Abstract
Due to the increasing number of structures of unknown function accumulated by ongoing structural genomics projects, there is an urgent need for computational methods for characterizing protein tertiary structures. As functions of many of these proteins are not easily predicted by conventional sequence database searches, a legitimate strategy is to utilize structure information in function characterization. Of a particular interest is prediction of ligand binding to a protein, as ligand molecule recognition is a major part of molecular function of proteins. Predicting whether a ligand molecule binds a protein is a complex problem due to the physical nature of protein-ligand interactions and the flexibility of both binding sites and ligand molecules. However, geometric and physicochemical complementarity is observed between the ligand and its binding site in many cases. Therefore, ligand molecules which bind to a local surface site in a protein can be predicted by finding similar local pockets of known binding ligands in the structure database. Here, we present two representations of ligand binding pockets and utilize them for ligand binding prediction by pocket shape comparison. These representations are based on mapping of surface properties of binding pockets, which are compactly described either by the two dimensional pseudo-Zernike moments or the 3D Zernike descriptors. These compact representations allow a fast real-time pocket searching against a database. Thorough benchmark study employing two different datasets show that our representations are competitive with the other existing methods. Limitations and potentials of the shape-based methods as well as possible improvements are discussed.
Keywords: protein surface, structure-based function prediction, pocket shape, pseudo-Zernike moments, 3D Zernike descriptors, ligand binding site
Introduction
Characterization of protein function is one of the most important tasks in bioinformatics1–4. Taking advantage of accumulated knowledge of gene functions stored in databases5;6, computational function prediction methods typically search similar patterns in sequences or structures of a query protein against databases of known proteins. Traditionally, methods for sequence database searches7, including homology search8–10, functional domain search11;12, and motif search13;14, have been widely used for function prediction, since sequence information is almost always available for genes of unknown function. Recent years have observed development of advanced approaches for sequence-based function prediction15–19, which achieve an improved accuracy and coverage in genome-scale function assignment. Moreover, many function prediction methods have been developed that utilize other types of data, such as protein-protein interaction data20;21, gene expression data22, and text mining23;24, or combination of such heterogeneous data25;26. Of recent particular importance is functional characterization of proteins from their tertiary structures since an increasing number of protein structures of unknown function have been solved by ongoing structural genomics projects27;28. As of this writing, there are currently more than 3000 protein structures of unknown function in the Protein Data Bank (PDB)29 that are awaiting functional characterization.
Roughly speaking, tertiary structure information can be used for function prediction either by considering global fold or local structure of proteins. The former approach utilizes the observation that the evolutionary relationships of proteins could be better tracked by considering overall protein fold similarity to reach a further evolutionary distance where proteins share barely detectable sequence similarity30–32. FINDSITE33 is one such method that utilizes global structure information to predict function. It uses groups of template structures of distant homologs of a target protein identified by threading. However, caution is needed in inferring function from the global structure similarity since there are protein folds which are adopted by many different proteins34. On the other hand, the latter approaches aim to capture local geometry of known functional sites or small ligand molecule binding sites. Since local methods directly search for geometrical and/or physicochemical properties of functional sites, it could be possible to predict molecular functions of proteins which lack homology to proteins of known function35.
Local structure-based function prediction can be logically divided into two parts: 1) detection of characteristic local sites, such as pockets, in a given protein surface, and 2) matching the local sites against a database of known functional site patterns. Of a particular interest in the first step is to detect pocket regions since binding of small ligand molecules occurs at pocket regions in many cases36. Therefore, if a protein is known to bind a ligand molecule, the binding site itself can be well predicted by just identifying pockets 37;38. We have shown in our previous work that ligand binding sites of proteins can be identified as one of the three largest pockets in the protein surface in 95% of the cases36.
Toward the goal of identifying potential ligand binding sites in proteins, several methods have been developed. SURFNET39 searches for a gap in a protein surface by fitting spheres inside the convex hull. POCKET40 and PHECOM41 also use probe spheres. PocketPicker42 and LIGSITE43 locate a protein onto a three dimensional (3D) grid and scan it for protein-void-protein events in many directions, whereas VisGrid36 uses the visibility of surface points to find pockets. PocketDepth44 clusters grid cells using information of the depth of the grid cells. CAST37 computes a Voronoi diagram of a protein and identifies pockets as void tetrahedrons. Several methods consider additional information, such as sequence conservation45–47 and energetics48–50, which are often combined while considering geometrical shape.
Algorithms used for matching local sites are closely interrelated with the representation of the local sites. In Catalytic Site Atlas51, AFT52, and SURFACE53, where a local site is represented as a set of few residue positions, the root mean square deviation (RMSD) of equivalent amino acid residues is computed. In SitesBase54, atoms in ligand binding sites are compared using geometric hashing. Another functional local site database, eF-site55, represents protein surface as a graph with nodes characterized by local geometry and the electrostatic potential, and hence uses a maximum subgraph algorithm for seeking similar sites. Recently, Thornton and her colleagues explored the use of spherical harmonics in representing and comparing protein pockets56;57. They compared ligand surface shape with pocket sizes56 and also did pocket to pocket comparison. Garutti and Bock proposed a 2D representation of binding sites by computing a collection of 2D histograms (spin-images) associated to surface points58. Comparing two sites consists in finding highly-correlated pairs of spin-images that also satisfy geometric criteria.
In this paper, we introduce two approaches for representation and comparison of properties of ligand binding pockets. In the first approach, the shape and the electrostatic potential of a binding pocket is mapped on a two dimensional (2D) picture, which is then represented as the pseudo-Zernike moments. The pseudo-Zernike moments are series expansion of a 2D function; hence a pocket is represented compactly by a vector of coefficients assigned to terms in the series. This representation is conceptually very different from, for example, the spin-image representation58: the spin-images require many 2D images per pocket, a simple correlation coefficient to compare images and computationally expensive geometric matching procedure, whereas our method only uses one 2D image, with mathematically more elaborate descriptors and inexpensive pocket matching. In the second approach, we employ the 3D Zernike descriptors59;60, a series expansion of a 3D function, to directly represent 3D pocket surface properties. These two compact representations allow a fast real-time search against a database of known pockets. For example, a search against all pockets in PDB would take only a few seconds. Employing two different datasets of ligand binding pockets, we compare performance of pocket retrieval by the 2D and 3D pocket representations as well as the other existing methods. We also investigated how well our methods perform when ligand-free binding pockets or predicted binding pockets are used as queries. Limitations and possible improvements of the shape-based methods are discussed.
Materials and Methods
In this work, we propose two binding pocket description models. The first model uses 2D moments to represent mapping of pocket surface properties on a 2D image. We compare three different 2D moments, the pseudo-Zernike, 2D Zernike, and Legendre moments in terms of invariance upon rotation of pockets and the accuracy of pocket retrieval from a database. The second one uses the 3D Zernike descriptors, 3D moments-based descriptors which represents 3D shape and properties of binding pockets.
2D pocket model using 2D moments
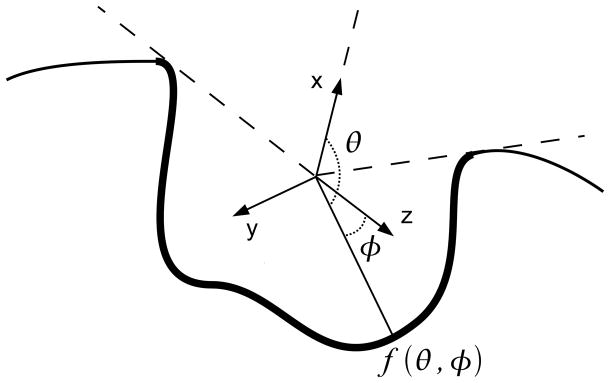
This binding pocket description model is based on ray-casting and 2D moments. Intuitively, a binding pocket is represented as a spherical panoramic picture viewed from its center of gravity. We then compute the pseudo-Zernike moments, 2D image descriptors, of the panoramic picture. Throughout this paper, the surface of a protein refers to the Connollysurface61, which is a commonly used definition in proteins surface visualization and surface-related computations. Following the Interact Cleft Model used in Kahraman’s work56, a ligand binding pocket (BP) is the surface of protein heavy atoms (i.e. atoms other than hydrogen) which are within 8Å to any heavy atom of the bound ligand. We define G as the center of gravity of BP, provided it does not lie inside the protein volume; otherwise, G is defined as any of the closest points outside of BP. The opening of BP is defined as the set of rays starting at G and not intersecting BP. In total of 64800 (=180 × 360) rays are shot from G to each (θ,φ) direction.
Ray-casting of outermost binding pocket surface
We now describe a ray-casting strategy62 to represent a BP as seen from G. A 3D Cartesian coordinate system (x⃗, y⃗, z⃗) specific to a BP is defined as follows (Figure 1): the point G is the origin of the coordinate system and the unit vector of the x-axis x⃗ is defined as a collinear vector to the average vector that define the opening of BP. In cases where the opening is empty, the x-axis is arbitrary defined. In the first Kahraman dataset, 19 out of 100 pockets have an empty opening. However, defining their x-axes arbitrarily still produces robust descriptors: the mean AUC of shape-only descriptors of these pockets is only 0.7% lower than the mean dataset AUC. We will later use 2D rotationally invariant moments on the (y⃗, z⃗) plane. Therefore, the remaining two vectors, y⃗ and z⃗, can be defined arbitrarily, as long as the basis (x⃗, y⃗, z⃗) is orthogonal. This choice of coordinate system provides a good approximation of rotation invariance for binding pockets descriptors, as seen in the section of pseudo-Zernike moments below.
Figure 1.
Mapping of a ligand binding pocket (shown in bold line) from its center of gravity. The x-axis is aligned with the center of the pocket opening, and the X-Y plane is arbitrarily oriented.
Using spherical coordinates, we define a spherical function f(θ,φ) that describes the outermost surface of BP on [0,2π]×[0,π]:
| (1) |
In this equation, the subscript i is used in the event that a ray intersects BP multiple times, but such situation is very rare. Figure 1 sketches the definition of f in two dimensions by projecting the scene on a fictional plane containing G. The function f is a piecewise continuous spherical function. Since it is only used to describe the shape of the pocket, f can be normalized such that its highest value is 1. In order to compute 2D moments, the function f has to be mapped to a 2D plane.
The protein surface electrostatic potential can also be mapped to the protein surface in the same fashion by defining the value f(θ,φ) as the surface electrostatic potential at the outermost intersection between the ray and the protein surface. We used the Finite Difference Poisson Boltzman (FDPB) solver of the BALL library63 version 1.2 (http://www.ball-project.org/) for computing the electrostatic potential. The grid spacing set to 0.8Å, solvent dielectric constant is 78.0, and the PARSE force field64 is used to assign atomic charges and radii, all of which are the default parameters for calculating the electrostatic potential with the FDPB solver in the BALL library.
Projection of 3D surface to 2D plane
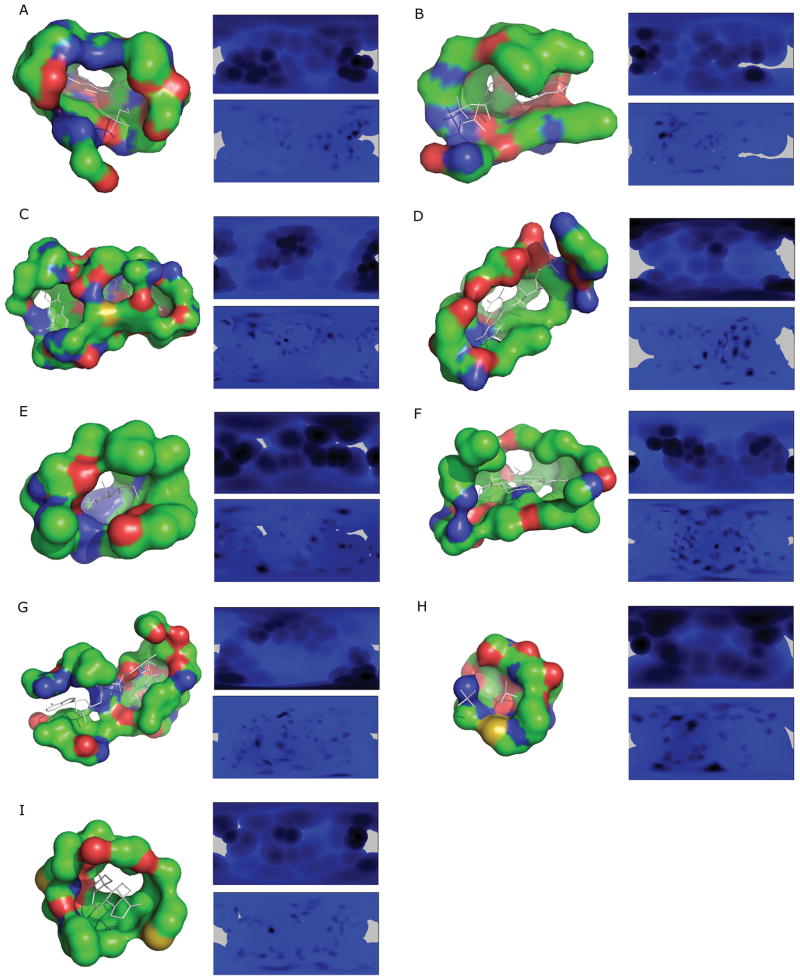
Numerous methods exist for spherical function projection, because no construction preserves the following three spherical properties altogether, the area, shape, and the distance. We choose to use a scheme, which is a special case of the equi-rectangular (distance preserving) projection named plate-carrée projection. This consists of mapping the surface representation, f(θ,φ), to a 2D plane (Fig. 1). By this mapping, the opening of the pocket corresponds to θ=0 and the bottom of the pocket (θ =π) is projected to the center of the image. Hence, rotations around the x-axis of the pocket (changes to θ) correspond to rotations around the center of the image, modulo distortions due to the projection. Computing 2D moments that are invariant around the center of the image compensates for the lack of reference for theta (i.e. arbitrary definition of the z-axis). Empirically, this projection is satisfactory because it does not distort shapes of a binding pocket beyond recognition by image descriptors (see Results). Projected surfaces and electrostatics of sample binding pockets are shown in Figure 2. The resolution of these pictures is 360×180, since the coordinates are mapped to integer values of (θ, φ). In the followings we describe the projections with 2D image descriptors, which we have examined in this study.
