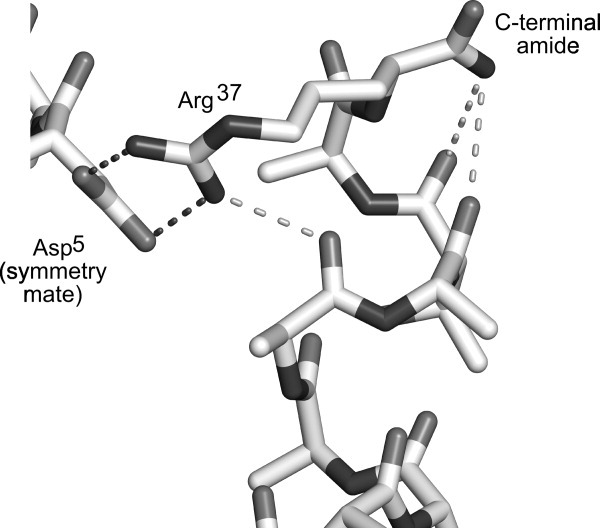
Cap structure of Type I AFP. The C-terminal region of Type I AFP from the crystal structure (PDB 1WFB, chain B) is shown in stick representation. Asp5 of a nearby symmetry mate is also shown. Intramolecular hydrogen bonds of the Arg37 side chain and the amide group are shown as light gray dotted lines, whereas intermolecular hydrogen bonds are shown in dark gray. The figure was drawn using Pymol (Version 1.2r1, Schrödinger, LLC).
The difference in TH between the two nonamidated proteins (14%) at 3 mM is likely due to aggregation of rHPLC6-Ala37 at higher concentrations of the protein, an observation also reported for other Type I AFP mutants.36,46 One explanation for this self-association is that the increased flexibility of rHPLC6-Ala37 allows the Ala-rich regions to form optimal hydrophobic interactions, whereas the presence of Arg37 causes both steric blockage and charge repulsion, thus preventing the proteins from binding to each other.
Effect of cap mutants on AFP activity
As a model AFP, HPLC6 has been extensively mutated to elucidate its mechanism of ice growth inhibition.29,31–36,46 The effects of the mutations can be broadly classified into those that (a) disrupt favorable contacts between the protein and ice because of the substitution of a residue to one with a long side chain (e.g., Ala to Leu mutation),35 (b) result in a loss of a favorable contact between the protein and ice because of the deletion of a functional group (e.g., Thr to Ser mutations),31,32,37 or (c) disrupt the backbone structure of the protein (e.g., loss of the salt bridge).29
In the cases involving the nonamidated rHPLC6-Arg37 and rHPLC6-Ala37, none of these possibilities are fully consistent with the experimental observations made here. In neither mutant is a residue introduced with a long side chain, eliminating explanation a. The rHPLC6-Ala37 mutant does involve the deletion of the Arg37 side chain, but our results with rHPLC6-Ala37-NH2 and those of Wen and Laursen29 show that there is no loss in TH activity or change in structure. Therefore, the Arg37 side chain is not important in the ice-binding interaction, and the terminal amide group is not likely to be involved in ice binding. Otherwise, all other atoms are present, making explanation b not an answer. Lastly, the CD analysis suggests that explanation c is not likely, as the helical structure for the protein has not been perturbed.
We propose that rHPCL6-Arg37 and rHPCL6-Ala37 lose activity because of their increased flexibility and not because of a loss of a functional group or a change in secondary structure. The current model of AFP/ice interactions is proposed to involve van der Waals contacts between the protein and the ice surface, which are rather weak in nature. The gain in flexibility of rHPLC6-Arg37 and rHPLC6-Ala37 disrupts this rigidity and would weaken these interactions. Should this dynamics occur while the protein is bound to ice, the movement of the C-terminal region away from the ice surface would have to be very limited; otherwise, the >10,000-fold molar excess of water over AFP would allow the ice to grow rapidly.8 This wriggling may therefore represent the transient loss of a small number of contacts between the protein and ice, such that the protein is never fully dissociated from ice but sufficiently enough to lower the TH barrier.
