Abstract
BACKGROUND:
In March 2009, global surveillance started detecting cases of influenza-like illness in Mexico. By mid-April 2009, two pediatric patients were identified in the United States who were confirmed to be infected by a novel influenza A (H1N1) strain. The present article describes the first identified severe respiratory infection and the first death associated with pandemic H1N1 (pH1N1) in Canada.
METHODS:
Enhanced public health and laboratory surveillance for pH1N1 was implemented throughout Alberta on April 24, 2009. Respiratory specimens from all patients with a respiratory illness and travel history or those presenting with a severe respiratory infection requiring hospitalization underwent screening for respiratory viruses using molecular methods. For the first severe case identified and the first death due to pH1N1, histocompatibility leukocyte antigens were compared by molecular methods.
RESULTS:
The first death (a 39-year-old woman) occurred on April 28, 2009, and on May 1, 2009, a 10-year-old child presented with severe respiratory distress due to pH1N1. Both patients had no travel or contact with anyone who had travelled to Mexico; the cases were not linked. Histocompatibility antigen comparison of both patients did not identify any notable similarity. pH1N1 strains identified in Alberta did not differ from the Mexican strain.
CONCLUSION:
Rapid transmission of pH1N1 continued to occur in Alberta following the first death and the first severe respiratory infection in Canada, which were identified without any apparent connection to Mexico or the United States. Contact tracing follow-up suggested that oseltamivir may have prevented ongoing transmission of pH1N1.
Keywords: Influenza, Pandemic, pH1N1
Abstract
HISTORIQUE :
En mars 2009, le réseau de surveillance mondial a commencé à détecter des cas de maladie pseudogrippale au Mexique. En date de la mi-avril 2009, on a confirmé aux États-Unis deux cas pédiatriques d’infection par une nouvelle souche du virus de la grippe A H1N1. Le présent article décrit la première infection respiratoire grave et le premier décès associés à la pandémie de grippe A H1N1 (H1N1pdm) au Canada.
MÉTHODE :
Le 24 avril 2009, sur tout le territoire de l’Alberta, les autorités de santé publique ont instauré un système amélioré de surveillance par analyses de laboratoire pour le dépistage de la grippe H1N1pdm. On a soumis des spécimens respiratoires de tous les patients atteints de maladies respiratoires et ayant voyagé ou présentant une grave infection respiratoire nécessitant une hospitalisation à un dépistage des virus respiratoires à l’aide de techniques moléculaires. Ces mêmes techniques ont servi à comparer les systèmes HLA du premier cas grave recensé et du premier décès attribués au H1N1pdm.
RÉSULTATS :
Le premier décès (une femme de 39 ans) est survenu le 28 avril 2009, et le 1er mai 2009, un enfant de dix ans a présenté une grave maladie respiratoire causée par le virus H1N1pdm. Aucun de ces deux patients ne s’était rendu au Mexique ni n’avait eu de contact avec qui que ce soit en provenance de ce pays et les deux cas n’étaient pas reliés. La comparaison des systèmes HLA des deux patients n’a pas permis d’établir une similarité notable. Les souches H1N1pdm identifiées en Alberta n’étaient pas différentes de la souche mexicaine.
CONCLUSIONS :
La transmission rapide du H1N1pdm a continué de se produire en Alberta après le premier décès et le premier cas grave d’infection respiratoire recensés au Canada, qui se sont révélés sans lien apparent avec les cas recensés au Mexique ou aux États-Unis. La recherche des contacts donne à penser que l’oseltamivir peut avoir empêché la propagation du H1N1pdm.
In March 2009, an influenza outbreak that originated in Mexico rapidly spread to other countries including the United States (US) and Canada (1–3). A novel strain of influenza A initially termed swine-origin influenza virus and later referred to as pandemic H1N1 (pH1N1) was identified. It has since been characterized as having a unique combination of gene segments that have not been previously identified among human or swine influenza A viruses (4). The first confirmed case in the US occurred on April 15, 2009, in California; the first few identified cases in Canada (four from Nova Scotia and two from British Columbia) were reported by the Public Health Agency of Canada (PHAC) on April 26, 2009 (2,4). Within days, more cases were identified across Canada including throughout Alberta. Over the subsequent months, adults and children with critical illnesses (often young adults with severe hypoxia and prolonged ventilation) due to pH1N1 were cared for across Canada. Similar findings have been reported in the US, Australia and New Zealand (5–7). The first laboratory-confirmed case identified in Alberta was a mild influenza-like illness (ILI) that occurred on April 24, 2009, in a five-year-old boy from a small northern town who had recently returned from Mexico. Similar to the experience in the US, most Alberta cases were described as a mild self-limited ILI. The present article describes the first two Canadian pH1N1 cases that occurred in 2009, which were associated with severe respiratory infection (SRI). SRI was defined as per the PHAC definition (8). On May 1, 2009, the first SRI case was identified in a previously healthy 10-year-old girl (patient 1A). The first pH1N1-associated death was in a 39-year-old woman (patient 1B). Although she died on April 28, 2009, the cause of death was not suspected to be influenza related until her autopsy case was reviewed on May 1, 2009.
