Abstract
NADPH oxidase complexes are multiprotein assemblies that generate reactive oxygen species in a variety of mammalian tissues. The canonical phagocytic oxidase consists of a heterodimeric, enzymatic core comprised of the transmembrane proteins, CYBB and CYBA and is regulated, in part, by an “organizing” function of NCF1 and an “activating” activity of NCF2. In contexts outside of the phagocyte, these regulatory functions may be encoded not only by NCF1 and NCF2, but alternatively by their respective paralogues, NOXO1 and NOXA1. To allow tissue-specific dissection of Noxa1 function in mouse, we have generated an allele of Noxa1 suitable for conditional inactivation. Moreover, by crossing Noxa1 conditional allele carriers to B6.129S4-Meox2tm1(Cre)Sor/J mice, we have generated first, Noxa1-null heterozygotes, and ultimately, Noxa1-null homozygotes. Through the thoughtful use of tissue-specific, Cre-expressing mouse strains, the Noxa1 conditional allele will offer insight into the roles of NOXA1 in the variety of tissues in which it is expressed.
Keywords: Cre-loxP, gene targeting, conditional inactivation, FLP-FRT, NADPH oxidase, Noxa1
NADPH oxidase (NOX) complexes are multiprotein assemblies with the enzymatic function of transferring electrons across biological membranes to oxygen (O2) thereby creating superoxide (O2•−) and a cascade of subsequent reactive oxygen species (ROS). Due to their reactivity, ROS interact with a variety of small inorganic molecules and biological macromolecules and can cause irreversible oxidative damage to these targets specifically, as well as to cells and organisms more generally. Nonetheless, systems have evolved to harness this reactivity into beneficial biological processes including host defense and a myriad of other reversible regulatory processes modulating such events as protein processing, cellular signaling, gene expression and cell differentiation. As a result of these opposing phenomena, NOX family NADPH oxidases are involved in a variety of physiological and pathophysiological processes (Bedard and Krause, 2007).
NADPH oxidase was initially discovered in phagocytic cells (phagocytic oxidase or phox) where it plays a vital role in host defense against bacterial and fungal pathogens. In fact, functional deficits within various components of the phagocytic oxidase complex underlie a genetically heterogeneous family of primary immune deficiencies known as the chronic granulomatous diseases (CGDs, OMIM #s 233690, 233700, 233710 and 306400) (Holland, 2010; Matute et al., 2009; Segal et al., 2009).
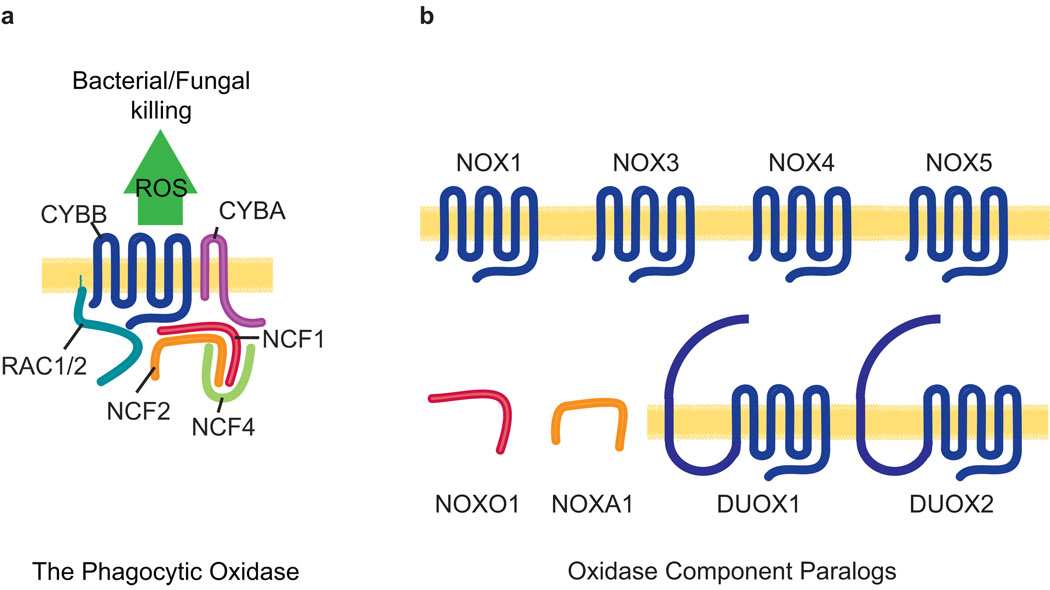
The phagocytic oxidase (Figure 1a) has been extensively studied and is now known to consist of a heterodimeric flavocytochrome at its enzymatic core. In turn, the heterodimer is comprised of CYBB, an NADPH-, FAD- and iron- (heme-) binding, six-span transmembrane protein; and, CYBA, a two-span transmembrane protein (for other naming conventions see Methods). Activation of the heterodimer, that is normally quiescent, requires the recruitment of at least two cytosolic factors including an “organizing” function of NCF1; and an “activating” activity of NCF2.
Figure 1. The phagocytic NADPH oxidase complex and its paralogues.
a. Schematic representation of the canonical NADPH oxidase of phagocytes. b. Paralogues of phagocytic oxidase components include the CYBB paralogues NOX1, NOX3, NOX4, NOX5, and the dual oxidase proteins DUOX1 and DUOX2; the NCF1 paralogue, NOXO1; and the NCF2 paralogue, NOXA1.
Expansion of the NOX family of NADPH oxidases occurred with the identification of a number of CYBB paralogues in the mammalian genome that are expressed in a wide variety of cells and tissues (Figure 1b). In humans, the NOX family includes six additional members — NOX1, NOX3, NOX4, NOX5 and the dual oxidase (NADPH oxidase and peroxidase) members, DUOX1 and DUOX2. Nox5 is absent in rodents (Nauseef, 2008). With regard to the organizing cytosolic factor NCF1, here too a paralogue has been identified, known as NOXO1 (Banfi et al., 2003).