Rigidity has been suggested to be a common property of AFPs,24 with the unusual structure of Tenebrio molitor being an extreme example of this.47,48 It has been stated that rigidity is important for maintaining a protein's structure in supercooled water.49 The hydrophobic effect is weaker at lower temperatures so that the core of a globular AFP is less able to keep the structure intact. It is possible that an additional role of rigidity is to keep an AFP in intimate contact with a rigid ice surface. It should be noted that the experiments reported here were performed at 3°C, and it is possible that the dynamics of HPLC6 could be different when bound to an ice surface.
We have produced isotopically labeled Type I AFP HPLC6 and three mutants to study the role of the C-terminal cap in the protein's structure and dynamics. Our results show that the loss of the C-terminal amide increases the flexibility of several C-terminal residues, which leads to a loss of thermal hysteretic activity, and that the terminal arginine residue is not important for helical stability or antifreeze activity. To perform the NMR experiments, we created a bacterial recombinant expression system that produces fully native and isotopically labeled Type I AFP, which has not been previously possible. This new system can be exploited to further understand how AFPs are able to bind to ice.
Materials and Methods
Cloning, expression, and protein purification
The Type I AFP, HPLC6 gene was a generous gift from Daniel Yang (McMaster University).41 A description of the material and methods used for the cloning and preparation of the nonamidated protein is included in the Supporting Information.
C-terminal amidation of rHPLC6-Ala37 and rHPLC6-Arg37
The transacylation reaction as outlined by Zhang et al.39 was used to convert Ala37 in rHPLC6-Ala37 into amidated Arg37 (rHPLC6). Briefly, 25 μL of a 40 mM stock of rHPLC6-Ala37 was mixed with 975 μL of a nucleophile solution [1M argininamide-HCl (Sigma Aldrich, St. Louis, MO) and 5 mM EDTA (pH 8.5)]. The reaction was initiated by the addition of 10 μg of carboxypeptidase Y (Sigma Aldrich, St. Louis, MO). The solution was incubated for 3 h at 45°C before the mixture of products was separated using reversed-phase HPLC with a C18 column. Conversion of rHPLC6-Ala37 to rHPLC6 was confirmed using mass spectrometry (Biological Mass Spectrometry Facility, University of Guelph). Fractions containing rHPLC6 were lyophilized for storage. The same procedure was used to produce rHPLC6-Ala37-NH2, except that 1M alaninamide-HCl (Sigma Aldrich, St. Louis, MO) was used with rHPLC6-Arg37 as the substrate.
Antifreeze protein activity measurement
Measurement of TH activity was carried out according to the method of Chakrabartty and Hew9 using an Otago nanoliter osmometer (Otago Osmometers, Dunedin, New Zealand). A ∼20 nL drop of protein solution in 10 mM ammonium bicarbonate was inserted into a Cargille oil Type B droplet in the cold-stage sample holder. The temperature was lowered to less than −20°C until the protein solution froze. Next, the temperature was raised and lowered until only a single crystal remained. The temperature was then slowly lowered (0.02°C decrements over ∼10 s) to see whether the ice crystal would change from a constantly growing plate with poorly defined edges into one with sharply defined boundaries and arrested growth. Digital photographs of ice crystals within the TH gap were taken using a Canon G10 digital camera with automatic exposure and focusing.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra of the four proteins were collected using a Jasco-815 CD spectropolarimeter (Easton, MD). Protein samples were dissolved in 10 mM ammonium bicarbonate at a concentration of 0.22 mg/mL (rHPLC6-Ala37-NH2), 0.25 mg/mL (rHPLC6 and rHPLC6-Ala37), or 0.5 mg/mL (rHPLC6-Arg37). A quartz cuvette with a 0.1-mm path length (Hellma, Concord, ON, Canada) was scanned from 250 to 190 nm. The spectra were averaged over eight scans, and all experiments were performed at 3°C.