METHODS
Public health and laboratory response in Alberta
Following the identification of pH1N1 infections in Mexico and the southern US in mid-April 2009, the PHAC alerted provincial public health authorities regarding the outbreak. A province-wide epidemiological investigation tracking number was initiated on April 26, 2009, to assess all respiratory specimens collected from patients with ILI and travel history or from patients with SRIs requiring hospitalization. The most common specimen types submitted for laboratory diagnosis of a respiratory infection were nasopharyngeal (NP) swabs/aspirates and throat swabs collected in universal transport medium (Copan Diagnostics Inc, USA). In patients presenting with SRIs, a direct fluorescent antibody test screening for influenza A and B, parainfluenza and respiratory syncytial virus was immediately performed, followed by initial respiratory viral screening using the xTag Respiratory Viral Panel (RVP) on the Luminex platform (Luminex Molecular Diagnostics Inc, Canada) that assessed for respiratory syncytial virus A and B, human metapneumovirus, influenza B, parainfluenza virus types 1 to 4, adenovirus, enterovirus and rhinovirus. The xTag RVP assay also facilitates simultaneous detection and typing of influenza A subtypes H1, H3 and H5. Respiratory specimens were concurrently tested for influenza A using a real-time reverse transcriptase polymerase chain reaction (RT-PCR) assay with hydrolysis probes targeting the matrix (M) gene that was developed by the US Centers for Disease Control and Prevention (CDC-M assay) (4). The RT-PCR assay was performed on the SDS 7500 system (Applied Biosystems, USA). Influenza subtype determination was performed on all specimens that tested positive for influenza A using an assay targeting the hemagglutinin (HA) gene for H1 and H3 subtypes (4). Specimens that tested positive for influenza A by either method and for which subtype determination could not be made were classified as ‘nontypeable’. Nontypeable and confirmed cases were immediately reported to the medical health officers and ordering physicians, so they could initiate follow-up and contact tracing. Nontypeable cases were confirmed or excluded as influenza A pH1N1 using gel-based PCR and HA and M gene sequencing (9,10). For patient 1B, described below, autopsy specimens were placed in formalin for six days before investigation. Total nucleic acid was extracted from formalin-fixed tissues using the QIAamp DNA Mini Kit (Qiagen Canada). Retrospective confirmation was performed using a real-time RT-PCR assay designed by the CDC targeting the HA gene (CDC-H1 swine) (4). All respiratory viral testing was centralized within the Provincial Laboratory for Public Health (Calgary, Alberta); selected pH1N1 results, including those for patients 1A and 1B, were confirmed by the National Microbiology Laboratory (Winnipeg, Manitoba). Low to medium resolution histocompatibility leukocyte antigen (HLA) class I and II typing for HLA-A, B, Cw, DRB1, DRB3, DRB4, DRB5 and DQ was performed using the One Lambda Micro SSP assay (One Lambda Inc, USA) according to the manufacturer’s instructions.
The two case investigations were submitted for review by the local health research ethics board biomedical panel and it was concluded that publication of the information did not require approval from the Human Research Ethics Board.
CASE PRESENTATIONS
Patient 1A
On May 1, 2009, a 10-year-old girl of Aboriginal descent presented to a tertiary pediatric hospital emergency department (ED) in Edmonton, Alberta, with a one-day history of fever, chills, nausea, abdominal pain, headache, myalgias, rhinorrhea, cough and weakness. She had been examined the day before at a peripheral community hospital ED with symptoms of a ‘sore stomach’, nausea, headache and chills, and was diagnosed with a viral illness and later discharged. The following day, she developed progressive shortness of breath and ‘sore bones’, prompting the visit to the tertiary pediatric ED. After a night in the ED, she was admitted to the pediatric intensive care unit (ICU) due to increased respiratory distress. She had no underlying comorbidities, no history of any vaccinations, no travel outside of Edmonton in the preceding weeks, and no known contacts with persons with ILI or contact with anyone who had travelled to Mexico or the US.