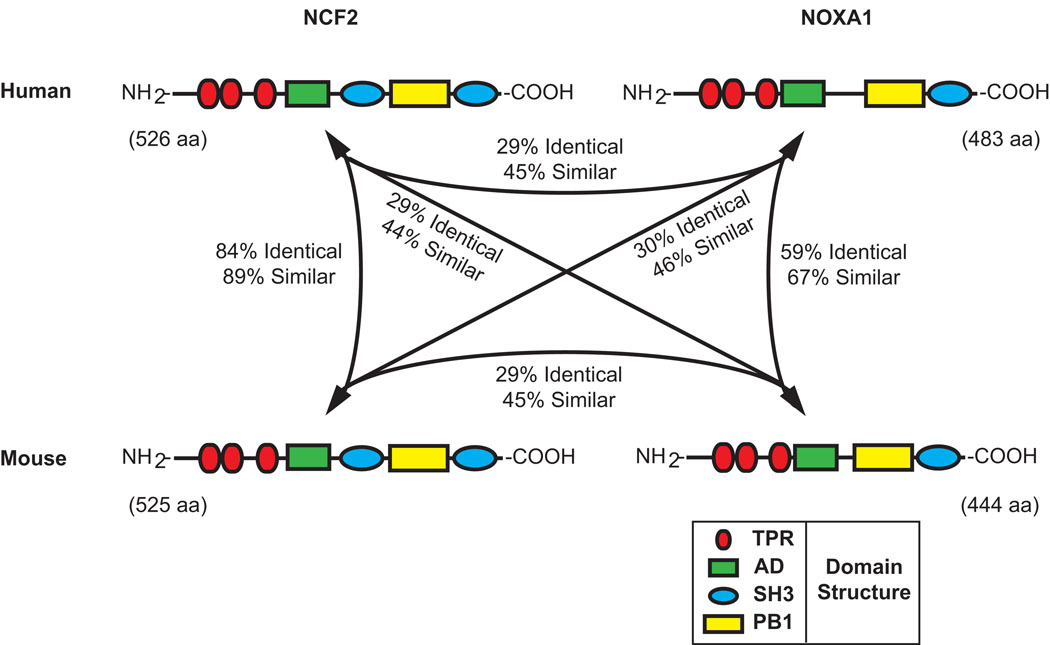
Likewise, an activator subunit paralogous to NCF2 has been identified and is known as NOXA1 (Banfi et al., 2003). NOXA1 is expressed in a variety of cell lines including guinea pig gastric mucosal cells, basilar artery epithelial cells, airway-like normal human bronchial epithelial cells and vascular smooth muscle cells (Ago et al., 2005; Ambasta et al., 2006; Bedard and Krause, 2007; Kawahara et al., 2005; Lavigne and Eppihimer, 2005). It is expressed in a number of organs including the spleen, stomach, colon, small intestine, lung, and uterus as well as the prostate, thyroid, and salivary glands (Banfi et al., 2004; Bedard and Krause, 2007; Geiszt et al., 2003). Interestingly, NOXA1 is also expressed in the inner ear (Banfi et al., 2004) where it may play a role in an emerging complex consisting of NOX3, CYBA and NOXO1 (Banfi et al., 2004; Bergstrom et al., 2004; Kiss et al., 2006; Nakano et al., 2008; Paffenholz et al., 2004). In human, the NOXA1 transcription unit spans 11,010 base pairs (bp) from nucleotides 140,317,847 to 140,328,857 on human Chromosome 9 (9q34.3, February 2009, GRCh37/hg19 genome assembly) in a region of conserved linkage synteny with the proximal end of mouse Chromosome 2. The domain structures of human and mouse NCF2 and NOXA1 proteins are similar (Figure 2) with the N-terminal halves of each protein containing multiple tetratricopeptide (TPR) repeats, the central region containing conserved activation domains (AD) and the C-terminal end of each protein containing both phox and Bem1 (PB1) and Src homology 3 (SH3) domains. In NCF2 additional SH3 domains are present centrally, adjacent to the AD.
Figure 2. Domain structure and sequence conservation among human and mouse NCF2 and NOXA1 proteins.
Schematic representations of the conserved domain structures of the human and mouse NCF2 and NOXA1 proteins are shown, as are the percentage of amino acid identity and similarity among the four proteins. Tetratricopeptide repeat (TPR) domains, red ellipses; activation domains (AD), green rectangles; Src homology 3 (SH3) domains, blue ellipses; phox and Bem1 (PB1) domains, yellow rectangles.
Although a number of studies have been conducted in vitro to assess the interaction of NOXA1 with other factors, its wide expression profile, ability to interact with multiple NOX partners and functional similarity to NCF2 make it difficult to assess the physiological and phenotypic significance of specific interactions in whole animals.