Figure 2.
Examples of the binding pocket representation by the 2D pocket model. The ligand binding pocket of a protein is sphere-mapped from its center of gravity and projected to a two dimensional plane. Blue to black colors indicate the Euclidean distance from the center to the pocket surface of that direction, and purple shows aperture. A, AMP binding site of 12AS; B, 1ESQ (ATP); C, 1HSK (FAD); D, 1P4M (FMN); E, 1K1W (GLC); F, 1NP4 (HEM); G, 1OG3 (NAD); H, 1GYP (PO4); I, 1E3R (STR).
Pseudo-Zernike moments
The pseudo-Zernike (p-Z) moments65 are commonly used in optics and are shown to be less sensitive to noise than conventional (two dimensional) Zernike moments66;67. The p-Z moments use a set of complete and orthogonal basis functions defined over the unit circle (x2+y2 ≤1) as follows:
| (2) |
where , θ= tan−1 (y/x), and n≥0, |m|≤n. Using the polynomials, the p-Z moments of the order n and the repetition m for a 2D image f(x, y) are defined as:
| (3) |
The asterisk (*) denotes the complex conjugate. Please refer the previous papers65;67 for more mathematical details. In this study, we used n = 5 for most of the computation. Among some other moments used in image processing, we chose the p-Z moments for 2D binding pocket representation because of the following reasons. First, they are orthogonal over the unit circle, thus information is not redundant between moments. Second, from the mathematical point of view these moments are rotationally invariant around the center of the image, which is a required property of the coordinate system we used in the model of binding pockets. Third, previous comparative studies show that these moments are one of the most tolerant to noise for shape description67–69.
2D Zernike moments
For comparison with the p-Z moments, we also employ the 2D Zernike moments and the Legendre moments, both of which are common alternative choices in the field of image analysis. The difference of the 2D Zernike moments and the p-Z moments is the radial function Rnm(r) in the Eqn. 2. The 2D Zernike moments use the following radial function in the polynomials:
| (4) |
with |m|≤n and n − |m| = even.
Legendre moments
The Legendre moments of order (m+n) for an 2D image f(x, y) are defined as
| (5) |
where m, n = 0, 1, 2, .. ∞. Pm(x) is the Legendre polynomials:
| (6) |
The 2D image function f(x, y) can be written as a series expansion in terms of the Legendre polynomials over the square [−1 ≤ x, y ≤ 1]. For more mathematical details of the 2D Zernike and the Legendre moments, refer elsewhere67. The 2D Zernike and the Legendre moments are computed for the same 2D picture of pockets (Fig. 2).
3D pocket model using 3D Zernike descriptors
In this model, binding pockets are extracted in the same way as the previous 2D moments-based pocket model and are represented by the 3D Zernike descriptors (3DZD). It was previously shown by our group and others that the 3DZD are effective in comparing global surface shape70–72, local surface regions73;74, and surface physicochemical properties75 of proteins. Naturally, the 3DZD can be also applied for comparing shape of small ligand molecule76. Recently we have developed surface shape-based protein docking prediction method named LZerD which uses the 3DZD for detecting complementarity of surface shapes77. In this work we examine how well the 3DZD perform in representing and comparing local shapes (binding pockets) of proteins.
3D Zernike descriptors
3DZD is a series expansion of a 3D function, which allows a compact representation of a 3D object (i.e. a 3D function). Mathematical foundation of the 3DZD was laid by Canterakis60 then Novotni and Klein59 have applied it to 3D shape retrieval. Below we provide brief mathematical derivation of the 3DZD. See the two papers59;60 for more details.
Pocket surface is extracted in the same way as the 2D moments-based pocket models but with a different distance threhosld value of 8Å to identify ligand binding atoms in the protein, since 8Å gave better results for the 3DZD than 5Å. Then, the pocket surface are placed on a 3D grid. To represent a surface shape, each grid cell (voxel) is assigned 1 if it is on the surface and 0 otherwise. Values of other physicochemical properties, such as the electrostatic potentials, are also assigned only to the surface voxels. The resulting voxels with values on them are considered as a 3D function, f(x), which is expanded into a series in terms of Zernike-Canterakis basis59 defined by the collection of functions
| (7) |
with −l < m < l, 0≤ l ≤ n, and (n− l) even. Spherical harmonics78, , is the angular portion of an orthogonal set of solutions to Laplace’s equation, which is given by:
| (8) |
where is a normalization factor,
| (9) |
and is the associated Legendre function. Rnl(r) are radial functions defined by Canterakis, constructed so that are polynomials when written in terms of Cartesian coordinates. , which are currently written in spherical coordinates, are converted into Cartesian coordinate functions in the following three steps:
- The conversion between spherical coordinates, (r, ϑ, ϕ), and Cartesian coordinates, x = (x, y, z), is defined as
(10) -
Using Eqn. 4, we define a function in Cartesian coordinates, which is later used for rewriting the 3D Zernike function (Eqn. 1) into Cartesian coordinates. The harmonics polynomials are defined as
(11) where are normalization factors(12) -
Using the harmonics polynomials , 3D Zernike functions (Eqn. 1) can be rewritten in Cartesian coordinates:
(13) where 2k = n − l and the coefficient are determined as follows to guarantee the orthonormality of the functions within the unit sphere,(14)
Now 3D Zernike moments of f (x) are defined as the coefficients of the expansion in this orthonormal basis, i.e. by the formula
| (15) |
Finally, the moments are collected into (2l+1) dimensional vectors and the rotational invariance is obtained by defining 3DZD, Fnl, as norms of vectors Ωnl:
| (16) |
The parameter n is called the order of 3DZD. The order determines the resolution (i.e. the number of terms in the series expansion) of the descriptor. n defines the range of l. And a 3DZD is a series of invariants (Eqn. 16) for each pair of n and l, where n ranges from 0 to the specified order. We use n = 20, which yields a total of 121 invariants, because it is shown to provide sufficient accuracy in a previous works of shape comparison59;70.
As for the surface electrostatic potentials, 3DZD is computed separately for the pattern of positive values and for the negative values and later combined in the following way75: First, voxels with a positive electrostatic potential value are kept but all the other voxels with a negative electrostatic potential value are reset with a value of zero. Then 3DZD of the pattern of the positive values in the cubic grid is computed. Next, similarly, voxels with a negative electrostatic potential value are kept but all the other voxels are reset with a value of zero. Then 3DZD of the pattern of the negative values is computed. Then, the two 3DZDs, one for voxels with a positive value and another one for voxels with a negative value are combined, yielding a descriptor with 2×121= 242 invariants. This is because Eqn. 16 does not differentiate positive and negative values, but only a pattern of non-zero values in the 3D space. Finally, we normalize numbers in a descriptor by the norm of the descriptor. This normalization is found to reduce dependency of 3DZD on the number of voxels used to represent a protein.
Scoring function for binding ligand prediction
The proposed binding pocket model is tested in terms of performance of retrieving pockets of the same binding ligand type as a query pocket. For a given query protein pocket of a protein, k “closest” pockets in a benchmark dataset (described below) are retrieved. The closeness (i.e. distance) of two pockets is defined as either by the Manhattan distance, the Euclidean distance, or the correlation coefficient-based metric of the descriptors of the two pockets. The Manhattan distance of two pockets, Pa and Pb, is defined as:
| (17) |
The Euclidean distance:
| (18) |
The correlation coefficient-based distance:
| (19) |
Here, and are the i-th value of the descriptors of pocket, Pa and Pb. N is the total number of values of the descriptors. The correlation coefficient-based metric, dc, equals zero when two descriptors correlate perfectly.
Using the k closest pockets to a query based on one of the distances (Eqns. 17, 18, 19) described above, the scoring function for a binding pocket of a ligand type F is defined as
| (20) |
where l(i) denotes the ligand type (AMP, FAD, etc.) of the i-th closest pocket to the query, n is the number of pockets of the type F in the database, and the function δX,Y equals to 1 if X is of type Y, and is null otherwise. The first term is to consider top k closest pockets to the query, with a higher score assigned to a pocket with a higher rank. The second term is to normalize the score by the number of pockets of the same type F included in the database. The ligand with the highest Pocket_score is predicted to bind to the query pocket.
Volumetric representation of pockets by spherical harmonics
In addition, our pocket representation is compared with a 3D volumetric representation of pockets by spherical harmonics, which was developed by the authors of the benchmark dataset of ligand pockets56 we use in this study. Among the three pocket shape approximation models proposed in their paper56, we compare our results with the Interact Cleft Model. The Interact Cleft Model defines the volume of a ligand binding pocket by SURFNET39, which places trial spheres of a certain range of sizes within 0.3 Å of protein atoms interacting with the bound ligand. The interacting atoms with the ligand are determined by HBPLUS79. The model uses spherical harmonics functions for representing the volume of a pocket. Since spherical harmonics are not invariant to rotation, a pocket needs to be pose normalized (An alternative to the prior pose normalization is to store amplitudes of frequencies of spherical harmonics to achieve rotation invariance 80). A pocket volume is first shifted so that its center of gravity is placed at the origin of the coordinate system. Then the pocket volume is rotated so that its moment of inertia tensor becomes diagonal with maximal values in x followed by y then followed by z. Now the outermost surface points of the surface volume is considered as a spherical function f(θ,φ) on a unit sphere and it is expanded as a series of spherical harmonics:
| (21) |
where the order lmax is set to 16, Re[Ylm(θ,φ)] is the real part of the spherical harmonic functions, and clm is the associated coefficients. The similarity of two pockets are measured by the Euclidean distance (Eqn. 18) of the vectors of coefficients clm of the two pockets. For more details of the procedure, refer to their papers56;57.
Benchmark datasets of ligand binding pockets
We used two datasets for benchmarking pocket retrieval performance of the methods. The first dataset compiled by Kahraman et al.56 is used to compare the performance of our methods with the previous 3D volumetric representation of pockets by spherical harmonics (the Kahraman set). This dataset consists of 100 proteins, each of which binds one of the following nine different ligands: adenosine monophosphate (AMP), adenosine-5′-triphosphate (ATP), flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), glucose (GLC), heme (HEM), nicotinamide adenine dinucleotide (NAD), phosphate (PO4), or steroid (STR). In the parentheses abbreviations of the ligand names are shown. The PDB IDs of ligand binding proteins in the dataset are listed in Table 1A. The tertiary structures of these proteins have been solved by X-ray crystallography and only structures which bind their cognate ligand are used. The proteins are each selected from different homologous families in the CATH database81 (i.e. H-level in CATH) so that they are not closely evolutionary related.
Table 1A.