On examination in the ICU, the patient weighed 61 kg, had an axillary temperature of 39°C, a heart rate (HR) of 142 beats/min, a respiratory rate (RR) of 55 breaths/min and blood pressure (BP) of 110/35 mmHg (mean 60 mmHg). She was lethargic, but rousable and oriented. She was in severe respiratory distress with shallow breaths, was able to speak only two to three words in one breath, and had costal indrawing. Chest auscultation revealed poor air entry bilaterally, with bibasilar crepitations. The remainder of the examination was unremarkable. She had a pulse oximeter oxygen saturation of 99% using nasal mask bilevel positive airway pressures of 12 cm/7 cm H2O and a fraction of inspired oxygen of 0.60.
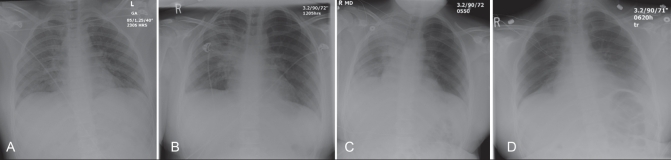
A chest roentgenogram showed bilateral lung infiltrates, with confluent opacification in the right perihilar region (Figure 1). Laboratory investigations showed an arterial partial pressure of CO2 of 33 mmHg, partial pressure of O2 of 130 mmHg (giving a partial pressure of O2 to fraction of inspired oxygen ratio of 216, compatible with acute lung injury), bicarbonate of 19.4 mmol/L, pH of 7.39 and lactate of 2.8 mmol/L. Her creatinine kinase was minimally elevated at 162 U/L (normal range 30 U/L to 160 U/L), phosphate was low at 0.49 mmol/L and platelet count was low at 112×109/L. Routine laboratory investigations included creatinine, urea, electrolytes, international normalized ratio, aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase, bilirubin, lipase, white blood cell count and differential, and hemoglobin, which were all normal. Blood cultures were negative after five days, and sputum Gram stain showed more than 25 polymorphonuclear leukocytes/low-power field, less than 10 epithelial cells/low-power field and no bacteria visualized with a culture report of mixed oropharyngeal flora.
Figure 1).
Chest roentgenograms from patient 1A showing the following: bilateral infiltrates with right lung consolidation on admission to the emergency department (A), progression of consolidation on admission to the pediatric intensive care unit (B), continued progression the day after admission to pediatric intensive care (C), and marked improvement on discharge from the pediatric intensive care unit on day 7 of hospitalization (D)
During the first hours in the ICU, the patient was treated with normal saline fluid boluses of 35 mL/kg for hypotension (mean arterial pressure below 55 mmHg), antibiotics, an intravenous phosphate infusion and noninvasive ventilation. The day after admission, zanamivir was started after receiving a laboratory report of a positive influenza A direct fluorescent antibody result from the patient’s NP swab. This result was verified by molecular assays that demonstrated a nontypeable influenza A infection. Over the first 48 h in the ICU, her respiratory distress gradually improved and she was weaned off of bilevel positive airway pressure at 56 h. Her hemodynamic status remained stable, and she developed no new organ dysfunction. Her chest roentgenogram initially worsened on day 2, followed by steady improvement after that time (Figure 1). She was discharged from the ICU and transferred to the ward on day 7 while on 6 L/min of oxygen by mask; she was on room air with an oxygen saturation of 94% on day 10 after admission. She had no respiratory distress and was discharged home the following day. She was treated with zanamivir for one day followed by oseltamivir for five days, cefotaxime and azithromycin for seven days, and vancomycin for two days.