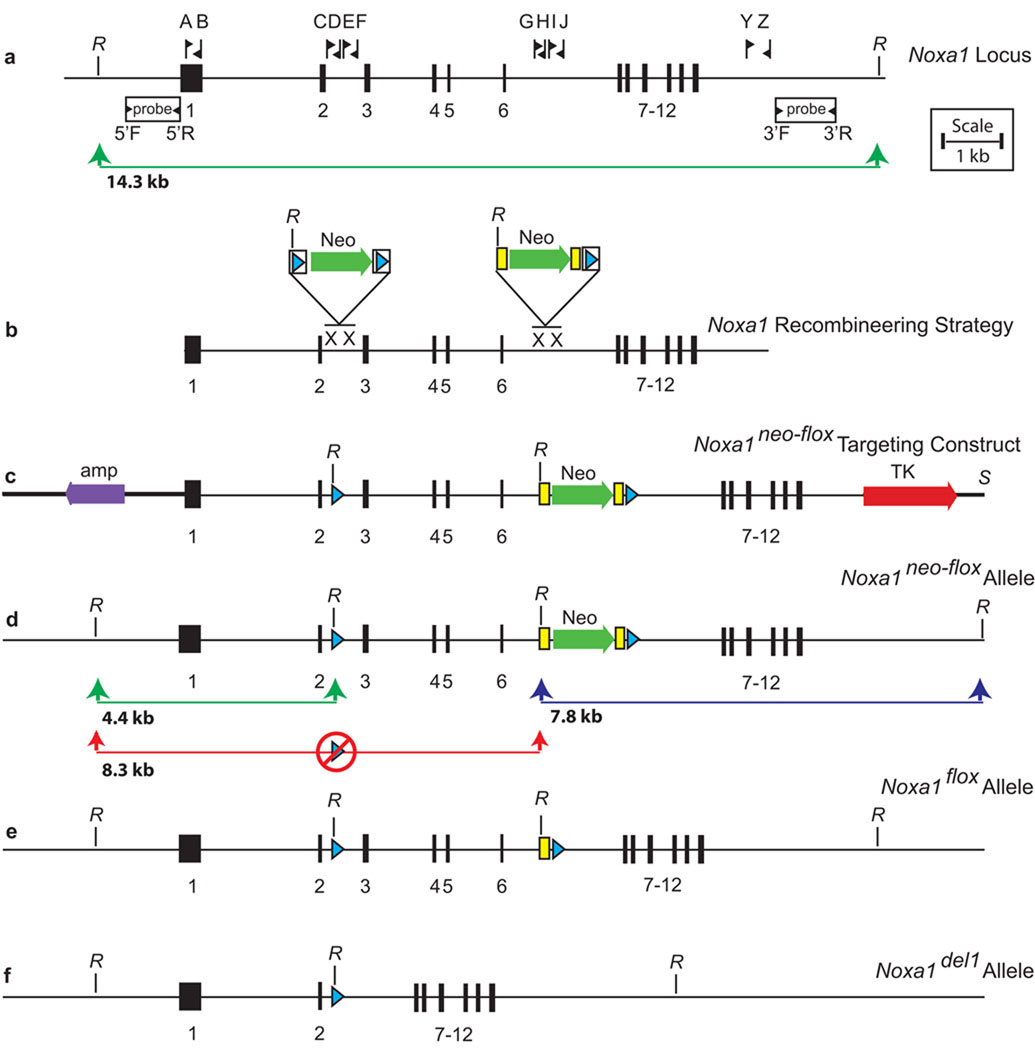
Accordingly, in order to allow tissue-specific dissection of the phenotypes caused by Noxa1 deficiency, we used the Cre-loxP based conditional mutagenesis approach (Furuta and Behringer, 2005) to generate a loxP-flanked (floxed) allele of Noxa1 suitable for conditional inactivation (Figure 3). The mouse Noxa1 transcription unit spans 9,478 bp from nucleotides 24,941,190 to 24,950,668 on mouse Chromosome 2 and consists of 12 exons (NCBI 37 Mouse Genome Assembly, Figure 3a). To generate the Noxa1neo-flox targeting vector, we used a recombineering approach (Figure 3b) (Copeland et al., 2001; Liu et al., 2003; Warming et al., 2005). Initially, a retrieval vector was prepared using 300–500 bp arms generated by PCR from the 5’ and 3’ regions of the Noxa1 gene. This retrieval vector was then used to capture, from a C57BL/6J-derived BAC, a 10.6 kb fragment of the Noxa1 gene extending from within exon 1 to beyond exon 12. A mini-targeting vector, constructed from two small homology arms in intron 2 and the floxed PGK/EM7 neomycin/kanamycin selection cassette from PL452, was introduced into the retrieved Noxa1 segment by recombination. Following transfer of the construct into SW105 and the induction of Cre recombinase with arabinose, the selection cassette was excised leaving only a single loxP site remaining in intron 2. Likewise, a second mini-targeting vector, constructed from two small homology arms in intron 6 and the FRT-flanked PGK/EM7 neomycin/kanamycin selection cassette and loxP site of PL451, was also introduced into the retrieved Noxa1 segment by recombination. The design of the completed targeting construct (Figure 3c) permits removal of the PGK/EM7 neomycin/kanamycin selection cassette from the Noxa1neo-flox allele (Figure 3d) by mating to a FLPe deleter mouse line (generating the Noxa1flox allele, Figure 3e) or removal of both Noxa1 genomic sequence (including exons 3 through 6) and the selection cassette by mating to a Cre deleter line (generating the Noxa1del1 allele, Figure 3f). This excision event results in the deletion of amino acids 88 to 220 encoding multiple TPR domains and the AD, and an ensuing frameshift in amino acids 221 to 444 causing functional loss of the PB1 and Src homology-3 domains of the NOXA1 protein.
Figure 3. Construction of a conditional allele of Noxa1 using a recombineering approach.