NMR data collection and analysis
The lyophilized protein was resuspended in 600 μL of NMR buffer [0.01% sodium azide, 0.1 mM 2,2′-dimethyl-2-silapentane-5-sulfonate (DSS), and 10% D2O (v/v)] at a concentration of 90 μM (rHPLC6-Ala37-NH2) or 0.5 mM (all others), with the pH adjusted to 7.0 for a series of NMR experiments.50 All experimental data were collected on a Bruker Avance DRX600 spectrometer equipped with a cryogenic triple-resonance probe at a temperature of 3.0°C. 1H and 15N referencing was performed relative to DSS as described.51 For the 15N-heteronuclear single-quantum coherence (HSQC) experiments, 1024 (1HN) and 128 (15N) complex data points were acquired with a total of 32 (rHPLC6-Ala37-NH2) or 16 (all others) transients per t1 increment using spectral widths of 8370.5 (1HN) and 1338 (15N) Hz. For the 15N-edited NOESY and 15N-edited TOCSY experiments, 1024 (1HN), 20 (15N), and 64 (1H) complex data points were acquired with a total of 16 transients per t1 increment using spectral widths of 9615.0 (1HN), 1338 (15N), and 9615.0 (1H) Hz. R1, R2, and {1H}-15N-NOE relaxation data were collected using the same protein sample described above at 3.0°C. For all three experiments, 1024 (1HN) and 128 (15N) complex data points were acquired with a total of 16 transients per t1 increment using spectral widths of 8370.5 (1HN) and 1338 (15N) Hz. For the R1 experiments, relaxation delays of 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, and 2.0 s were applied. For the R2 experiments, relaxation delays of 16.064, 32.128, 48.192, 64.256, 80.320, 112.450, 144.580, 176.700, 208.830, and 240.960 ms were applied. The {1H}-15N steady-state NOE values represent the intensity ratio from the two HSQC spectra with and without 1H saturation applied before the 15N excitation pulse.
Spectra were processed using NMRPipe,52 and chemical shift assignments were made using the CCPNMR analysis software version 2.1.53 Chemical shifts of rHPLC6 1H and 15N atoms were obtained from previously published studies on the synthetic protein,28,54 and 15N-TOCY and 15N-NOESY experiments were used to assign resonances that underwent large chemical shifts in the rHPLC6-Arg37 and rHPLC6-Ala37 mutants. The R1 and R2 relaxation times were analyzed by fitting the cross-peak intensity decay to a two parameter, single exponential decay function I(t) = I0e−tR1,2, where I0 is the intensity at time t = 0 and I(t) is the intensity after a delay time t for the R1 or R2 experiment. The errors were estimated from the uncertainty of the nonlinear fits.
Acknowledgments
The authors thank the University of Guelph NMR Centre for use of the facility and help with setting up the experiments. The authors also thank Brent Sinclair (University of Western Ontario) for access to his nanoliter osmometer for several activity measurements. Infrastructure support to S.P.G. comes from the Canadian Foundation for Innovation and the Ontario Innovation Trust.