Patient 1B
On April 26, 2009, a 39-year-old Aboriginal woman living in northwest Alberta was admitted to her local ED with a one-day history of dyspnea. She had a history of type 2 diabetes mellitus, hypertension and asthma, and was on medications for each of these conditions. Her vital signs on presentation at 9:00 were the following: temperature of 37.2°C, HR of 106 beats/min, RR of 30 breaths/min, BP of 121/73 mmHg and oxygen saturation of 96%. A chest roentgenogram was reported as unremarkable. Nebulizer therapy was started, and she was admitted to hospital. At 14:00, her vital signs were temperature of 37.4°C, HR of 81 beats/min, RR of 22 breaths/min and oxygen saturation of 94%. The following morning, her vital signs were HR of 98 beats/min, RR of 22 breaths/min, BP of 118/71 mmHg and oxygen saturation of 96%; she continued to complain of dyspnea and had wheezes on chest auscultation. She was on oxygen, but refused to wear her nasal prongs in the evening. Early on April 28, 2009, she was found to be grunting with an RR of 50 breaths/min, and an unreadable oxygen saturation, which after being placed on 8 L/min mask oxygen, rose to 94%. Oxygen flow was subsequently reduced to 4 L/min. She took her medications and ate lunch without distress. A few hours later, she was found lying in her hospital bed without any vital signs and could not be resuscitated. A limited autopsy was requested because the cause of death was deemed to be an uncertain pulmonary event. The autopsy on patient 1B performed on April 30, 2009 (there was no suspicion of influenza infection) did not reveal heavy lungs that would suggest bacterial infection or edema; no mucous plugs were noted in the airways and there was no evidence of pulmonary embolism. CDC-M RT-PCR demonstrated influenza A RNA in specimens from all formalin-fixed organs (heart, liver, trachea and lung) and this was confirmed as pH1N1 in the lung and trachea by gel-based PCR and sequencing, as well as by CDC-H1 swine (Table 1). Microscopic hematoxylin and eosin stain examination revealed diffuse moderate to severe alveolar damage and severe tracheal bronchitis without any inclusion bodies. The patient’s heart and liver tissue did not demonstrate evidence of inflammation (images not shown).
TABLE 1.
Results of molecular assays performed on tissues from patient 1B
| Tissue | CDC-M Ct | CDC-H1, H3 typing | CDC-H1 swine Ct | Identity to published pH1N1 M gene sequences |
|---|---|---|---|---|
| Lung | 33.53 | Nontypeable | 37.46 | 100% |
| Trachea | 26.19 | Nontypeable | 30.02 | 100% |
| Liver | 31.64 | Nontypeable | 36.39 | Not performed |
| Heart | 35.47 | Nontypeable | Negative | Not performed |
The crossing threshold (Ct) value indicates the number of amplification cycles required for the specimen to be reported as positive on the SDS 7500 system (Applied Biosystems, USA). Threshold refers to the point at which a reaction reaches a fluorescent intensity above background. CDC Centers for Disease Control and Prevention; H1 Hemagglutinin subtype 1; H3 Hemagglutinin subtype 3; M Matrix; pHINI Pandemic HINI
EPIDEMIOLOGY INVESTIGATION
Patient 1A
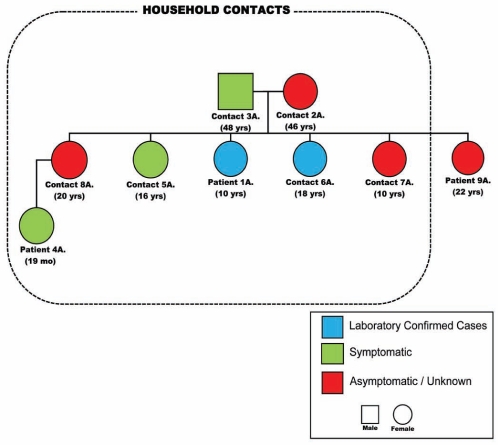
Communicable disease control nurses made contact with the family of patient 1A on May 3, 2009, after the laboratory confirmed human H1N1 (pH1N1) from her NP swab. The results of the investigation into the three adults and five children living in the household are shown in Figure 2. Patient 1A was the index case and became ill on April 29, 2009; the male adult member of the household (contact 3A) became symptomatic the next day as did the 19-month-old niece of patient 1A (patient 4A). Patient 4A presented to the ED on April 30, 2009, with fever, cough, nausea and vomiting, but was signed out that night against medical advice. After being assessed on May 2, 2009, by the department of public health, she began treatment with oseltamivir and has since recovered. On May 1, 2009, two sisters of patient 1A became symptomatic (contacts 5A and 6A) with symptoms of fever, cough and dyspnea that did not require a visit to a health care provider and that resolved within two days. Patient 6A was confirmed to be infected with pH1N1. The mother (contact 2A) and two other sisters of patient 1A remained healthy (contacts 7A [twin sister] and 8A). At the time of the investigation, it was noted that another sister of patient 1A (patient 9A), who is a brittle diabetic residing in northern Alberta, had been admitted to the ICU at a local hospital with diabetic ketoacidosis and coma but no previous history of ILI could be obtained. She and her three children had visited patient 1A on April 23 to 28, 2009. An NP swab was collected from patient 10A on May 5, 2009, which tested negative by RVP and by CDC-M assay. Inquiries at the school attended by patient 1A failed to discover unusual frequencies of ILIs or illnesses in any close contacts. Neither attendees of the school nor any of the family members had any history of travel to Mexico or contact with someone who had travelled to Mexico.