Schematic representations of the endogenous Noxa1 locus, the Noxa1 conditional allele construction strategy and construct, and the Noxa1 conditional (and derivative) alleles are shown. a. The endogenous Noxa1 locus of mouse. b. Recombineering strategy used to generate the Noxa1 targeting construct. c. The Noxa1 targeting construct. d. The properly targeted Noxa1neo-flox allele detected by Southern analysis as hybridizing 4.4 kb (5’ probe, green line) and 7.8 kb (3’ probe, blue line) fragments. Improperly targeted clones lacking the intron 2 loxP site (red circle) were detected by a hybridizing 8.3 kb band (5’ probe, red line) e. The Noxa1flox allele following FLPe-mediated removal of the neomycin resistance cassette. f. The Noxa1del1 allele following Cre-mediated removal of exons 3 through 6. Fine horizontal lines, genomic DNA; bold horizontal lines, vector DNA; numbered black rectangles, exons; lettered flags, oligonucleotide primers; white rectangles, 5’ and 3’ probes; green arrows, neomycin resistance cassettes (Neo); bold red arrow, negatively selectable thymidine kinase (TK) cassette; purple arrow, vector ampicillin resistance gene; blue triangles, loxP sites; yellow rectangles, FRT sites; X, sites of recombination; R, EcoRI.
The Noxa1neo-flox targeting vector was electroporated into B6(Cg)-Tyrc-2J/J-PRX-B6-albino #1 ES cells (The Jackson Laboratory Item 000058C02) and 384 colonies were picked for analysis. Initial screening was performed by Southern analysis using a 5’ probe on EcoRI-digested DNA (Figure 4a). For the wild type-allele of Noxa1, this probe identifies a 14.3 kb fragment (Figure 3a, green line). In targeted clones in which recombination occurred as desired within the outer half of the 5’ arm, the retention of the intron two loxP site and the presence of an EcoRI site within this site result in the generation a novel 4.4 kb band (Figure 3d, green line). Among the 307 scored clones, four such clones were identified. In targeted clones in which recombination occurred inopportunely within the inner half of the 5’ arm, the loss of the distal loxP site in intron 2 (Figure 3d, red circle) and the retention of the novel EcoRI site within the selection cassette result in the generation a novel 8.3 kb band (Figure 3d, red line). Among 307 scored clones, twenty such clones were identified. Of the four properly prepared clones, one was confirmed to be properly targeted using a 3’ probe (Figure 3d, blue line, data not shown) and injected into C57BL/6J host blastocysts. Resulting albino/black male chimeras were mated to B6(Cg)-Tyrc-2J/J females. ES-cell derived pups were identified by their albino coat color. Germline transmission of the Noxa1neo-flox allele was confirmed by PCR with primers flanking the distal (intron 2) loxP site (See Methods). All subsequent crosses were inter se or to C57BL/6J mice not harboring the Tyrc-2J (albino) allele of the original ES cells. Male and female Noxa1neo-flox/+ heterozygotes (Figure 4b, lane 2) were bred to each other and Noxa1neo-flox/Noxa1neo-flox homozygotes were identified by PCR amplification across the region of the distal loxP site, scoring for the absence of the wild type-band (data not shown).
Figure 4. Southern, genotyping and RT-PCR assays to characterize the Noxa1 conditional allele.
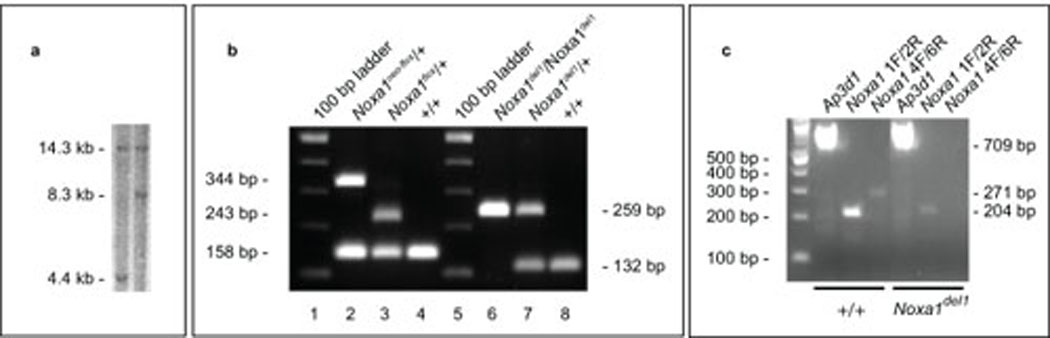
a. Southern analysis of targeted ES cell colonies identifies clones showing retention (left lane) or loss (right lane) of the loxP site in intron 2. b. Genotyping assays. Lanes 1–4 and lanes 5–8 showing the amplification products of two genotyping assays used to distinguish among the Noxa1+, Noxa1neo-flox, Noxa1flox and Noxa1del1 alleles (for details see Methods). c. RT-PCR analysis of Noxa1 transcripts from +/+ and Noxa1del1/Noxa1del1 temporal bone/inner ear RNA. Amplification of cDNA sequences upstream of the Cre-mediated deletion (primers 1F and 2R) is reduced in Noxa1del1 homozygotes. Amplification of cDNA sequences within the Cre-mediated deletion (primers 4F and 6R) is absent in Noxa1del1 homozygotes. Amplification of cDNA sequences from Ap3d1 serves as a control for the quality of the RNA.
Mice both heterozygous and homozygous for the Noxa1neo-flox allele were viable, fertile and did not exhibit any obvious, overt phenotypic abnormalities; indicating that neither the loxP site in intron 2, nor the FRT-PGK/EM7 Neo-FRT-loxP site in intron 6 has any obvious, adverse effects on any critical Noxa1 function.
The Noxa1flox allele was generated by mating Noxa1neo-flox mice to the B6.Cg-Tg(ACTFLPe)9205Dym/J deleter strain, which expresses FLPe recombinase in the male and female germlines (Rodriguez et al., 2000). Excision of the PGK/EM7 Neo cassette was demonstrated by PCR spanning the FRT-loxP site, which results in an 85 bp increase in the size of the Noxa1flox product (Figure 4b, lane 3) when compared to the wild type-allele (Figure 4b, lane 4) and a 101 bp decrease when compared to the Noxa1neo-flox allele (Figure 4b, lane 2).