The ligand pocket benchmark dataset from Kahraman et al.
| Binding ligand molecule | Number of PDB entries | PDB entries |
|---|---|---|
| AMP | 9 | 12asA, 1amuA, 1c0aA, 1ct9A, 1jp4A, 1khtB, 1qb8A, 1tb7B, 8gpbA |
| ATP | 14 | 1a0iA, 1a49A, 1aylA, 1b8aA, 1dv2A, 1dy3A, 1e2qA, 1e8xA, 1esqA, 1gn8B, 1kvkA, 1o9tA, 1rdqE, 1tidA |
| FAD | 10 | 1cqxA, 1e8gB, 1eviB, 1h69A, 1hskA, 1jqiA, 1jr8B, 1k87A, 1poxA, 3grsA |
| FMN | 6 | 1dnlA, 1f6vA, 1ja1A, 1mvlA, 1p4cA, 1p4mA |
| GLC | 5 | 1bdgA, 1cq1A, 1k1wA, 1nf5C, 2gbpA |
| HEM | 16 | 1d0cA, 1d7cA, 1dk0A, 1eqgA, 1ew0A, 1gweA, 1iqcA, 1nazE, 1np4B, 1po5A, 1pp9C, 1qhuA, 1qlaC, 1qpaB, 1soxA, 2cpoA |
| NAD | 15 | 1ej2B, 1hexA, 1ib0A, 1jq5A, 1mewA, 1mi3A, 1o04A, 1og3A, 1qaxA, 1rlzA, 1s7gB, 1t2dA, 1toxA, 2a5fB, 2npxA |
| PO4 | 20 | 1a6q, 1b8oC, 1brwA, 1cqjB, 1d1qB, 1dakA, 1e9gA, 1ejdC, 1eucA, 1ew2A, 1fbtB, 1gypA, 1h6lA, 1ho5B, 1l5wA, 1l7mA, 1lbyA, 1lyvA, 1qf5A, 1tcoA |
| STR | 5 | 1e3rB, 1fdsA, 1j99A, 1lhuA, 1qktA |
The list of the proteins are compiled by Kahraman et al.55 and shown as Table 1 in their paper with detailed information of the proteins. Here only PDB IDs are provided for readers’ convenience. Underlined entries in Tables 1A and 1B are used in the performance comparison with eF-Seek and SiteBase, as these are commonly included in the database of the two methods. The entries with double underline are used as queries.
The second dataset contains in total of 175 proteins, each of which binds one of the 12 ligand molecules (Table 1B). This dataset is constructed based on the ligand bound and unbound protein pairs listed in Table 4 in the paper by Huang & Schroeder46. They used the dataset for benchmarking pocket identification methods. Their original list can be found also at http://kiharalab.org/visgrid_suppl/, as we have also used it in our previous study36. From this list, first we discarded proteins which bind non-natural ligands. Then, we consulted the PDBsum database82 and removed entries if they do not have sufficient number of the other non homologous PDB entries (with a sequence identity of less than 30%) that bind the same ligand molecule. This set is called the Huang dataset. The Kahrman set and the Huang set do not have overlap neither in terms of proteins nor ligand types. The purpose of this dataset is twofold: to test the proposed methods on another dataset and also to investigate the performance of the methods when unbound pockets are used as queries.
Table 1B.
The bound and unbound pocket benchmark dataset (Huang dataset).
| Ligand molecule a) | Number of PDB entries | PDB entries | Unbound Structure |
|---|---|---|---|
| ADN | 11 | 1bx4A, 1fmoE, 1pg2A, 1vhwA, 2evaA, 2fqyA, 2pgfA, 2pkmA, 2zgwA, 3ce6A, 3fuuA | 1lioA |
| BTN | 12 | 1bdoA, 1hxdA, 1stpA, 1swgA, 2b8gA, 2c4iA, 2f01A, 2jgsA, 2zscA, 3d9lA, 3ew2A, 3g8cA | 1swb |
| F6P | 12 | 1lbyA, 1nuyA, 1tipA, 1uxrA, 2axnA, 2bifA, 2cxsA, 2r66A, 3bxhA, 3h1yA, 3iv8A, 4pfkA | 2fbpA |
| FUC | 14 | 1k12A, 1lfgA, 1lslA, 1onqA, 1uzvA, 2a2qL, 2bs6A, 2ha2A, 2j1tA, 2nzyA, 2volA, 3cigA, 3cqoA, 3kmb1 | 1nna |
| GAL | 36 | 1axzA, 1gcaA, 1hwmB, 1iszA, 1jz7A, 1kwkA, 1muqA, 1nsxA, 1okoA, 1r47A, 1rdk1, 1rvtJ, 1s5dD, 1tlgA, 1v3mA, 1w8nA, 1xc6A, 1z45A, 1zizA, 2b3fA, 2dqvA, 2e9mA, 2ehnB, 2eukA, 2galA, 2j1aA, 2j5zA, 2rjoA, 2v72A, 2vjjA, 2vnoB, 2zgnB, 3a23A, 3c69A, 3dh4A, 5abpA | 1gcg |
| GUN | 12 | 1a95C, 1d6aA, 1it7A, 1sqlA, 1wetA, 1xe7A, 2i9uA, 2o74A, 2oodA, 2pucA, 2pufA, 3bp1B | 1ula |
| MAN | 18 | 1bvwA, 1bxoA, 1g12A, 1jndA, 1js8A, 1kdgA, 1kza1, 1m3yA, 1nhcA, 1qmoA, 1rinA, 1xxrB, 2duqA, 2e3bA, 2vn4A, 3c6eC, 3d2uF, 3d87D | 3app |
| MMA | 10 | 1kiuB, 1kwuA, 1lobA, 1msaA, 1mvqA, 1rdl1, 1s4pA, 2bv4A, 2g93A, 3g81A | 2ctvA |
| PIM | 5 | 1e9xA, 1f4tA, 1phdA, 1s1fA, 2d0tA | 1phc |
| PLM | 26 | 1e7hA, 1eh5A, 1gxaA, 1hxs1, 1lv2A, 1m66A, 1mzmA, 1pz4A, 1sz7A, 1u19A, 2debA, 2dt8A, 2e9lA, 2fikA, 2go3A, 2jafA, 2nnjA, 2qztA, 2uwhA, 2w3yA, 2z73A, 3bfhA, 3bkrA, 3cueE, 3eglA, 3epyA | 1ifb |
| RTL | 5 | 1fbmA, 1fmjA, 1gx8A, 1kt6A, 2rctA | 1brq |
| UMP | 13 | 1f7nA, 1o26A, 1ouqB, 1q3uB, 1q3vB, 1r2zA, 1sehA, 2bsyA, 2g8oA, 2jarA, 2qchA, 3cl9A, 3dl5A | 3tms |
Abbreviations of hetero atoms used in PDB: ADN, adenosine; BTN, biotin; F6P, fructose 6-phosphate; FUC, fucose; GAL, galactose; GUN, guanine; MAN, mannose; MMA, O1-methyl mannose; PIM, 2-phenylimidazole; PLM, palmitic acid; RTL, retinol; UMP, 2′-deoxyuridine 5-monophosphate.
Table 4A.
Summary of binding ligand prediction by the 2D pocket model using Pseudo-Zernike moments on Kahraman dataset.
| Descriptora) | Rank | AMP | ATP | FAD | FMN | GLC | HEM | NAD | PO4 | STR | Averageb) | Totalc) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pocket shape + pocket size (G) | Top1 | 11.1(%) | 57.1 | 60.0 | 0.0 | 60.0 | 50.0 | 33.3 | 100.0 | 0.0 | 41.2 | 51 |
| Top3 | 77.8(%) | 100.0 | 80.0 | 33.3 | 100.0 | 68.8 | 86.7 | 100.0 | 40.0 | 76.3 | 82 | |
| Average distance | 5.01 | 3.8 | 5.02 | 7.28 | 4.4 | 4.47 | 5.73 | 30.6 | 3.78 | 4.73 | - | |
| Elec. Potential + pocket size (E) | Top 1 | 11.1 | 0.0 | 70.0 | 0.0 | 20.0 | 0.0 | 46.7 | 100.0 | 0.0 | 27.5 | 36 |
| Top 3 | 33.3 | 64.3 | 80.0 | 0.0 | 40.0 | 56.2 | 66.7 | 100.0 | 20.0 | 51.2 | 62 | |
| Avg. distance | 11.64 | 11.14 | 10.14 | 16.27 | 17.35 | 11.06 | 9.14 | 7.63 | 15.16 | 12.17 | - | |
| Pocket shape+ size + electrostatic potential (G+E) | Top 1 | 22.2 | 57.1 | 70.0 | 0.0 | 40.0 | 50.0 | 33.3 | 100.0 | 0.0 | 41.4 | 52 |
| Top 3 | 77.8 | 92.2 | 80.0 | 50.0 | 100.0 | 86.2 | 86.7 | 100.0 | 40.0 | 75.9 | 80 | |
| Avg. distance | 5.88 | 4.76 | 5.52 | 8.3 | 5.76 | 5.51 | 6.41 | 3.84 | 5.52 | 5.7 | - | |
| Pocket size d) | Top1 | 22.2 | 7.1 | 50.0 | 0.0 | 0.0 | 0.0 | 26.7 | 100.0 | 0.0 | 22.8 | 32 |
| Top3 | 55.6 | 78.6 | 80.0 | 0.0 | 0.0 | 81.2 | 60.0 | 100.0 | 0.0 | 50.6 | 66 | |
| Avg. Distance | 0.20 | 0.13 | 0.15 | 0.30 | 0.47 | 0.11 | 0.10 | 0.06 | 0.30 | 0.20 | - | |
| Random retrieval e) | Top 1 | 9.8 (σ=10.1) | 13.2 (9.4) | 9.7 (9.2) | 6.3 (9.8) | 5.4 (9.5) | 15.4 (8.7) | 14.4 (9.0) | 19.4 (9.1) | 6.2 (11.2) | 11.0 | 15.2 |
| Top 3 | 28.0 (σ=14.7) | 39.7 (12.7) | 30.7 (14.5) | 20.8 (16.3) | 17.0 (16.4) | 45.0 (12.8) | 42.1 (13.0) | 54.5 (11.2) | 19.0 (18.0) | 33.0 | 34.9 |
The Euclidean distance is used for measuring similarity of pockets, and the Equation 7 is used for finally predicting bound ligands. The Top1 and Top3 success rate by random chance is shown in the parentheses in the rows of the geometry-based descriptor.
The geometry-based descriptor (G) denotes the pseudo-Zernike moments combined with the pocket size using an optimal weighting factor (Eqns. 12, 13). The electrostatic potential-based descriptor (E) uses the pseudo-Zernike moments of the electrostatic potential of the pocket surface combined with the pocket size using an optimal weighting factor. G+E uses optimally weighted Euclidean distance of G and E.
The average value of the success rate of the nine ligands.
The number of pockets which successfully retrieved pockets of the same type within Top1 or Top3.
The pocket size information only (Table 3) is used to retrieve pockets in the dataset.
The average and the standard deviation (in the parentheses) of 500 random trials are shown.
Results
Effect of rotation to the three 2D moments
To begin with, we examine the effect of rotation of pockets to the 2D moments-based pocket models, namely, the p-Z, the 2D Zernike, and the Legendre moments. In projecting the pocket geometry to a 2D plane, a degree of freedom still exist around the x-axis, which is defined as the direction from the center to the opening of the pocket (Fig. 1). It should be also noted that the rotation invariance in the projected 2D space does not ensure rotation invariance in the original 3D space. Thus, rotation should not alter the pocket descriptors to the level that the recognition of pockets of the same ligand type becomes impractical.
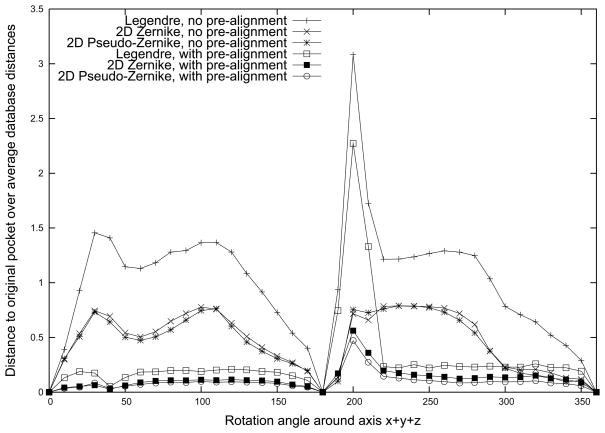
Here, a ligand binding pocket is rotated arbitrarily and the difference of the moments caused by the rotation (the rotation error) is evaluated. Concretely, the AMP binding pocket of asparagine synthetase (PDB: 12AS) is rotated around the axis x⃗+ y⃗+ z⃗ of an arbitrary coordinate set locating its origin at the center of gravity of the pocket. We computed and compared the moments of the pocket at each rotated position with and without pre-alignment: Firstly, we simply computed the moments of the pocket at each rotated position and compared them with the ones computed at the original position (i.e. without pre-alignment of pockets). Secondly, for each rotated pocket, we aligned it with the pocket at the original position before computing the moments (i.e. with pre-alignment). The pre-alignment consists of the following steps. The x-axis of the two pockets are aligned, then the z-axis is defined such that its principle moment of inertia (PMI) is maximized over all posisble directions on the plane orthogonal to the x-axis. From the mathematical point of view, the 2D Zenrike and the p-Z moments are invariant upon rotation around the axis while the Ledendre moments are not. Thus, it is expected that the comparison without pre-alignment gives better results for the 2D Zernike and p-Z than the Legendre moments. The comparison with the pre-alignment is performed to see whether the Ledendre moments shows comparable performance with the other two moments. The error is defined as the ratio of the Euclidean distance between the moments of the pocket at a rotated position and at the original position relative to the average Euclidean distance of the pocket (at the original position) to the other types pockets in the Kahraman dataset.