References
- 1.Scholander PF, van Dam L, Kanwisher J, Hammel T, Gordon MS. Supercooling and osmoregulation in arctic fish. J Cell Comp Physiol. 1957;49:5–24. [Google Scholar]
- 2.DeVries AL, Wohlschlag DE. Freezing resistance in some Antarctic fishes. Science. 1969;163:1073–1075. doi: 10.1126/science.163.3871.1073. [DOI] [PubMed] [Google Scholar]
- 3.Graham LA, Liou YC, Walker VK, Davies PL. Hyperactive antifreeze protein from beetles. Nature. 1997;388:727–728. doi: 10.1038/41908. [DOI] [PubMed] [Google Scholar]
- 4.Xu H, Griffith M, Patten CL, Glick BR. Isolation and characterization of an antifreeze protein with ice nucleation activity from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can J Micro. 1998;44:64–73. [Google Scholar]
- 5.Hoshino T, Kiriaki M, Ohgiya S, Fujiwara M, Kondo H, Nishimiya Y, Yumoto I, Tsuda S. Antifreeze proteins from snow mold fungi. Can J Botany. 2003;81:1175–1181. [Google Scholar]
- 6.Duman JG, Olsen TM. Thermal hysteresis protein-activity in bacteria, fungi, and phylogenetically diverse plants. Cryobiology. 1993;30:322–328. [Google Scholar]
- 7.Raymond JA, DeVries AL. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc Natl Acad Sci USA. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight CA, Cheng CC, DeVries AL. Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes. Biophys J. 1991;59:409–418. doi: 10.1016/S0006-3495(91)82234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabartty A, Hew CL. The effect of enhanced alpha-helicity on the activity of a winter flounder antifreeze polypeptide. Eur J Biochem. 1991;202:1057–1063. doi: 10.1111/j.1432-1033.1991.tb16470.x. [DOI] [PubMed] [Google Scholar]
- 10.Harding MM, Ward LG, Haymet AD. Type I ‘antifreeze’ proteins. Structure-activity studies and mechanisms of ice growth inhibition. Eur J Biochem. 1999;264:653–665. doi: 10.1046/j.1432-1327.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 11.Sicheri F, Yang DS. Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature. 1995;375:427–431. doi: 10.1038/375427a0. [DOI] [PubMed] [Google Scholar]
- 12.Gronwald W, Loewen MC, Lix B, Daugulis AJ, Soennichsen FD, Davies PL, Sykes BD. The solution structure of type II antifreeze protein reveals a new member of the lectin family. Biochemistry. 1998;37:4712–4721. doi: 10.1021/bi972788c. [DOI] [PubMed] [Google Scholar]
- 13.Nishimiya Y, Kondo H, Takamichi M, Sugimoto H, Suzuki M, Miura A, Tsuda S. Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus. J Mol Biol. 2008;382:734–746. doi: 10.1016/j.jmb.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 14.Sonnichsen FD, Sykes BD, Chao H, Davies PL. The nonhelical structure of antifreeze protein type III. Science. 1993;259:1154–1157. doi: 10.1126/science.8438165. [DOI] [PubMed] [Google Scholar]
- 15.Jia Z, DeLuca CI, Chao H, Davies PL. Structural basis for the binding of a globular antifreeze protein to ice. Nature. 1996;384:285–288. doi: 10.1038/384285a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng G, Laursen RA. Isolation and characterization of an antifreeze protein from the longhorn sculpin, Myoxocephalus octodecimspinosis. Biochim Biophys Acta. 1998;1388:305–314. doi: 10.1016/s0167-4838(98)00180-0. [DOI] [PubMed] [Google Scholar]
- 17.Graether SP, Kuiper MJ, Gagne SM, Walker VK, Jia Z, Sykes BD, Davies PL. Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature. 2000;406:325–328. doi: 10.1038/35018610. [DOI] [PubMed] [Google Scholar]