Mice heterozygous for the Noxa1del1 allele were produced by mating Noxa1neo-flox mice to B6.129S4-Meox2tm1(Cre)Sor/J mice, which express Cre recombinase widely in the early embryo (Tallquist and Soriano, 2000). Excision was shown by PCR spanning the entire deleted region of the allele, which results in a novel 259 bp product (Figure 4b, lane 7). Noxa1del1/+ heterozygotes show no obvious phenotypic abnormalities.
Mice homozygous for the Noxa1del1 allele were created by intercrossing Noxa1del1 heterozygotes. Homozygosity of the Noxa1del1 allele was demonstrated by PCR, scoring for the absence of the Noxa1neo-flox allele (Figure 4b, lane 6). RT-PCR analysis of temporal bone/inner ear RNA confirms an absence of Noxa1-derived sequences from within the Cre-mediated deletion; and moreover, when compared to +/+ samples, weak amplification from exons 1 and 2 upstream of the deletion suggests that the deleted, frameshifted Noxa1del1 transcript is subject to nonsense-mediated decay (Figure 4c). Noxa1del1/Noxa1del1 homozygotes were viable, showed no overt phenotype and were produced in the expected (1:2:1) Mendelian ratio. Possible more subtle phenotypes of Noxa1del1 homozygotes or tissue-specific Noxa1 ablation studies are beyond the scope of this manuscript. Mice bearing the Noxa1neo-flox allele are available for distribution by contacting the corresponding author or from The Jackson Laboratory’s Genetic Resource Science Repository (http://research.jax.org/repository/index.html).
Through the thoughtful use of specific recombinase-expressing mouse strains, the conditional allele of Noxa1 will provide greater insight into the roles of NOXA1 in the variety of cells and organs where it is expressed.
METHODS
Mice
All mice were obtained from The Jackson Laboratory including strains C57BL/6J (Stock 000664), B6(Cg)-Tyrc-2J/J (Stock 000058), B6.Cg-Tg(ACTFLPe)9205Dym/J (Stock 005703), B6.129S4-Meox2tm1(Cre)Sor/J (Stock 003755) and CByB6F1/J (Stock 100009). All mice were housed on a bedding of white pine shavings and fed NIH-31 5K54 (4% fat) or NIH-31 5K52 (6% fat) diet and acidified water, ad libitum. All experiments were performed with the approval of The Jackson Laboratory Institutional Animal Care and Use Committee (IACUC) and in compliance with all applicable laws and guidelines.
Nomenclature
All mouse strains are referred to by official nomenclature. All mouse gene symbols (first letter upper case, italicized) and human gene symbols (all letters upper case, italicized) and proteins (all letters upper case) are referred to by official nomenclature throughout the body of the text. Relationships to other naming conventions and full names are shown here: Cytochrome b-245, beta polypeptide=CYBB=NOX2=gp91phox; Cytochrome b-245, alpha polypeptide=CYBA=p22phox; Neutrophil cytosolic factor 1=NCF1=p47phox; and Neutrophil cytosolic factor 2=NCF2=p67phox.
The unaltered Noxa1 conditional allele containing the FRT-flanked neomycin/kanamycin resistance cassette and loxP-flanked Noxa1 exons is named Noxa1tm1Brg by official nomenclature, but is referred to throughout this publication as Noxa1neo-flox. The FLP-altered allele having lost the neomycin/kanamycin resistance cassette is named Noxa1tm1.1Brg by official nomenclature, but is referred to throughout this publication as Noxa1flox. The Cre-altered allele having lost the neomycin/kanamycin resistance cassette and Noxa1 exons 3 through 6 is named Noxa1tm1.2Brg by official nomenclature, but is referred to throughout this publication as Noxa1del1.
Generation of the Noxa1 conditional gene-targeting vector
The Noxa1 conditional gene-targeting vector was constructed from BAC clone RP23-261J20 (Strain: C57BL/6J) using a recombineering approach (Copeland et al., 2001; Liu et al., 2003; Warming et al., 2005). Genomic DNA from the 5’ and 3’ ends of the Noxa1 gene was amplified from the BAC with oligonucleotide pairs A & B and Y & Z, digested with NotI and HinDIII (A & B) or HinDIII and SpeI (Y & Z) and cloned together into the NotI and SpeI digested targeting vector, PL253.
This plasmid was linearized with HinDIII and used to retrieve a portion of the Noxa1 gene (from mouse Chromosome 2, position 24,939,966 to 24,950,622; UCSC Genome Browser, July 2007 assembly) contained on the same BAC.
To insert a single loxP site into intron 2, a mini-targeting vector was constructed using arms that were amplified from the BAC using oligonucleotide pairs C & D and E & F, digested with NotI and EcoRI (C & D) or BamHI and SalI (E & F) and cloned along with the EcoRI/BamHI fragment of PL452 into pBluescript II KS(+). After introducing the mini-targeting vector into the retrieval product by recombination, the neomycin/kanamycin selection cassette and one of the loxP sites were removed (leaving a single loxP site remaining) by electrotransforming the construct into the recombineering strain, SW105, and inducing Cre recombinase expression with arabinose (Warming et al., 2005).
An FRT-flanked neomycin/kanamycin selection cassette and an additional loxP site were then introduced into intron 6 by recombination using a similar approach. In this instance a second mini-targeting vector was constructed using arms that were amplified from the BAC using oligonucleotide pairs G & H and I & J, digested with NotI and EcoRI (G & H) or BamHI and SalI (I & J) and cloned along with the EcoRI/BamHI fragment of PL451 into pBluescript II KS(+). This second mini-targeting vector was then introduced into the modified retrieval product by recombination to complete the conditional Noxa1 gene-targeting vector.