In Figure 3, the rotation error of the three moments is plotted with and without the pre-alignment of the pockets. First, as expected, the p-Z and 2D Zernike moments show lower error than the Legendre moments when pockets are not pre-aligned. Next, when pockets are pre-aligned, the error of all three moments is reduced remarkably. However, still the p-Z and 2D Zernike show a smaller error than the Legendre moments. A closer look at the results of the three moments with pre-alignment by computing the sum of the error values at each rotation angle (X-axis) shows that the p-Z has the smallest error with the value of 3.49, while the values of the 2D Zernike and the Legendre moments are 4.47 and 10.47, respectively.
Figure 3.
Quantification of rotation invariance of the descriptors. The AMP binding pocket of 12AS is rotated around the axis x⃗ + y⃗ + z⃗ of an arbitrary coordinate set from the pocket geometric center. The angle of rotation is shown on the x axis and the y axis shows the rotation error as compared with the pocket at the original orientation. The rotation error is defined as the L2 distance of a rotated binding pocket divided by the average distance to the other pockets in the benchmark dataset.
Pocket retrieval performance by the three 2D moments and the 3DZD
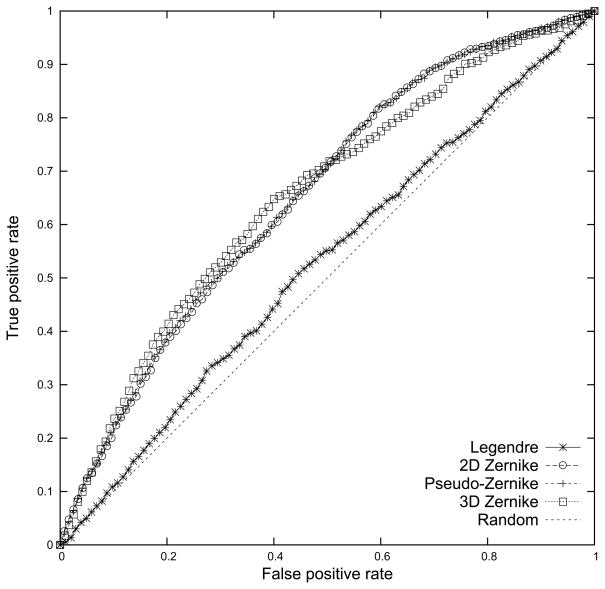
Next, we compare the 2D pocket models using the three 2D moments and the 3D pocket model using the 3DZD in terms of actual performance of identifying binding pockets of the same ligand. Note that ligands are pre-aligned for computing the Legendre moments. Figure 4 shows the Receiver Operating Characteristic (ROC) curve of the three 2D moments and the 3DZD averaged over searching results of different ligand binding pockets in the benchmark dataset. Concretely, given a query pocket, pockets in the database which are within a threshold Euclidean distance (Eqn. 5) are retrieved, and are then subject to evaluation by computing the false positive (x-axis) and the true positive (y-axis) rate. Varying the threshold value from strict to more permissive values yields the ROC curve. The false positive rate of a set of retrieved pockets for a query is defined as the ratio of the number of retrieved pockets of a different ligand (i.e. false positives) relative to the total number of pockets of a different ligand (i.e. false positives and true negatives) in the dataset. The true positive rate is the ratio of the number of correctly retrieved pockets (i.e. true positives) relative to the total number of pockets of the same type in the dataset.
Figure 4.
The ROC for the three moments, the Legendre, the 2D Zernike, the pseudo-Zernike moments, and the 3D Zernike descriptors. The Euclidean distance is used. The average value of nine different ligand types is plotted.
The results are shown in Figure 4. Firstly, all the four moments perform better than random retrieval. Secondly, the p-Z and the 2D Zernike moments show almost identical performance on this plot, which is significantly better than the Legendre moments with the pre-alignment of the pockets. The 3D pocket model with the 3DZD has slightly higher AUC values than the p-Z and 2D Zernike when the false positive rate is small (0 to around 0.5) and has lower values for the latter half of the false positive rate (around 0.5 to 1.0). Quantitative computation of the Area Under the Curve (AUC) of the ROC curve (upper half of Table 2, results using the “Pocket shape only” descriptor) shows that the p-Z, the 2D Zernike moments, and 3DZD have an identical AUC value of 0.66 when the Euclidean distance is used. Note that this value is larger than the results by the spherical harmonics (0.64). The p-Z moments perform slightly better than the 2D Zernike moments and the 3DZD when the Manhattan distance (dM) is used. Since the p-Z moments show a better performance among the three 2D moments (Legendre, p-Z, and 2D Zernike), we decided to use the p-Z moments with the Euclidean distance in the subsequent experiments, and further compared the performance with the 3D pocket model using the 3DZD. We have also tested the p-Z moments with the pocket pre-alignment but the improvement was not significant (0.75% improvement in the AUC value). Therefore, the pocket pre-alignment is not used in the following experiments.
Table 2.
Average area under ROC curve for different metrics and descriptors.
| Descriptor a) | Metric b) | Legendre (with PMI) | Pseudo-Zernike | 2D Zernike | 3D Zernike | Spherical Harmonics c) |
|---|---|---|---|---|---|---|
| Pocket shape only | Manhattan (dM) | 0.53 | 0.66 | 0.65 | 0.64 | - |
| Euclidean (dE) | 0.53 | 0.66 | 0.66 | 0.66 | 0.64 | |
| Correlation Coefficient-based (dC) | 0.55 | 0.52 | 0.53 | 0.64 | - | |
| Pocket shape and size | dM | 0.76 | 0.76 | 0.75 | 0.79 | - |
| dE | 0.77 | 0.79 | 0.78 | 0.81 | 0.77 | |
| dC | 0.55 | 0.55 | 0.55 | 0.72 | - |
The “pocket shape only” descriptor uses the moments and the “pocket shape and volume” descriptor uses the moments and the volume of pockets, with optimal size/shape weighting factors for each descriptor. Pseudo-Zernike moments are computed with order 4, Legendre and 2D Zernike moments with order 6 and 3D Zernike with order 20.
The values are taken from Table 3 in the paper by Kahraman et al. Only the Euclidean distance was reported in the paper.
Combining pocket size information
Kahraman et al. reported that pocket retrieval accuracy improves when the shape descriptor by spherical harmonics is combined with pocket volume information56. Inspired by their idea, we explore combinations of pocket shape by the p-Z moments or the 3DZD and the pocket size using a weighting factor, w. These two pieces of information are combined in the descriptor of a pocket, Pa, as the following vector:
| (22) |
where SPa is the size of the pocket Pa, AkPa is the k-th value of the moments of the pocket Pa (the pseudo-Zernike, the 2D Zernike, the Legendre, or the 3DZD), and N is the total number of values of the moments. Thus, using the vector above, the Euclidean distance between the descriptors of two pockets, Pa and Pb, becomes:
| (23) |
where Sa and Sb are the size of the two pockets. As the size of a pocket, we use the average distance from the center of gravity G of the pocket to the pocket surface. Table 3 shows the size of the nine different types of pockets in the Kahraman set and twelve ligand types in the Huang set. The average distance has a significant correlation coefficient of 0.853 to the molecular mass of ligands (g/mol).
Table 3.
Ligand binding pocket size.
| Ligand Type | Average Sizea) (Å) | Standard Deviation | Minimum value | Maximum value | Molecular mass (g/mol) |
|---|---|---|---|---|---|
| AMP | 8.8 | 1.0 | 6.8 | 9.8 | 347.22 |
| ATP | 9.5 | 0.8 | 7.1 | 11.0 | 507.18 |
| FAD | 11.0 | 0.4 | 10.4 | 11.8 | 785.55 |
| FMN | 9.7 | 0.6 | 8.6 | 10.4 | 456.34 |
| GLC | 8.5 | 0.3 | 8.1 | 9.0 | 180.16 |
| HEM | 10.2 | 0.7 | 9.2 | 11.4 | 616.49 |
| NAD | 10.1 | 1.1 | 7.6 | 11.5 | 663.43 |
| PO4 | 7.4 | 0.4 | 6.2 | 7.8 | 94.97 |
| STR | 9.2 | 0.5 | 8.6 | 10.1 | 288.42/272.38 b) |
| ADN | 9.1 | 0.6 | 8.1 | 9.8 | 267.24 |
| BTN | 8.4 | 1.0 | 6.5 | 9.6 | 244.31 |
| F6P | 8.7 | 0.4 | 7.9 | 9.2 | 262.15 |
| FUC | 6.7 | 1.1 | 4.5 | 8.3 | 164.16 |
| GAL | 7.8 | 0.8 | 5.0 | 9.1 | 180.08 |
| GUN | 8.1 | 0.2 | 7.7 | 8.4 | 151.13 |
| MAN | 6.3 | 1.0 | 4.5 | 7.8 | 180.16 |
| MMA | 7.6 | 0.8 | 5.8 | 8.7 | 194.08 |
| PIM | 8.1 | 0.2 | 7.9 | 8.5 | 144.18 |
| PLM | 9.0 | 0.7 | 6.8 | 10.2 | 256.42 |
| RTL | 9.3 | 0.5 | 8.7 | 9.7 | 286.45 |
| UMP | 8.7 | 0.8 | 7.6 | 9.6 | 308.18 |
The pocket size is defined as the average distance from the center of gravity of the pocket to the pocket surface. The Kahraman set is shown in the first half and the Huang set is shown in the latter half.
The average is taken over all the pockets of the same type.
Values for 3-β-hydroxy-5-androsten-17-one (AND) and estradiol (EST) are shown. 1e3r and 1j99 bind AND, while 1fds, 1lhu, and 1qkt bind EST.
In Figure 5, the weighting factor w of the pocket size term (Eqn. 23) is searched from 1.0 to 8.0(with an interval of 0.5) for the p-Z moments and 0.01 to 0.08 (with an interval of 0.01) for the 3DZD and the average AUC value over different pocket types is computed. In addition to the value for weighting factor w, we also examined different resolution of the p-Z moments and the 3DZD, i.e. the number of terms in the moments (x-axis). Mathematically, a target function is perfectly described by an infinite number of terms in the moments. However, practically using too many terms is inefficient and may even be harmful for our purpose, because the primary goal of this work is to compare and retrieve pockets of the same type that are not exactly identical in shape, rather than to describe a pocket’s shape as accurately as possible.
Figure 5.
The Area Under the Curve value relative to the number of Zernike coefficients (the x axis) and the pocket volume weight (the y axis). A, the pseudo-Zernike moments; B, the 3DZD.
For the p-Z moments (Fig. 5A), we find that fifteen terms (which correspond to the order of up to n = 4 in Eqn. 2) give a sufficient AUC value and using more terms does not improve the results. In terms of the weight w, 4.5 gives the highest AUC value. For the 3DZD (Fig. 6A), the order of 20 with the weight of 0.04 gave the highest AUC value. The optimal weight is much smaller for the 3DZD because the average norm of 3DZD is two orders of magnitude lower than the p-Z moments.
Figure 6.
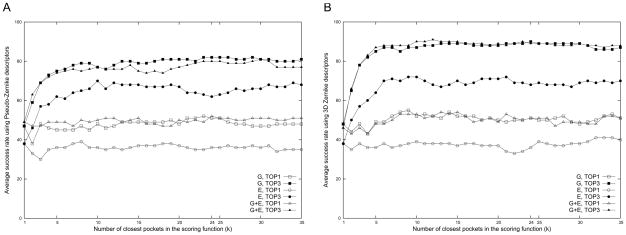
The success rate of binding ligand prediction as a function of the number of closest pockets considered in the scoring function. The x-axis is the parameter k in the Pocket_score (Eqn. 7). The success rate in the TOP1 and TOP3 by the three pocket descriptors, the shape (G), the electrostatic potential (E), and the shape and the electrostatic potential combined (G+E), are plotted. A, the pseudo-Zernike moments; B, the 3DZD.
The bottom half of Table 2 summarizes the effect of adding the pocket size information using the weight of 4.5 for the three 2D moments and 0.04 for the 3DZD. It is shown that the AUC value increases consistently by adding the pocket size in all the combinations of different moments and the distance metrics tested. Among all tested in Table 2, the best AUC value, 0.81, is achieved by the 3DZD with the pocket size using the Euclidean distance. The descriptor with the p-Z moments comes to the second with 0.79 followed by the 2D Zernike (0.78) and Legendre moments (0.77). The values of the 3DZD, the p-Z and the 2D Zernike moments are higher than the AUC value achieved by a spherical harmonics-based descriptor combined with the pocket volume proposed by Kahraman et al.56 (the right most column).