- 18.Liou Y, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a α-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- 19.Zhang DQ, Liu B, Feng DR, He YM, Wang SQ, Wang HB, Wang JF. Significance of conservative asparagine residues in the thermal hysteresis activity of carrot antifreeze protein. Biochem J. 2004;377:589–595. doi: 10.1042/BJ20031249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SB. X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J Am Chem Soc. 2008;130:9695–9701. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham LA, Davies PL. Glycine-rich antifreeze proteins from snow fleas. Science. 2005;310:461. doi: 10.1126/science.1115145. [DOI] [PubMed] [Google Scholar]
- 22.Jia Z, Davies PL. Antifreeze proteins: an unusual receptor-ligand interaction. Trends Biochem Sci. 2002;27:101–106. doi: 10.1016/s0968-0004(01)02028-x. [DOI] [PubMed] [Google Scholar]
- 23.Davies PL, Baardsnes J, Kuiper MJ, Walker VK. Structure and function of antifreeze proteins. Philos Trans R Soc Lond B Biol Sci. 2002;357:927–935. doi: 10.1098/rstb.2002.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graether SP, Sykes BD. Cold survival in freeze-intolerant insects. Eur J Biochem. 2004;271:3285–3296. doi: 10.1111/j.1432-1033.2004.04256.x. [DOI] [PubMed] [Google Scholar]
- 25.Patel SN, Graether SP. Structures and ice-binding faces of the alanine-rich type I antifreeze proteins. Biochem Cell Biol. 2010;88:223–229. doi: 10.1139/o09-183. [DOI] [PubMed] [Google Scholar]
- 26.Evans RP, Fletcher GL. Isolation and characterization of type I antifreeze proteins from Atlantic snailfish (Liparis atlanticus) and dusky snailfish (Liparis gibbus) Biochim Biophys Acta. 2001;1547:235–244. doi: 10.1016/s0167-4838(01)00190-x. [DOI] [PubMed] [Google Scholar]
- 27.Graether SP, Slupsky CM, Davies PL, Sykes BD. Structure of type I antifreeze protein and mutants in supercooled water. Biophys J. 2001;81:1677–1683. doi: 10.1016/S0006-3495(01)75821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graether SP, Slupsky CM, Sykes BD. Effect of a mutation on the structure and dynamics of an alpha-helical antifreeze protein in water and ice. Proteins. 2006;63:603–610. doi: 10.1002/prot.20889. [DOI] [PubMed] [Google Scholar]
- 29.Wen D, Laursen RA. Structure-function relationships in an antifreeze polypeptide. The role of charged amino acids. J Biol Chem. 1993;268:16396–16400. [PubMed] [Google Scholar]
- 30.Doig AJ. Stability and design of alpha-helical peptides. Prog Mol Biol Trans Sci. 2008;83:1–52. doi: 10.1016/S0079-6603(08)00601-6. [DOI] [PubMed] [Google Scholar]
- 31.Chao H, Houston ME, Hodges RS, Kay CM, Sykes BD, Loewen MC, Davies PL, Soennichsen FD. A diminished role for hydrogen bonds in antifreeze protein binding to ice. Biochemistry. 1997;36:14652–14660. doi: 10.1021/bi970817d. [DOI] [PubMed] [Google Scholar]
- 32.Haymet AD, Ward LG, Harding MM, Knight CA. Valine substituted winter flounder ‘antifreeze’: preservation of ice growth hysteresis. FEBS Lett. 1998;430:301–306. doi: 10.1016/s0014-5793(98)00652-8. [DOI] [PubMed] [Google Scholar]
- 33.Wen D, Laursen RA. Structure-function relationships in an antifreeze polypeptide. The role of neutral, polar amino acids. J Biol Chem. 1992;267:14102–14108. [PubMed] [Google Scholar]
- 34.Wen D, Laursen RA. Structure-function relationships in an antifreeze polypeptide. The effect of added bulky groups on activity. J Biol Chem. 1993;268:16401–16405. [PubMed] [Google Scholar]
- 35.Baardsnes J, Kondejewski LH, Hodges RS, Chao H, Kay C, Davies PL. New ice-binding face for type I antifreeze protein. FEBS Lett. 1999;463:87–91. doi: 10.1016/s0014-5793(99)01588-4. [DOI] [PubMed] [Google Scholar]