As constructed, the final vector is designed to delete the selection cassette upon interaction with FLP recombinase and exons 3 through 6 upon interaction with Cre recombinase. The Cre-induced deletion causes the ablation of amino acids 88 to 220 from any hypothetical, altered NOXA1 protein and, due to a frame shift, would direct the incorporation of 19 novel amino acids prior to a premature termination codon.
Generation of mice carrying the Noxa1 conditional allele
The Noxa1 conditional gene-targeting vector was linearized with SalI, electroporated into B6(Cg)-Tyrc-2J/J-PRX-B6-albino #1 ES cells (The Jackson Laboratory Item 000058C02) and cultured under G418 and FIAU selection.
Clones were assayed for proper targeting by Southern analysis. Oligonucleotides 5’F & 5’R and 3’F & 3’R were used to amplify unique probe sequences from mouse genomic DNA. Next, the genomic DNAs of 384 ES clones were digested with EcoRI, blotted and analyzed with the 5’ probe. Of 307 scored clones, four clones contained a hybridizing 4.4 kb band expected for properly targeted clones that had retained the 5’ loxP site. An additional 20 clones contained a hybridizing 8.3 kb band expected for targeted clones that had lost the 5’ loxP site. All clones contained a hybridizing 14.3 kb band arising from the wild type-allele.
All four clones that retained the 5’ loxP site were injected into C57BL/6J blastocysts and transferred into the oviducts (on day 0.5 post coitus) or the uterine horns (on day 2.5 post coitum) of CByB6F1/J pseudopregnant females. The resulting male black and albino chimeras were mated to B6(Cg)-Tyrc-2J females. Germline transmission was observed for one clone and the resulting offspring were identified by their albino coat color. Transmission of the targeted allele, and retention of the loxP in intron 2, was confirmed by Noxa1 conditional allele-specific PCR with oligonucleotides, Noxa1-loxPF and Noxa1-loxPR, which flank the loxP site in intron 2.
Genotyping
The presence or absence of the Meox2-Cre knockin, Meox2tm1(Cre)Sor (Tallquist and Soriano, 2000), was ascertained using the three primers oIMR1542, oIMR1871, and oIMR3671 as recommended by the distributor. Using this assay, the knockin allele gives rise to an approximately 300 bp product. The wild type-allele gives rise to an approximately 410 bp product. All products were resolved on 2% SeaKem LE agarose gels (Lonza).
The presence or absence of the actin-FLPe transgene, TgN(ACTFLPe)9205Dym (Rodriguez et al., 2000), was ascertained using the four primers oIMR7338, oIMR7339, oIMR1348, and oIMR1349 as recommended by the distributor. Using this assay, an internal control amplicon derived from the interleukin 2 (Il2) gene gives rise to a 324 bp product. The transgene gives rise to a 725 bp product. All products were resolved on 1.5% SeaKem LE agarose gels (Lonza).
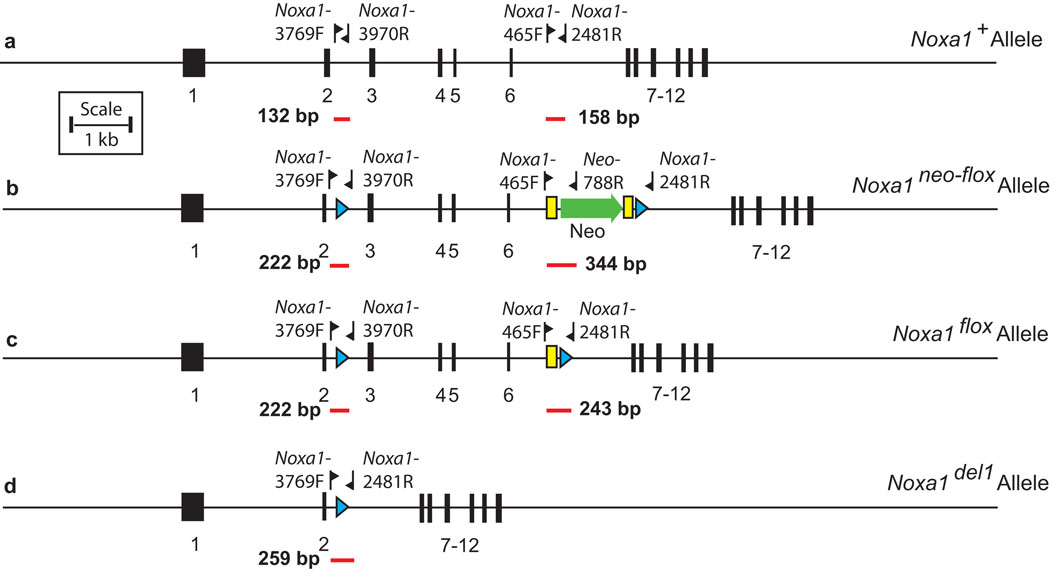
After exposure to FLP recombinase, mice carrying the Noxa1neo-flox, Noxa1flox or Noxa1+ (wild type-) alleles were distinguished with a genotyping assay containing the three primers Noxa1-465F, Neo-788R, and Noxa1-2481R (Figure 4b, lanes 1–4; Figure 5). Using this assay the Noxa1neo-flox allele gives rise to a 344 bp product, the Noxa1flox allele gives rise to a 243 bp product and the Noxa1+ allele gives rise to a 158 bp fragment. All products were resolved on 2% SeaKem LE agarose gels (Lonza).
Figure 5. Genotyping strategies to distinguish among the Noxa1+, Noxa1neo-flox, Noxa1flox and Noxa1del1 alleles.
a. The Noxa1+ allele. b. The Noxa1neo-flox allele. c. The Noxa1flox allele. d. The Noxa1del1 allele. Fine horizontal lines, genomic DNA; numbered black rectangles, exons; labeled flags, oligonucleotide primers; green arrow, neomycin resistance cassette (Neo); blue triangles, loxP sites; yellow rectangles, FRT sites; labeled red lines, amplification products.