The number of top scoring pockets to consider in the Pocket_score
For a given query pocket, the Euclidean distance is computed against all pockets in the dataset and then the final prediction of the binding ligand is made using Pocket_score (Eqn. 7). Since the final prediction depends on the number of closest pockets (the parameter k in Eqn. 20) to consider, we examined the effect of the value of k on the resulting success rate. In Figure 6, the average success rate of the nine ligands in the Kahraman set for k = 1 to 35 is plotted. The plot in Fig 6A is the results of the 2D pocket model with the p-Z moments while Fig 6B shows results by the 3DZD. Three pocket descriptors are tested: either the surface shape (G in Fig. 6) or the surface electrostatic potential (E) combined with the pocket size (w = 4.5 for the p-Z and 0.04 for the 3DZD as determined in Fig. 5) and the average distance of those by the two descriptors (G+E). In the Top1 results, the rate is measured with the highest scoring ligand being the correct one, while the Top3 allows the correct answer to lie in the first three highest scoring ligands.
When the top scoring ligand is counted (curves of TOP1 in Fig. 6), increasing the number of closest pockets to consider in the scoring function does not help much to improve the accuracy. However, it does make dramatic improvement when top three ligands are considered (TOP3). In the case of TOP3 prediction, the success rate of sharply improves by roughly over forty to fifty percentage points when k is set to 10 or higher as compared with the results with k = 1. The improvement is more significant in the 3DZD as compared with the p-Z 2D pocket model. In all three pocket descriptors, the success rate gradually increases until k is from about 15 to 25. We decided to use k = 24 for both p-Z and 3DZD for the subsequent analysis, because it gives the second best success rate by the pocket shape descriptor (G) in the TOP3 prediction (82.0% for the p-Z and 90.0% for the 3DZD) and also gives good performance by the combination of the shape and the electrostatic potential descriptor (G+E).
Retrieval accuracy of individual pocket types in the Kahraman dataset
Up to the previous sections, we examined the pocket retrieval performance of three different 2D moments and a 3D pocket model using the 3DZD and determined the parameters for the pocket retrieval. Here we further discuss the retrieval accuracy of individual pocket types. We first show the results on the Kahraman dataset as it was used to examine the types of moments and the parameters. Then, later we show the results on the Huang dataset, which is an independent dataset from the Kahraman set. For the both datasets, performance of the p-Z moments and the 3DZD are compared.
Table 4 gives the success rate of retrieving individual binding ligands using k = 4.5 for the p-Z moments (Table 4A) and k = 0.04 for the 3DZD (Table 4B). For both p-Z and the 3DZD, all the three pocket descriptors (G, E, G+E) perform far better than the random retrieval. The pocket shape descriptor (G) shows the best average success rate in the TOP3 prediction consistently for the p-Z (76.3%) and for the 3DZD (82.7%). Adding the electrostatic information to the shape information, i.e. G+E descriptor, makes a small improvement in the TOP1 prediction in the case of the p-Z moments (Table 4A, from G: 41.2% to G+E: 41.4%), but give the same success rate for the 3DZD (Table 4B, both G and G+E give 36.1%). In terms of the TOP3 prediction, adding the electrostatic information slightly deteriorates the success rate for both p-Z moments and the 3DZD. This is consistent with a recent observation by Thornton group which reports that the electrostatic potential in ligand binding pockets are highly variable within families83. Comparing the performance of the p-Z moments and the 3DZD, the p-Z moments show slightly higher success rate in the TOP1 prediction while the 3DZD show higher value in the TOP3 prediction success rate. The success rate differs from ligand to ligand and these trends are quite consistent for both for the p-Z moments and the 3DZD. For example, in TOP3 with the shape and the size descriptor (G), ATP, GLC, and FAD performs well while FMN and STR show poorer results. This implies that the difference in performance for each ligand is attributed not to the characteristics of our 2D/3D pocket models but to the actual similarity or divergence of pocket shapes of particular ligand types. The low retrieval accuracy of FMN can be explained by two main factors: Among the three smallest ligand types (GLC, FMN and STR), FMN is the most flexible one with an average RMSD of 1.08 Angstrom. Also, it is the third under-represented ligand type in the Kahraman dataset (6 structures), hence random retrieval will be relatively less accurate.
Table 4B.
Summary of the binding ligand prediction using 3D Zernike descriptors on Kahraman dataset.
| Descriptora) | Rank | AMP | ATP | FAD | FMN | GLC | HEM | NAD | PO4 | STR | Averageb) | Totalc) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pocket shape + pocket size (G) | Top1 | 0.0(%) | 21.4 | 50.0 | 0.0 | 0.0 | 87.5 | 60.0 | 100.0 | 0.0 | 36.1 | 51 |
| Top3 | 77.8(%) | 100.0 | 90.0 | 16.7 | 80.0 | 100.0 | 100.0 | 100.0 | 80.0 | 82.7 | 90 | |
| Average distance | 0.05 | 0.04 | 0.04 | 0.05 | 0.07 | 0.04 | 0.03 | 0.05 | 0.07 | 0.05 | - | |
| Elec. Potential + pocket size (E) | Top 1 | 11.1 | 21.4 | 60.0 | 0.0 | 20.0 | 6.2 | 33.3 | 95.0 | 0.0 | 28.2 | 36 |
| Top 3 | 44.4 | 85.7 | 80.0 | 16.7 | 40.0 | 68.8 | 66.7 | 100.0 | 0.0 | 55.8 | 68 | |
| Avg. distance | 4.07 | 2.86 | 2.7 | 5.08 | 5.71 | 3.17 | 2.18 | 4.69 | 6.06 | 4.06 | - | |
| Pocket shape+ size + electrostatic potential (G+E) | Top 1 | 0.0 | 21.4 | 50.0 | 0.0 | 0.0 | 87.5 | 60.0 | 100.0 | 0.0 | 36.1 | 51 |
| Top 3 | 66.7 | 100.0 | 90.0 | 16.7 | 80.0 | 100.0 | 100.0 | 100.0 | 80.0 | 81.5 | 89 | |
| Avg. distance | 0.05 | 0.04 | 0.04 | 0.05 | 0.07 | 0.04 | 0.03 | 0.05 | 0.07 | 0.05 | - |
Table 4A also gives the retrieval results by solely using the pocket size (given in Table 3). It turned out that PO4 can be perfectly retrieved by just using the size, as it is the smallest ligand in the Kahraman set. The overall retrieval accuracy with the pocket size is 50.6 for the Top3 value, which might seem relatively high. But this is qualitatively consistent with the observation by Kahraman et al.56, who made the dataset and reported that the AUC value by using size information is as high as 0.73 as compared with 0.77 achieved by combining the pocket shape and the size information (Table 2). In comparison with the performance by the pocket shape and size descriptor (G) of the p-Z moments (Table 4A) and the 3DZD (Table 4B), the addition of the shape information makes improvement or tie in all the cases of Top1 and Top3 values expect for three cases (the TOP1 value for NAD, the Top 3 value for HEM by the p-Z moments and the Top 1 value for AMP by the 3DZD). Thus overall the pocket shape information makes effective contribution to improving the retrieval accuracy.
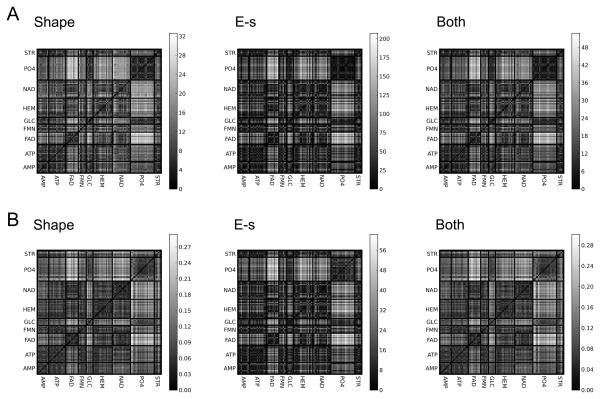
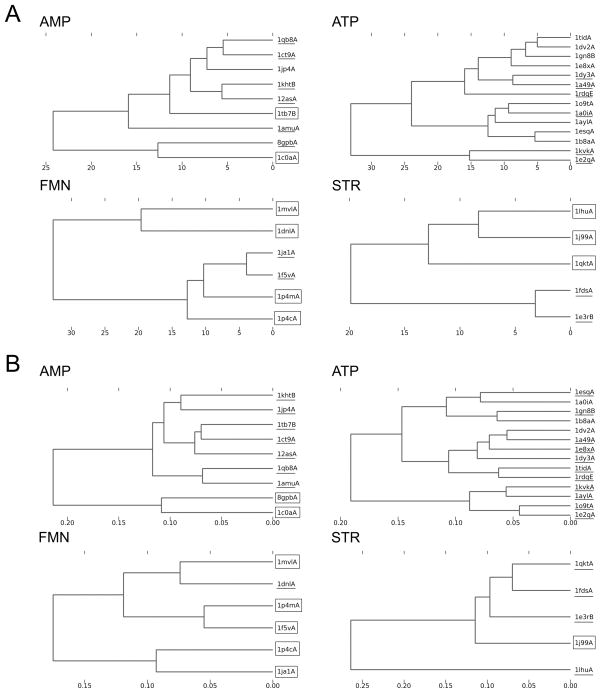
The pocket retrieval success rate in Table 4 roughly agrees with the distance of all against all pockets shown in Figure 7, which visualizes the Euclidean distance of all the pocket pairs. Figure 7A and 7B show the distance by the p-Z moments and the 3DZD, respectively. It can be seen that all the ligand binding pockets have a close distance to the other pockets of the same type (i.e. diagonal squares are in darker gray), however, ligands with a poor retrieval success rate also show similarity to the other ligand types. For example, AMP binding pockets seem to be close to some of ATP binding pockets, and FMN binding pockets have a close distance to ATP binding pockets. HEM and NAD binding pockets also seem to be similar. Figure 8 examines mutual distance of four individual ligand binding pockets, AMP, ATP, FMN, and STR. In all the ligand cases, binding pockets which are relatively distant from the other members of the same ligand type tend to fail in binding ligand prediction. For example, in the case of AMP binding pockets with the 3DZD, 8gpbA and 1c0aA failed in the TOP3 results.
Figure 7.
The Euclidean distance of all-against-all pocket pairs using the pseudo-Zernike descriptors. The gray scale represents log-transformed distance with a darker color indicating a closer distance. Distances by the three pocket descriptors, the shape (G), the electrostatic potential (E), and the shape and the electrostatic potential combined (G+E) are shown. A, the pseudo-Zernike moments; B, the 3DZD.
Figure 8.
Complete linkage clustering of ligand binding pockets. The Euclidean distance of the pocket shape descriptor (G) is used. The PDB IDs underlined are those for which its binding ligand is not correctly predicted within the TOP1 but within the TOP3 while those surrounded by a rectangle are those for which its ligand is not correctly predicted even within the TOP3. Pockets of four ligands are shown, AMP; ATP; FMN; STR. A, the pseudo-Zernike moments; B, the 3DZD.
To have a better understanding of the pocket prediction process, we closely examined examples of search results for individual cases of FAD binding pockets by the p-Z moments as examples (Table 5). Table 5 shows two successful and two failed cases. 1eviB is a very successful case where five other FAD binding pockets are retrieved within top 5 ranks. In the case of 1e8gB, the search retrieved three FAD binding pockets contaminated with HEM binding pockets, which resulted in the second rank in the final prediction. On the other hand, 1cqxA did retrieve FAD binding sites within top 25 but the top hits are dominated by four other ligand types. No FAD binding pocket is retrieved within the top 25 in the case of 1jr8B.
Table 5.