Figure 2).
Pedigree of the epidemiologically linked cases to patient 1A. Mo Months; Yrs Years
Patient 1B
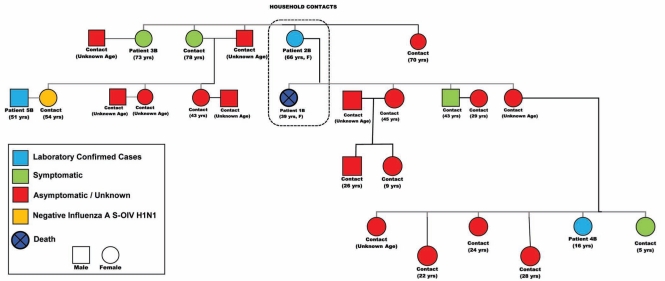
On April 30, 2009, the mother of patient 1B (patient 2B), a 66-year-old woman who was staying in the same residence as her deceased daughter, was brought to the local hospital ED by another daughter (not living with her) with complaints of dyspnea, fever and cough. She suffered from chronic obstructive pulmonary disease and hypertension. Significant findings on examination were a temperature of 39°C with a RR of 36 breaths/min, prolonged expiratory effort and wheezing. The patient remained under observation overnight, refused admission and was discharged on oseltamivir 75 mg orally twice daily; however, she returned to the ED with dyspnea both on May 2 and 3, 2009, for which she received oxygen but refused admission. An NP swab on the first visit confirmed infection with pH1N1, reported on May 4, 2009. A 73-year-old Aboriginal woman (patient 3B), also a family contact to patient 1B, required admission to hospital on May 5, 2009, with acute dyspnea that was confirmed as pneumonia by chest roentgenography. She was admitted under close observation, received antibiotic therapy and was discharged on May 9, 2009, with an uneventful recovery. Her NP swab did not reveal any respiratory viruses, and therapy did not include oseltamivir. Figure 3, an epidemiological pedigree centred around patient 1B, demonstrates the multiple contacts. Many of these nonhousehold family and community contacts occurred at patient 1B’s funeral. Symptomatic patients were offered oseltamivir, while asymptomatic patients were instructed to seek medical attention if respiratory symptoms developed. Two patients who developed symptoms (patient 4B and 5B) were confirmed to be pH1N1 infected. No pregnant contacts were identified in either of these investigations.
Figure 3).
Pedigree of the epidemiologically linked cases to patient 1B. F Female; S-OIV Swine-origin influenza virus; Yrs Years
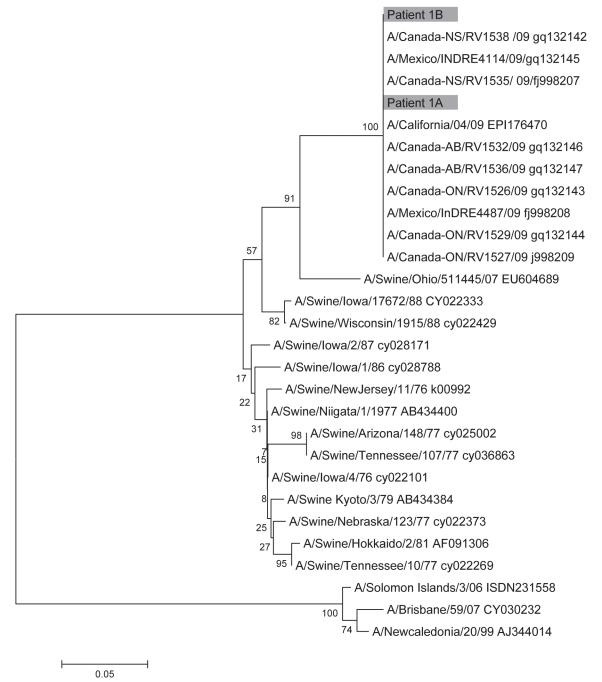
Sequencing of the pH1N1 HA and M genes from patient 1A and 1B showed that they were closely related to other known pH1N1 strains. Phylogenetic analysis showed that the HA genes from patient 1A and 1B clustered with other Canadian, American and Mexican pH1N1 strains and belonged to the North American swine lineage (Figure 4). Similarly, the M genes from patients 1A and 1B clustered with other pH1N1 strains and belonged to the Eurasian swine lineage (data not shown). Review of the HLA typing on each patient did not reveal any shared antigens (data not shown).