After exposure to Cre recombinase, mice carrying the Noxa1neo-flox, Noxa1del1or Noxa1+ (wild type-) alleles were distinguished with a genotyping assay containing the three primers Noxa1-3769F, Noxa1-2481R, and Noxa1-3970R (Figure 4b, lanes 5–8; Figure 5). Using this assay the Noxa1neo-flox allele gives rise to a 222 bp product, the Noxa1del1allele gives rise to a 259 bp product and the Noxa1+ allele gives rise to a 132 bp fragment. All products were resolved on 2% SeaKem LE agarose gels (Lonza).
All PCR reactions were performed using the following standard cycling conditions: Preheating at 95°C for 3 min; 40 cycles of amplification at 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec; and a final extension of 72°C for 9 min 30 sec.
RT-PCR
Total RNA was isolated from individual Noxa1del1/Noxa1del1 and +/+ temporal bone explants (containing the inner ear) using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s directions. First strand synthesis reactions (8 µL RNA, 200–300 ng) were run in accordance with the SuperScript III (Invitrogen) random hexamer protocol. PCR reactions (30 µL) were performed on each cDNA and on no-reverse transcriptase controls to amplify two segments of the Noxa1 transcript (Noxa1 primers ex-1F and ex-2R, upstream; and primers ex-4F and ex-6R, within; the Cre-mediated deletion) and the Ap3d1 transcript (cDNA quality control). Each reaction used 2 µL cDNA, 5 Prime Master Mix (5 Prime, final concentration 1X) and the appropriate primers (200 nM) (Table 1). Amplification was performed on a MJ Research PTC-200 Thermocycler (Bio-Rad) and involved a 2:30 minute denaturing step at 95°C followed by 40 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and a 72°C extension step for 90 seconds. Products were resolved on a 2% SeaKem LE Agarose gel (Lonza).
Table 1. Oligonucleotides.
Oligonucleotides (Integrated DNA Technologies) used for construction of the Noxa1 conditional gene targeting vector, amplification of Southern analysis probes, genotyping and RT-PCR are shown. Each oligonucleotide used in the vector construction consists of an arbitrary CGCAT sequence at the 5’ end (italicized), any of various restriction enzyme recognition sites internally (bold) and sequences derived from within the Noxa1 gene at the 3’ end (plain text). The remaining oligonucleotides do not contain the CGCAT sequence or restriction enzyme recognition sites.
| Used for construction of the Noxa1 conditional gene-targeting vector |
| A, CGCATGCGGCCGCTCAAGGACCGACTGTCACAA |
| B, CGCATAAGCTTATGCACACAGCCCCTGTTA |
| C, CGCATGCGGCCGCGTCTCTGCAAACCCATGCTT |
| D, CGCATGAATTCCTCCTCCCTTCCTCCTCTTG |
| E, CGCATGGATCCAGGGTTACAGGCCCCAGTG |
| F, CGCATGTCGACAGGGGACCATGCCTATATCC |
| G, CGCATGCGGCCGCCAAGCCCTTGTGAAGAGGAG |
| H, CGCATGAATTCGCTGGCAGATGCAGGTTTA |
| I, CGCATGGATCCCTTCATAAGGGTCACACAGTGAA |
| J, CGCATGTCGACCCTGCATCAGTGGAGAGTCA |
| Y, CGCATAAGCTTTCTTAGGGCAGTCACCTGCT |
| Z, CGCATACTAGTGTGGGGTTTTCTTGGACAGA |
| Used for the generation of Southern probes |
| 5’ probe-F, TTAGCTGTCCTGCGTCACAC |
| 5’ probe-R, TTCGTCAGGCCTATCTGTCC |
| 3’ probe-F, ACGTGAGGTTCAGGAAATGG |
| 3’ probe-R, GCTTTCCTCTGCTCATGGTC |
| Used for genotyping |
| To confirm germline transmission of the Noxa1 conditional allele, Noxa1neo-flox, and presence of the loxP site in intron 2 |
| Noxa1-loxPF, ACCAGGGCTTATCCCAAGAT |
| Noxa1-loxPR, TCCCCAAACCCCTTCTAACT |
| To test for the presence of Meox2tm1(Cre)Sor knockin |
| oIMR1542, GGGACCACCTTCTTTTGGCTTC |
| oIMR1871, AAGATGTGGAGAGTTCGGGGTAG |
| oIMR3671, CCAGATCCTCCTCAGAAATCAGC |
| To test for the presence of the Tg(ACTFLPe)9205Dym transgene |
| oIMR7338, CTAGGCCACAGAATTGAAAGATCT |
| oIMR7339, GTAGGTGGAAATTCTAGCATCATCC |
| oIMR1348, CACTGATATTGTAAGTAGTTTGC |
| oIMR1349, CTAGTGCGAAGTAGTGATCAGG |
| To distinguish among the Noxa1neo-flox, Noxa1del1 and Noxa1+ alleles after exposure to Cre recombinase |
| Noxa1-3769F, ACCAGGGCTTATCCCAAGAT |
| Noxa1-2481R, CCTCTACCCTCTGTGGGACA |
| Noxa1-3970R, ACTTTCCCCAGTCACCCTTC |
| To distinguish among the Noxa1neo-flox, Noxa1flox or Noxa1+ alleles after exposure to FLP recombinase |
| Noxa1-465F,CCCTAGGACTTAAACCTGCATC |
| Noxa1-2481R,CCTCTACCCTCTGTGGGACA |
| Neo-788R,GGGGAACTTCCTGACTAGGG |
| Used for RT-PCR |
| Ap3d1-ex1F, TCCGCAACCACAAGGAGGACGA |
| Ap3d1-ex8R, TCGATCAGCTTCTTGCCCAGCC |
| Noxa1 cDNA ex-1F, AGGGGATCAGATACGGGACT |
| Noxa1 cDNA ex-2R, GCCATGCAGGTGTCCTTAGT |
| Noxa1 cDNA ex-4F, TACAACATGGCATCAGCACA |
| Noxa1 cDNA ex-6R, CGTTGTGGTCATCAGGAATG |
ACKNOWLEDGMENTS
The authors acknowledge the support of the Molecular Biology, Cell Biology, Microinjection and Cryopreservation Services of The Jackson Laboratory. The authors also thank Dr. Gregory Cox, Dr. Stephen Murray, Dr. Gareth Howell, Dr. Laura Reinholdt and Ms. Alicia Valenzuela for critical review of the manuscript and Ms. Leslie Haynes and The Jackson Laboratory Research Animal Facility staff for animal husbandry. This work was supported by NIH/NIDCD grant DC007431 to D.E.B. and NIH/NCI CORE grant CA34196 to The Jackson Laboratory.