Examples of database search results for FAD binding pocket prediction using 2D Pseudo-Zernike moments.
| A. Two successful cases. | ||||||
|---|---|---|---|---|---|---|
| Query | 1eviB | 1e8gB | ||||
| Rank | Ligand type | PDB ID | Euclidean Distance | Ligand type | PDB ID | Euclidean Distance |
| 1 | FAD | 1jqi | 3.032 | HEM | 2cpo | 4.982 |
| 2 | FAD | 3grs | 4.489 | HEM | 1np4 | 5.128 |
| 3 | FAD | 1k87 | 5.032 | FAD | 1k87 | 5.189 |
| 4 | FAD | 1h69 | 6.287 | FAD | 1h69 | 5.469 |
| 5 | FAD | 1pox | 6.501 | FAD | 1jqi | 5.998 |
| 6 | NAD | 1t2d | 6.840 | NAD | 1o04 | 6.521 |
| 7 | FAD | 1e8g | 7.457 | HEM | 1gwe | 6.696 |
| 8 | NAD | 1ej2 | 8.005 | HEM | 1d0c | 6.821 |
| 9 | HEM | 2cpo | 8.382 | HEM | 1pp9 | 6.875 |
| 10 | HEM | 1gwe | 8.540 | NAD | 1t2d | 6.922 |
| 11 | HEM | 1np4 | 8.657 | ATP | 1rdq | 7.026 |
| 12 | NAD | 1o04 | 8.705 | NAD | 1s7g | 7.148 |
| 13 | FAD | 1hsk | 8.731 | FAD | 1evi | 7.457 |
| 14 | NAD | 1jq5 | 9.309 | FAD | 1cqx | 7.504 |
| 15 | NAD | 1mi3 | 9.390 | NAD | 1qax | 7.564 |
| 16 | HEM | 1d0c | 9.580 | HEM | 1po5 | 7.847 |
| 17 | ATP | 1rdq | 10.206 | FMN | 1p4c | 8.008 |
| 18 | HEM | 1pp9 | 10.577 | FAD | 1pox | 8.298 |
| 19 | NAD | 1qax | 10.707 | HEM | 2bs2 | 8.394 |
| 20 | FAD | 1cqx | 10.712 | FAD | 1hsk | 8.478 |
| 21 | NAD | 1og3 | 10.936 | NAD | 1mi3 | 8.502 |
| 22 | HEM | 1po5 | 11.248 | HEM | 1eqg | 8.597 |
| 23 | HEM | 1iqc | 11.413 | HEM | 1naz | 8.976 |
| 24 | NAD | 1s7g | 11.617 | HEM | 1iqc | 9.125 |
| 25 | FMN | 1p4c | 11.771 | ATP | 1tid | 9.141 |
| Final Ligand prediction | ||||||
| Rank | Ligand type | Occurrence within top 25 | Score | Ligand type | Occurrence within top 25 | Score |
| 1 | FAD | 8 | 77.80 | HEM | 10 | 59.36 |
| 2 | NAD | 8 | 13.31 | FAD | 7 | 32.45 |
| 3 | HEM | 7 | 8.48 | NAD | 5 | 6.92 |
| 4 | FMN | 1 | 0.25 | ATP | 2 | 0.84 |
| 5 | ATP | 1 | 0.16 | FMN | 1 | 0.44 |
| B. Two failed cases. | ||||||
| Query | 1cqxA | 1jr8B | ||||
| Rank | Ligand type | PDB ID | Euclidean Distance | Ligand type | PDB ID | Euclidean Distance |
| 1 | HEM | 1eqg | 4.569 | ATP | 1ayl | 3.911 |
| 2 | ATP | 1e8x | 4.582 | NAD | 1og3 | 4.103 |
| 3 | HEM | 1d0c | 4.724 | NAD | 2a5f | 4.416 |
| 4 | ATP | 1dv2 | 4.866 | NAD | 1qax | 4.694 |
| 5 | ATP | 1rdq | 5.248 | NAD | 1mi3 | 5.079 |
| 6 | NAD | 2npx | 5.274 | FMN | 1p4c | 5.383 |
| 7 | NAD | 1s7g | 5.505 | ATP | 1tid | 5.443 |
| 8 | NAD | 2a5f | 5.596 | ATP | 1gn8 | 5.489 |
| 9 | ATP | 1tid | 5.617 | ATP | 1dv2 | 5.656 |
| 10 | FMN | 1ja1 | 5.839 | HEM | 2bs2 | 5.750 |
| 11 | FMN | 1f5v | 5.842 | HEM | 1eqg | 5.834 |
| 12 | HEM | 1np4 | 5.853 | FMN | 1p4m | 6.041 |
| 13 | FMN | 1p4m | 5.952 | NAD | 1o04 | 6.350 |
| 14 | AMP | 1tb7 | 6.379 | HEM | 1dk0 | 6.444 |
| 15 | ATP | 1gn8 | 6.836 | HEM | 1sox | 6.500 |
| 16 | HEM | 1dk0 | 7.275 | ATP | 1e8x | 6.532 |
| 17 | FAD | 1h69 | 7.309 | AMP | 1tb7 | 6.751 |
| 18 | NAD | 1og3 | 7.315 | HEM | 1d0c | 6.814 |
| 19 | FMN | 1p4c | 7.396 | AMP | 1jp4 | 6.949 |
| 20 | FAD | 1jr8 | 7.400 | HEM | 1po5 | 6.955 |
| 21 | ATP | 1ayl | 7.431 | FMN | 1ja1 | 7.008 |
| 22 | NAD | 1t2d | 7.498 | HEM | 1naz | 7.015 |
| 23 | FAD | 1e8g | 7.504 | STR | 1lhu | 7.049 |
| 24 | STR | 1lhu | 7.559 | STR | 1e3r | 7.076 |
| 25 | HEM | 2cpo | 7.712 | AMP | 1qb8 | 7.120 |
| Final Ligand prediction | ||||||
| Rank | Ligand type | Occurrence within top 25 | Score | Ligand type | Occurrence within top 25 | Score |
| 1 | ATP | 6 | 35.16 | ATP | 5 | 36.47 |
| 2 | HEM | 5 | 33.14 | NAD | 5 | 32.12 |
| 3 | FMN | 4 | 14.91 | HEM | 7 | 14.88 |
| 4 | NAD | 5 | 12.46 | FMN | 3 | 10.54 |
| 5 | FAD | 3 | 3.19 | AMP | 3 | 3.58 |
Raw pocket comparison results of four FAD binding pockets are shown, two cases for successful and the other two for failed cases when TOP3 predicted ligands are considered. Retrieved FAD binding pockets are underlined.
Retrieval Accuracy on the Huang dataset
We further tested the pocket models with the p-Z moments and the 3DZD on another independent dataset using the same parameters determined based on the Kahraman dataset (Table 6). This dataset, the Huang dataset, which consists of 175 proteins which bind one of the twelve ligands, have no overlap in the proteins and ligand types with Kaharaman set.
Table 6A.
Binding ligand prediction results on the Huang dataset with Pseudo-Zernike moments.
| Descriptora) | Rank | ADN | BTN | F6P | FUC | GAL | GUN | MAN | MMA | PIM | PLM | RTL | UMP | Average b) | Totalc) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pocket shape + pocket size (G) | Top1 | 40.0 | 38.5 | 16.7 | 14.3 | 44.4 | 50.0 | 77.8 | 0.0 | 60.0 | 69.2 | 20.0 | 0.0 | 35.9 | 41 |
| Top3 | 80.0 | 69.2 | 58.3 | 85.7 | 97.2 | 83.3 | 83.3 | 40.0 | 80.0 | 96.2 | 80.0 | 53.8 | 75.6 | 81 | |
| Average distance | 3.61 | 4.29 | 3.72 | 5.64 | 3.50 | 3.54 | 4.98 | 5.51 | 4.03 | 4.99 | 6.13 | 4,83 | 4.6 | - | |
| Elec. Potential + pocket size (E) | Top 1 | 40.0 | 0.0 | 0.0 | 0.0 | 47.2 | 25.0 | 66.7 | 0.0 | 60.0 | 38.5 | 40.0 | 15.4 | 27.7 | 31 |
| Top 3 | 70.0 | 7.7 | 66.7 | 78.6 | 77.8 | 66.7 | 83.3 | 30.0 | 100.0 | 65.4 | 40.0 | 38.5 | 60.4 | 64 | |
| Avg. distance | 9.7 | 14.0 | 7.3 | 14.7 | 8.3 | 10.9 | 12.6 | 13.9 | 8.3 | 10.9 | 13.1 | 11.1 | 11.2 | ||
| Pocket shape+ size + electrostatic potential (G+E) | Top 1 | 30.0 | 15.4 | 8.3 | 14.3 | 52.8 | 50.0 | 72.2 | 0.0 | 40.0 | 65.4 | 20.0 | 7.7 | 31.3 | 39 |
| Top 3 | 90.0 | 69.2 | 66.7 | 85.7 | 91.7 | 83.3 | 77.8 | 50.0 | 80.0 | 80.8 | 60.0 | 53.8 | 74.1 | 78 | |
| Avg. distance | 4.48 | 5.69 | 4.95 | 6.95 | 4.41 | 4.49 | 6.45 | 6.89 | 4.74 | 6.11 | 6.92 | 5.73 | 5.7 | ||
| Random retrievald) | Top 1 | 7.0 (σ=7.5) | 6.3 (7.2) | 7.7 (7.6) | 7.0 (5.8) | 16.4 (6.5) | 7.7 (8.5) | 9.7 (6.8) | 7.8 (9.0) | 3.6 (7.7) | 12.6 (6.9) | 3.2 (8.4) | 7.7 (7.1) | 8.1 | 10.0 |
| Top 3 | 18.6 (σ=11.0) | 24.5 (14.7) | 21.7 (12.8) | 25.9 (11.2) | 45.9 (8.1) | 23.2 (11.9) | 28.2 (9.1) | 19.8 (13.3) | 11.6 (13.3) | 34.8 (7.9) | 9.2 (12.8) | 27.6 (13.5) | 24.2 | 27.9 |
On this dataset, both p-Z moments and the 3DZD show lower success rate by the pocket shape descriptor (G) as compared to the results for the Kahraman set (Table 4). For example, the Top3 success rate by the pocket shape descriptor of the p-Z moments is 75.6%, which is a small decrease from 76.3% shown for the Kahraman set, while the shape descriptor of the 3DZD shows a larger decrease, from 82.7% (on the Kahraman set) to 59.8%. On the other hand, we observe higher success rate of the electrostatic potential descriptor (E) on this dataset relative to the Kahraman set both by the p-Z moments and the 3DZD. Of course both of our pocket models consistently show significantly better performance than random retrieval. In the case of the 3DZD (Table 6B), the combination of the shape and the electrostatic potential descriptors (G+E) shows improvement over the shape descriptor (G) due to the aforementioned two reasons, i.e. descrease in the success rate by the shape descriptor (G) and the good performance by the electrostatic potential descriptor (E) on this dataset.
Table 6B.
Binding ligand prediction results on the Huang dataset with 3D Zernike descriptors.
| Descriptor | Rank | ADN | BTN | F6P | FUC | GAL | GUN | MAN | MMA | PIM | PLM | RTL | UMP | Average | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pocket shape + pocket size (G) | Top1 | 9.1 | 33.3 | 33.3 | 21.4 | 38.9 | 75.0 | 38.9 | 0.0 | 0.0 | 92.3 | 0.0 | 23.1 | 30.4 | 40 |
| Top3 | 54.5 | 66.7 | 75.0 | 85.7 | 80.6 | 91.7 | 72.2 | 10.0 | 0.0 | 92.3 | 20.0 | 69.2 | 59.8 | 71 | |
| Average distance | 0.05 | 0.04 | 0.04 | 0.05 | 0.04 | 0.05 | 0.05 | 0.04 | 0.00 | 0.03 | 0.06 | 0.04 | 0.0 | - | |
| Elec. Potential + pocket size (E) | Top 1 | 27.3 | 0.0 | 33.3 | 0.0 | 50.0 | 58.3 | 27.8 | 10.0 | 0.0 | 80.8 | 0.0 | 53.8 | 28.4 | 38 |
| Top 3 | 63.6 | 33.3 | 41.7 | 64.3 | 91.7 | 100.0 | 88.9 | 60.0 | 20.0 | 96.2 | 40.0 | 69.2 | 64.1 | 74 | |
| Avg. distance | 8.2 | 3.4 | 4.3 | 4.6 | 3.2 | 3.6 | 3.6 | 4.3 | 4.7 | 2.0 | 3.0 | 2.1 | 3.9 | - | |
| Pocket shape+ size + electrostatic potential (G+E) | Top 1 | 18.2 | 33.3 | 33.3 | 14.3 | 44.4 | 83.3 | 33.3 | 0.0 | 0.0 | 92.3 | 0.0 | 30.8 | 31.9 | 41 |
| Top 3 | 63.6 | 66.7 | 75.0 | 85.7 | 83.3 | 91.7 | 72.2 | 10.0 | 0.0 | 92.3 | 20.0 | 76.9 | 61.5 | 72 | |
| Avg. distance | 0.05 | 0.04 | 0.04 | 0.05 | 0.04 | 0.05 | 0.05 | 0.05 | 0.00 | 0.03 | 0.06 | 0.04 | 0.0 |
Comparison with existing methods
Next, we compare the pocket retrieval performance of our methods with two other existing methods, eF-Seek84 (http://ef-site.hgc.jp/eF-seek/) and SitesBase54 (http://www.modelling.leeds.ac.uk/sb/). Both methods mainly use geometrical shape information for quantifying similarty of pockets: eF-Seek represents protein surface as triangle meshes and employs a graph matching algorithm to seek similar local sites stored in the eF-Site database55. On the other hand, SitesBase uses geometric hashing algorithm to identify equivalent atom constellations between pairs of ligand binding sites. Readers should be reminded that in principle an entirely fair comparison of existing servers is not possible, as each method has been trained on a different dataset and currently contains a different set of binding pockets in its own database. Hence this comparison is meant only to provide rough ideas of how our methods perform in comparison with others.