- 36.Loewen MC, Chao H, Houston ME, Baardsnes J, Hodges RS, Kay CM, Sykes BD, Soennichsen FD, Davies PL. Alternative roles for putative ice-binding residues in type I antifreeze protein. Biochemistry. 1999;38:4743–4749. doi: 10.1021/bi982602p. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Laursen RA. Structure-function relationships in a type I antifreeze polypeptide. The role of threonine methyl and hydroxyl groups in antifreeze activity. J Biol Chem. 1998;273:34806–34812. doi: 10.1074/jbc.273.52.34806. [DOI] [PubMed] [Google Scholar]
- 38.Breddam K, Widmer F, Meldal M. Amidation of growth hormone releasing factor (1-29) by serine carboxypeptidase catalysed transpeptidation. Int J Peptide Protein Res. 1991;37:153–160. doi: 10.1111/j.1399-3011.1991.tb00096.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang ZZ, Yang SS, Dou H, Mao JF, Li KS. Expression, purification, and C-terminal amidation of recombinant human glucagon-like peptide-1. Protein Expr Purif. 2004;36:292–299. doi: 10.1016/j.pep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Davies PL, Hew CL. Biochemistry of fish antifreeze proteins. FASEB J. 1990;4:2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- 41.Tong L, Lin Q, Wong WK, Ali A, Lim D, Sung WL, Hew CL, Yang DS. Extracellular expression, purification, and characterization of a winter flounder antifreeze polypeptide from Escherichia coli. Protein Expr Purif. 2000;18:175–181. doi: 10.1006/prep.1999.1176. [DOI] [PubMed] [Google Scholar]
- 42.Kwan AHY, Fairley K, Anderberg PI, Liew CW, Harding MM, Mackay JP. Solution structure of a recombinant type I sculpin antifreeze protein. Biochemistry. 2005;44:1980–1988. doi: 10.1021/bi047782j. [DOI] [PubMed] [Google Scholar]
- 43.Solomon RG, Appels R. Stable, high-level expression of a type I antifreeze protein in Escherichia coli. Protein Expr Purif. 1999;16:53–62. doi: 10.1006/prep.1999.1040. [DOI] [PubMed] [Google Scholar]
- 44.Peters ID, Hew CL, Davies PL. Biosynthesis of winter flounder antifreeze proprotein in E. coli. Protein Eng. 1989;3:145–151. doi: 10.1093/protein/3.2.145. [DOI] [PubMed] [Google Scholar]
- 45.Doig AJ, Baldwin RL. N- and C-capping preferences for all 20 amino acids in alpha-helical peptides. Protein Sci. 1995;4:1325–1336. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haymet AD, Ward LG, Harding MM. Hydrophobic analogues of the winter flounder ‘antifreeze’ protein. FEBS Lett. 2001;491:285–288. doi: 10.1016/s0014-5793(01)02213-x. [DOI] [PubMed] [Google Scholar]
- 47.Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- 48.Daley ME, Spyracopoulos L, Jia Z, Davies PL, Sykes BD. Structure and dynamics of a beta-helical antifreeze protein. Biochemistry. 2002;41:5515–5525. doi: 10.1021/bi0121252. [DOI] [PubMed] [Google Scholar]
- 49.Graether SP, Gagne SM, Spyracopoulos L, Jia Z, Davies PL, Sykes BD. Spruce budworm antifreeze protechanges in structure and dynamics at low temperature. J Mol Biol. 2003;327:1155–1168. doi: 10.1016/s0022-2836(03)00235-3. [DOI] [PubMed] [Google Scholar]
- 50.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR spectroscopy: principles and practice. London: Academic Press; 2007. [Google Scholar]
- 51.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1 H, 13 C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 54.Gronwald W, Chao H, Reddy DV, Davies PL, Sykes BD, Soennichsen FD. NMR characterization of side chain flexibility and backbone structure in the type I antifreeze protein at near freezing temperatures. Biochemistry. 1996;35:16698–16704. doi: 10.1021/bi961934w. [DOI] [PubMed] [Google Scholar]