Grant Support: DC007431/DC/NIDCD NIH HHS/USA and CA34196/CA/NCI NIH HHS/USA
REFERENCES
- Ago T, Kitazono T, Kuroda J, Kumai Y, Kamouchi M, Ooboshi H, Wakisaka M, Kawahara T, Rokutan K, Ibayashi S, Iida M. NAD(P)H oxidases in rat basilar arterial endothelial cells. Stroke. 2005;36:1040–1046. doi: 10.1161/01.STR.0000163111.05825.0b. [DOI] [PubMed] [Google Scholar]
- Ambasta RK, Schreiber JG, Janiszewski M, Busse R, Brandes RP. Noxa1 is a central component of the smooth muscle NADPH oxidase in mice. Free Radic Biol Med. 2006;41:193–201. doi: 10.1016/j.freeradbiomed.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Banfi B, Clark RA, Steger K, Krause KH. Two novel proteins activate superoxide generation by the NADPH oxidase NOX1. J Biol Chem. 2003;278:3510–3513. doi: 10.1074/jbc.C200613200. [DOI] [PubMed] [Google Scholar]
- Banfi B, Malgrange B, Knisz J, Steger K, Dubois-Dauphin M, Krause KH. NOX3, a superoxide-generating NADPH oxidase of the inner ear. J Biol Chem. 2004;279:46065–46072. doi: 10.1074/jbc.M403046200. [DOI] [PubMed] [Google Scholar]
- Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- Bergstrom DE, Bergstrom RA, Munroe RJ, Schimenti JC, Gagnon LH, Johnson KR, Heinzmann U, Stumm G, Paffenholz R. The Nox3 and Noxo1 genes, encoding presumptive members of an NADPH oxidase complex, are required for normal vestibular function and development. The 18th International Mammalian Genome Conference; Seattle, USA. 2004. [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Behringer RR. Recent innovations in tissue-specific gene modifications in the mouse. Birth Defects Res C Embryo Today. 2005;75:43–57. doi: 10.1002/bdrc.20036. [DOI] [PubMed] [Google Scholar]
- Geiszt M, Lekstrom K, Witta J, Leto TL. Proteins homologous to p47phox and p67phox support superoxide production by NAD(P)H oxidase 1 in colon epithelial cells. J Biol Chem. 2003;278:20006–20012. doi: 10.1074/jbc.M301289200. [DOI] [PubMed] [Google Scholar]
- Holland SM. Chronic granulomatous disease. Clin Rev Allergy Immunol. 2010;38:3–10. doi: 10.1007/s12016-009-8136-z. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Kohjima M, Kuwano Y, Mino H, Teshima-Kondo S, Takeya R, Tsunawaki S, Wada A, Sumimoto H, Rokutan K. Helicobacter pylori lipopolysaccharide activates Rac1 and transcription of NADPH oxidase Nox1 and its organizer NOXO1 in guinea pig gastric mucosal cells. Am J Physiol Cell Physiol. 2005;288:C450–C457. doi: 10.1152/ajpcell.00319.2004. [DOI] [PubMed] [Google Scholar]
- Kiss PJ, Knisz J, Zhang Y, Baltrusaitis J, Sigmund CD, Thalmann R, Smith RJ, Verpy E, Banfi B. Inactivation of NADPH oxidase organizer 1 results in severe imbalance. Curr Biol. 2006;16:208–213. doi: 10.1016/j.cub.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Lavigne MC, Eppihimer MJ. Cigarette smoke condensate induces MMP-12 gene expression in airway-like epithelia. Biochem Biophys Res Commun. 2005;330:194–203. doi: 10.1016/j.bbrc.2005.02.144. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute JD, Arias AA, Wright v, Wrobel I, Waterhouse CC, Li XJ, Marchal CC, Stull ND, Lewis DB, Steele M, Kellner JD, Yu W, Meroueh SO, Nauseef WM, Dinauer MC. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Longo-Guess CM, Bergstrom DE, Nauseef WM, Jones SM, Banfi B. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J Clin Invest. 2008;118:1176–1185. doi: 10.1172/JCI33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz R, Bergstrom RA, Pasutto F, Wabnitz P, Munroe RJ, Jagla W, Heinzmann U, Marquardt A, Bareiss A, Laufs J, Russ A, Stumm G, Schimenti JC, Bergstrom DE. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004;18:486–491. doi: 10.1101/gad.1172504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Segal BH, Romani L, Puccetti P. Chronic granulomatous disease. Cell Mol Life Sci. 2009;66:553–558. doi: 10.1007/s00018-009-8506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]