Since the databases used by these methods are different, we first identified proteins in the Tables 1A and 1B that are commonly included both in the databases of eF-Seek and SitesBase. Then, among the 21 ligand types in total in Table 1(nine in Table 1A and twelve in Table 1B), we selected twelve ligands for which most of the listed proteins are included in the eF-Seek and SitesBase databases. They are AMP, ATP, FAD, FMN, GLC, HEM, NAD, F6P, GAL, GUN, MMA, and PLM. The proteins for these ligands which are also found in eF-Seek and SitesBase are underlined with single or double line. For each of these ligands, randomly selected three proteins are used as queries (those underlined with double line in Table 1). eF-Seek and SitesBase return different number of pockets in a result file. Out of the proteins they return, we only selected underlined proteins and computed the evaluation values, i.e. Top1, Top3, and AUC values. The query protein which appear as the top hit is discarded in the evaluation. For eF-Seek results, we sorted the retrieved proteins by the distance from the ideal score, which we define as
| (24) |
We ranked the eF-Seek results in this way because eF-Seek does not rank the retrieved proteins but only provides a 2D plot of the Z-score and the coverage. The maximum Z-score value for the queries is 13.4 and the maximum coverage value is 1.0. The AUC value of the two methods is computed by randomly adding missing proteins from the selected proteins in Table 1 to the end of the list of retrieved proteins, assuming that these proteins are retrieved in that order. The AUC curve was computed ten times for eF-Seek and SitesBase and the average and the standard deviations are recorded.
The results are summarized in Table 7. The 3DZD pocket model shows the best performance among all in terms of all the metrics, i.e. Top1, Top3, and AUC. The p-Z pocket model comes to the second in terms of the AUC and ranked the third following SitesBase in the Top1 and Top3 value. Overall, our 2D and 3D pocket models show better or comparable performance to the two methods compared.
Table 7.
Comparison with eF-Seek and SitesBase.
| Methods | Top1 (%) | Top3 (%) | AUC |
|---|---|---|---|
| Pseudo-Zernike | 25.0 | 52.7 | 0.746 |
| 3D Zernike | 47.2 | 75.0 | 0.860 |
| eF-Seek | 19.4 | 36.1 | 0.502 (0.001) a) |
| SitesBase | 32.2 | 60.0 | 0.598 (0.014) |
Entries with double underline in Tables 1A and 1B are used as queries and evaluation values, AUC, Top1, and Top3, are computed by examining if entries with single or double underline are retrieved or not.
The AUC values are computed by appending missing pockets in the retrieved pocket list in random order to the end of the list. The AUC values are computed 10 times and the average value is shown with the standard deviation shown in the parenthesis.
Performance with ligand-free pockets
Shape of a ligand binding pocket will be slightly changed upon binding of the ligand molecule. To investigate how well our pocket models perform for ligand-free pockets, here we searched the Huang set (Table 1B) with ligand-free pocket shapes. The Huang set is very suitable for this type of testing since it orginates from a list of pairs of ligand-bound and ligand-free form of binding sites 46. Ligand binding amino acid residues in the ligand-free proteins are identified by the sequence and structural alignment between the ligand-bound proteins. The RMSD value of ligand-bound and ligand-free proteins ranges from 0.19Å to 2.48Å with an average value of 0.86Å. This is consistent with a through survey by Brylinski and Skolnick 85, which reports that the average RMSD of ligand-bound and ligand-free form is 0.74Å.
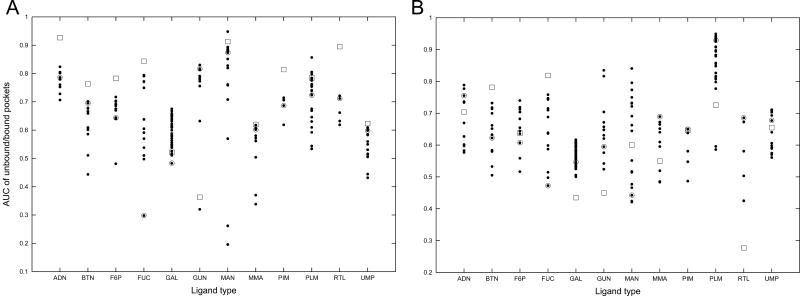
Figure 9 shows the AUC values of the ligand-bound pockets (filled dots) and ligand-free pockets (open squares) of the twelve ligand types using the shape desctriptor (G in Tables 4 and 5). Figure 9A is the results by the p-Z moments and 9B is those by the 3DZD. It is shown that the AUC values of ligand-free pockets are not particularly worse as compared with ligand-bound pockets. This may be because the shape of most of the ligand-free pockets do not differ too much as compared with the variation of shapes of the ligand-bound pockets (the largest average RMSD over all the pairs of ligand-binding pockets within the same ligand type is 3.19Å, which is in the case of RTL). In many cases, the AUC value of the ligand-free pockets is similar to the value of the closest ligand-bound pockets (shown in open circles).
Figure 9.
The AUC values by querying with ligand-bound and ligand-free form of pockets. The Huang dataset is used. Filled circles, ligand-bound pockets; open squares, ligand-free pockets; open circles, ligand-bound pocket which has the smallest RMSD to the ligand-free pocket of the ligand type. A, the pseudo-Zernike moments; B, the 3DZD.
Perfomance with predicted pockets
In the actual situation of binding ligand prediction, binding pockets may not be known beforehand and hence binding pockets need to be predicted. To simulate the actual scenario, we examined the retrieval accuracy by using a predicted pocket in a query protein. A pocket in a query protein is predicted by LIGSITE43, which identifies pocket regions solely by geometrical information. Table 8 shows the retrieval accuracy by the p-Z moments and the 3DZD on the Kahraman dataset. Compared to Tables 4A and 4B, the TOP3 value dropped to almost half both for the p-Z moments and the 3DZD. The AUC value also show a large decrease from 0.79 to 0.52 for the p-Z moments and 0.81 to 0.53 for the 3DZD. We have also constructed a database of predicted pockets by LIGSITE and queried against it by the predicted pockets, but this procedure did not improve the results (data not shown). The retrieval performance with predicted binding pockets largely relies on the accuracy of binding pocket predictions. We can expect that the results will improve by employing recent more accurate binding pocket prediction methods45;86, which combine geometrical information with sequence information.
Table 8.
Retrieval accuracy with predicted pocket regions.
| Pocket model | AUC | Top1 (%) | Top3 (%) |
|---|---|---|---|
| Pseudo-Zernike | 0.52 | 13.6 | 38.9 |
| 3D Zernike | 0.53 | 16.8 | 41.0 |
Benchmarked on the Kahraman dataset. The pocket region in a query protein is extracted (predicted) by the LIGSITE program. The shape descriptor (G) is used. The average AUC, Top1, Top3 values across different ligands are shown.
Implementation and computational speed of the algorithm
The programs for computing the pocket models and those for performing the pocket comparison were written in C. We implemented the program, named Pocket-Surfer, on a web page, http://kiharalab.org/pocket-surfer/. In the demo version, searches within the benchmark dataset by using the p-Z moments can be performed by specifying a PDB ID in Table 1 as a query, so that readers can experience how the method works and reproduce the results reported in this paper. In addition, we have also implemented an alpha version, from which a user-specified PDB entry can be used as a query. In this version, a pocket is detected by LIGSITE43 in a query protein and the detected pocket is scanned against the benchmark dataset of the 100 pockets. In both versions, pockets are compared in terms of shape and size.
The running speed of each subtask of the programs is shown in Table 9. These numbers are the average of five executions on a Linux computer with a Pentium 4 3.0 GHz processor. Here, the largest pocket on the surface of a protein, 1h2h, is extracted by LIGSITE43. Next, the p-Z moments and the 3DZD are computed for the pocket, which is then compared with the 100 pockets in the database used in this study and finally the 100 pockets are sorted by the distance to the query pocket of 1h2h to make binding ligand prediction for the query. It should be noted that the database search can be performed very rapidly with our method once the descriptor of the query pocket is computed. Searching against the database of the 100 pockets only took 0.0125 seconds for the p-Z moments and 0.023 seconds for the 3DZD. We also measured the search speed for a database of 200 pockets (by doubling the number of pockets in the dataset), which resulted in 0.014 and 0.034 seconds, respectively for the p-Z moments and the 3DZD. Extrapolating these two measured search speeds, a search against a larger database of 62200 pockets, which is the number of entries in the whole PDB database as of the writing of the paper, would take only 0.94 and 6.85 seconds by the p-Z moments and the 3DZD, respectively. Recall that pocket descriptors to be stored in the database can be pre-computed just once, as our pocket comparison method does not need pose normalization of pockets for comparison. Note that this speed is much faster than eF-Seek where a search on the website takes for at least a couple of hours and often a couple of days.
Table 9.
Running speed of the programs.
| Phase | Task | Duration (seconds) |
|---|---|---|
| Preparation | Extracting a pocket (by LIGSITE) | 4.3 |
| p-Z moments: Projecting the pocket with ray-tracing | 9.1 | |
| 3DZD: surface voxelization | 24.0 | |
| p-Z moments: Computing the pseudo-Zernike moments | 1.1 | |
| 3DZD: Computing 3DZD | 16 | |
| Database Search | Computing the distance against pockets in the database p-Z moments: | 0.0075 |
| 3DZD: | 0.018 | |
| Sorting and scoring p-Z moments: | 0.005 | |
| 3DZD: | 0.005 | |
The speed of each task is the average of 5 executions on a Linux machine with a Pentium 4, 3.0GHz processor. The protein PDB entry 1h2h is scanned by LIGSITE to identify its largest pocket. Then the p-Z moments and the 3DZD for the largest pocket is computed. The database scanned is the same as used in the benchmark of this study (i.e. the 100 pockets).
Discussion
In this paper, we have compared two methods for describing ligand binding pockets, one that uses the two dimensional p-Z moments and another one using the 3DZD. Our pocket representations are quite versatile in the sense that many different properties of pockets, not only the geometrical shape but also various physicochemical properties can be naturally represented and combined. The 2D pocket model with the p-Z moments and the 3D model with the 3DZD successfully retrieve pockets with the same binding ligand molecule in 76.3% and 82.7% of the cases within the top 3 closest hits (Tables 4A & 4B). The performance of our methods are favorably compared with similar methods including the spherical harmonics56-based method, eF-Seek, and SitesBase in terms of the pocket retrieval success rate.
A significant advantage of our method is its very fast computational speed for a database search. Using Pocket-Surfer, searching a binding pocket within the whole PDB database could be performed on a desktop computer in 1 second using the 2D pocket model and about 7 seconds by the 3D pocket model. Note that the aforementioned spherical harmonics-based method needs pose normalization of pockets as a preprocessing step of shape comparison, since the spherical harmonics vary when an object is placed in different orientations. Pose normalization does not only add more computational cost but could also errors in comparison of protein shapes, since they are almost globular and determining the principle axes may not be robust.
As discussed in Introduction, local structure-based function prediction procedure includes two steps, detection of a potential ligand binding pocket in a query protein followed by matching the query pocket against a database of known binding pockets. This study is focusing on the second step of database searching. We showed that the two pocket models we introduced, one using the p-Z moments and another one using the 3DZD, perform reasonably well and preferably compared with the other existing methods. However, the experiment with predicted pockets by LIGSITE (Table 8) indicates that performance also depends largely on the accuracy of binding pocket detection step. Therefore, establishing a well coordinated procedure of detecting (predicting) and searching binding pockets is left as an important future direction of this work. Another key development for the improment is to investigate combinations of other features of pockets into the descriptor, such as hydrophobicity, flexibility or the degree of residue conservation. An intrinsic weakness of any shape-based method is that it is sensitive to the change of pocket shape due to any reasons including flexible nature of pockets, prediction, and binding of water molecules. Thus combining other features is aimed to compensate the limitation of shape information.
The urgency and the importance of computational characterization of protein structures has become clear as an increasing number of solved protein structures have been remaining of unknown function. However, computational protein structure analysis significantly lags behind sequence analysis7 in a practical sense, since almost no methods for global/local structure comparison have been developed until recently that concern conveniently fast running speed for handling large scale data. In contrast, real-time sequence database search has been realized more than a decade ago by BLAST8 and FASTA10, and most of existing sequence analysis methods7 for homology, domain12;87, and motif searches14;88, can be performed in a real-time manner. As a solution for fast protein structure database search, we have recently proposed to represent proteins with their surface shape73;89 and employed the 3D Zernike descriptors that achieve real-time global protein structure database search70;71;75. Along the same line, here we developed a method for real-time protein local pocket shape search. We believe that the fast protein local shape comparison method together with the global shape comparison methods have paved the way for further developments of fast and convenient protein structure analysis methods.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01GM075004, U24 GM077905) and National Science Foundation (DMS0604776, DMS0800568, EF0850009, IIS0915801). The authors are grateful to Gregg Thomas for proof reading the manuscript.
Reference List
- 1.Hawkins T, Kihara D. Function prediction of uncharacterized proteins. J Bioinform Comput Biol. 2007;5:1–30. doi: 10.1142/s0219720007002503. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins T, Chitale M, Kihara D. New paradigm in protein function prediction for large scale omics analysis. Mol Biosyst. 2008;4:223–231. doi: 10.1039/b718229e. [DOI] [PubMed] [Google Scholar]
- 3.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Valencia A. Automatic annotation of protein function. Curr Opin Struct Biol. 2005;15:267–274. doi: 10.1016/j.sbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitale M, Hawkins T, Kihara D. Automated prediction of protein function from sequence. In: Bujnicki J, editor. Prediction of Protein Strucutre, Functions, and Interactions. John Wiley & Sons Ltd; 2009. pp. 63–86. [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, Copley R, Courcelle E, Das U, Durbin R, Fleischmann W, Gough J, Haft D, Harte N, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McDowall J, Mitchell A, Nikolskaya AN, Orchard S, Pagni M, Ponting CP, Quevillon E, Selengut J, Sigrist CJ, Silventoinen V, Studholme DJ, Vaughan R, Wu CH. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics. 2008;Chapter 2:Unit. doi: 10.1002/0471250953.bi0205s23. [DOI] [PubMed] [Google Scholar]
- 13.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulo N, Bairoch A, Bulliard V, Cerutti L, De CE, Langendijk-Genevaux PS, Pagni M, Sigrist CJ The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins T, Luban S, Kihara D. Enhanced automated function prediction using distantly related sequences and contextual association by PFP. Protein Sci. 2006;15:1550–1556. doi: 10.1110/ps.062153506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins T, Chitale M, Luban S, Kihara D. PFP: Automated prediction of gene ontology functional annotations with confidence scores using protein sequence data. Proteins. 2009;74:566–582. doi: 10.1002/prot.22172. [DOI] [PubMed] [Google Scholar]
- 17.Vinayagam A, del VC, Schubert F, Eils R, Glatting KH, Suhai S, Konig R. GOPET: a tool for automated predictions of Gene Ontology terms. BMC Bioinformatics. 2006;7:161. doi: 10.1186/1471-2105-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wass MN, Sternberg MJ. ConFunc--functional annotation in the twilight zone. Bioinformatics. 2008;24:798–806. doi: 10.1093/bioinformatics/btn037. [DOI] [PubMed] [Google Scholar]
- 19.Chitale M, Hawkins T, Park C, Kihara D. ESG: Extended similarity group method for automated protein function prediction. Bioinformatics. 2009;25:1739–1745. doi: 10.1093/bioinformatics/btp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua HN, Sung WK, Wong L. Using indirect protein interactions for the prediction of Gene Ontology functions. BMC Bioinformatics. 2007;8(Suppl 4):S8. doi: 10.1186/1471-2105-8-S4-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troyanskaya OG. Integrated analysis of microarray results. Methods Mol Biol. 2007;382:429–437. doi: 10.1007/978-1-59745-304-2_27. [DOI] [PubMed] [Google Scholar]
- 23.Si L, Yu D, Kihara D, Yi F. Combining sequence similarity scores and textual information for gene function annotation in the literature. Information Retrieval. 2008;11:389–404. [Google Scholar]
- 24.Rzhetsky A, Seringhaus M, Gerstein M. Seeking a new biology through text mining. Cell. 2008;134:9–13. doi: 10.1016/j.cell.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci U S A. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao XM, Chen L, Aihara K. Protein function prediction with high-throughput data. Amino Acids. 2008;35:517–530. doi: 10.1007/s00726-008-0077-y. [DOI] [PubMed] [Google Scholar]
- 27.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 28.Saqi MA, Wild DL. Expectations from structural genomics revisited: an analysis of structural genomics targets. Am J Pharmacogenomics. 2005;5:339–342. doi: 10.2165/00129785-200505050-00006. [DOI] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 31.Kihara D, Skolnick J. Microbial Genomes have over 72% structure assignment by the threading algorithm PROSPECTOR_Q. Proteins. 2004;55:464–473. doi: 10.1002/prot.20044. [DOI] [PubMed] [Google Scholar]
- 32.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure (Camb) 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orengo CA, Jones DT, Thornton JM. Protein superfamilies and domain superfolds. Nature. 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 35.Ausiello G, Peluso D, Via A, Helmer-Citterich M. Local comparison of protein structures highlights cases of convergent evolution in analogous functional sites. BMC Bioinformatics. 2007;8(Suppl 1):S24. doi: 10.1186/1471-2105-8-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Turuvekere S, Agrawal M, La D, Ramani K, Kihara D. Characterization of local geometry of protein surfaces with the visibility criterion. Proteins. 2007;71:670–683. doi: 10.1002/prot.21732. [DOI] [PubMed] [Google Scholar]
- 37.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–328. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 40.Levitt DG, Banaszak LJ. POCKET: a computer graphics method for identifying and displaying protein cavities and their surrounding amino acids. J Mol Graph. 1992;10:229–234. doi: 10.1016/0263-7855(92)80074-n. [DOI] [PubMed] [Google Scholar]
- 41.Kawabata T, Go N. Detection of pockets on protein surfaces using small and large probe spheres to find putative ligand binding sites. Proteins. 2007;68:516–529. doi: 10.1002/prot.21283. [DOI] [PubMed] [Google Scholar]
- 42.Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. 2007;1:7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–63. 389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 44.Kalidas Y, Chandra N. PocketDepth: a new depth based algorithm for identification of ligand binding sites in proteins. J Struct Biol. 2008;161:31–42. doi: 10.1016/j.jsb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Tseng YY, Dundas J, Liang J. Predicting Protein Function and Binding Profile via Matching of Local Evolutionary and Geometric Surface Patterns. J Mol Biol. 2009;387:451–464. doi: 10.1016/j.jmb.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ota M, Kinoshita K, Nishikawa K. Prediction of catalytic residues in enzymes based on known tertiary structure, stability profile, and sequence conservation. J Mol Biol. 2003;327:1053–1064. doi: 10.1016/s0022-2836(03)00207-9. [DOI] [PubMed] [Google Scholar]
- 48.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 49.Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–896. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 50.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arakaki AK, Zhang Y, Skolnick J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- 53.Ferre F, Ausiello G, Zanzoni A, Helmer-Citterich M. SURFACE: a database of protein surface regions for functional annotation. Nucleic Acids Res. 2004;32:D240–D244. doi: 10.1093/nar/gkh054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gold ND, Jackson RM. Fold independent structural comparisons of protein-ligand binding sites for exploring functional relationships. J Mol Biol. 2006;355:1112–1124. doi: 10.1016/j.jmb.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 55.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahraman A, Morris RJ, Laskowski RA, Thornton JM. Shape variation in protein binding pockets and their ligands. J Mol Biol. 2007;368:283–301. doi: 10.1016/j.jmb.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 57.Morris RJ, Najmanovich RJ, Kahraman A, Thornton JM. Real spherical harmonic expansion coefficients as 3D shape descriptors for protein binding pocket and ligand comparisons. Bioinformatics. 2005;21:2347–2355. doi: 10.1093/bioinformatics/bti337. [DOI] [PubMed] [Google Scholar]
- 58.Bock ME, Garutti C, Guerra C. Cavity detection and matching for binding site recognition. Theor Comput Sci. 2008;408:151–162. [Google Scholar]
- 59.Novotni M, Klein R. 3D Zernike descriptors for content based shape retrieval. ACM Symposium on Solid and Physical Modeling, Proceedings of the eighth ACM symposium on Solid modeling and applications; 2003. pp. 216–225. [Google Scholar]
- 60.Canterakis N. 3D Zernike moments and Zernike affine invariants for 3D image analysis and recognition. Proc 11th Scandinavian Conference on Image Analysis; 1999. pp. 85–93. [Google Scholar]
- 61.Connolly ML. Shape complementarity at the hemoglobin alpha 1 beta 1 subunit interface. Biopolymers. 1986;25:1229–1247. doi: 10.1002/bip.360250705. [DOI] [PubMed] [Google Scholar]
- 62.Roth SD. Ray Casting for Modeling Solids. Computer Graphics and Image Processing. 1982;18:109–144. [Google Scholar]
- 63.Moll A, Hildebrandt A, Lenhof HP, Kohlbacher O. BALLView: a tool for research and education in molecular modeling. Bioinformatics. 2006;22:365–366. doi: 10.1093/bioinformatics/bti818. [DOI] [PubMed] [Google Scholar]
- 64.Sitkoff D, Sharp K, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 65.Bhatia AB, Wolf E. On the Circle Polynomials of Zernike and Related Orthogonal Sets. Proceedings of the Cambridge Philosophical Society. 1954;50:40–48. [Google Scholar]
- 66.Zernike F. Beungungsthoerie des Schneiden-verfahrens und seiner verbesserten Form. Physica. 1934;1:689–701. [Google Scholar]
- 67.Teh CH, Chin RT. On Image-Analysis by the Methods of Moments. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1988;10:496–513. [Google Scholar]
- 68.Zhang D, Lu G. Content-based shape retrieval using different shape descriptors: A comparative study. ICME. 2001:1139–1142. [Google Scholar]
- 69.Mehtre BM, Kankanhalli MS, Lee WF. Shape measures for content based image retrieval: A comparison. Information Processing & Management. 1997;33:319–337. [Google Scholar]
- 70.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72:1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- 71.La D, Esquivel-Rodriguez J, Venkatraman V, Li B, Sael L, Ueng S, Ahrendt S, Kihara D. 3D-SURFER: software for high-throughput protein surface comparison and analysis. Bioinformatics. 2009;25:2843–2844. doi: 10.1093/bioinformatics/btp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mak L, Grandison S, Morris RJ. An extension of spherical harmonics to region-based rotationally invariant descriptors for molecular shape description and comparison. J Mol Graph Model. 2007;26:1035–1045. doi: 10.1016/j.jmgm.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 73.Venkatraman V, Sael L, Kihara D. Potential for protein surface shape analysis using spherical harmonics and 3D Zernike descriptors. Cell Biochem Biophys. 2009;54:23–32. doi: 10.1007/s12013-009-9051-x. [DOI] [PubMed] [Google Scholar]
- 74.Sael L, Kihara D. Characterization and classification of local protein surfaces using self-organizing map. International Journal of Knowledge Discovery in Bioinformatics (IJKDB) 2010 In press. [Google Scholar]
- 75.Sael L, La D, Li B, Rustamov R, Kihara D. Rapid comparison of properties on protein surface. Proteins. 2008;73:1–10. doi: 10.1002/prot.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venkatraman V, Chakravarthy PR, Kihara D. Application of 3D Zernike descriptors to shape-based ligand similarity searching. J Cheminformatics. 2009;1:19. doi: 10.1186/1758-2946-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venkatraman V, Yang YD, Sael L, Kihara D. Protein-protein docking using region-based 3D Zernike descriptors. BMC Bioinformatics. 2009;10:407. doi: 10.1186/1471-2105-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dym H, McKean H. Fourier series and integrals. San Diego: Academic Press; 1972. [Google Scholar]
- 79.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 80.Kazhdan M, Funkhouser T, Rusinkiewicz S. Rotation invariant spherical harmonic representation of 3D shape descriptors. Proc of the 2003 Eurographics/ACM SIGGRAPH symposium on Geometry processing. 2003;43:156–164. [Google Scholar]
- 81.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited--architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kahraman A, Morris RJ, Laskowski RA, Favia AD, Thornton JM. On the diversity of physicochemical environments experienced by identical ligands in binding pockets of unrelated proteins. Proteins. 2009 doi: 10.1002/prot.22633. in press. [DOI] [PubMed] [Google Scholar]
- 84.Kinoshita K, Murakami Y, Nakamura H. eF-seek: prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007;35:W398–W402. doi: 10.1093/nar/gkm351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brylinski M, Skolnick J. What is the relationship between the global structures of apo and holo proteins? Proteins. 2008;70:363–377. doi: 10.1002/prot.21510. [DOI] [PubMed] [Google Scholar]
- 86.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 89.Sael L, Kihara D. Protein surface representation and comparison: New approaches in structural proteomics. In: Chen J, Lonardi S, editors. Biological Data Mining. Boca Raton, Florida, USA: Chapman & Hall/CRC Press; 2009. pp. 89–109. [Google Scholar]