Figure 4).
Phylogenic analysis of the hemagluttinin gene. Nucleotide sequences were determined for patient 1A (nucleotides 100 to 590) and patient 1B (nucleotides 359 to 590). The corresponding gene sequences from previously reported swine-origin influenza virus isolates were also analyzed. Phylogenetic analysis was performed using the neighbour-joining method of the mega program
DISCUSSION
There have been thousands of laboratory-confirmed cases of human infection with the novel pH1N1 virus identified in Alberta. We report the first pH1N1-associated death (patient 1B) in Canada – a 39-year-old woman of Aboriginal descent living in a rural community, and the first confirmed SRI in a 10-year-old girl of Aboriginal descent (patient 1A) living and attending school in a city. The first US cases reported were a 22-month-old Mexican child suffering from myasthenia gravis, a ventriculoseptal defect, swallowing dysfunction and chronic hypoxia, and a 33-year-old woman with asthma, rheumatoid arthritis and psoriasis who was 35 weeks’ pregnant (4). Our patient 1B who died, certainly had chronic but stable medical conditions that had a very acute decline possibly due to a sudden onset respiratory failure or infection of the heart that may have triggered an arrythmia. Influenza A RNA was detected in the lungs, trachea, heart and liver of patient 1B, suggesting that pH1N1 may cause disseminated infection, although inflammatory cells were not observed in the heart and liver. Disseminated influenza A has been observed previously (11,12). Bacterial cultures and autopsy findings excluded a bacterial superinfection, and the RVP Luminex assay excluded coinfection with other viruses. Noteworthy, there were two patients with enterovirus and pH1N1 coinfection not linked in any way to patients 1A or 1B or to one another, and each recovered uneventfully. Many close contacts around patient 1B were promptly treated with oseltamivir after being identified, including patient 2B. Patient 2B was older and had chronic health conditions similar to patient 1B; oseltamivir may have prevented an adverse outcome. An alternative explanation is that, because of her older age (66 years) similar to patient 3B (73 years), she may have possessed some immunity from remote influenza infections. Some evidence exists to suggest that older individuals previously infected with influenza strains may possess protective antibodies (13). Patient 1A was a healthy child with multiple contacts who had ILI and a sister who was also admitted to the ICU for diabetic ketoacidosis for which a trigger was not identified.
Our observation at the outset raised the question of whether there may have been a disproportionate number of individuals with SRI or who died in Mexico who were Aboriginal North Americans, perhaps suggesting a genetic predisposition to be considered for planning public health responses. One-quarter of patients admitted to the ICU for critical illness in Canada and New Zealand were Aboriginal (5,6). Genetic predisposition for severe illness has been suggested in the setting of other respiratory viral infections. One study (14) demonstrated an association with certain HLA types and SRI from severe acute respiratory syndrome coronavirus in Hong Kong and Taiwan. However, a recent study (15) from Guangdong province in China, which examined HLA antigen frequencies in 95 individuals who recovered from severe acute respiratory syndrome, failed to show an association between HLA antigens and disease susceptibility. Of note, samples from individuals who were deceased were not available; consequently, it could not be determined whether there are HLA alleles associated with more severe disease. It would be of interest to assess the background immunogenomics to help predict which subpopulations may be at an increased risk of severe disease from pH1N1 (16). In patients 1A and 1B, the HLA was not concordant but may provide a reference point for future comparison if required.
The possibility of a mutation rendering this virus more virulent may explain the occurrence of SRI and death. Sequence analysis of the strains from both patient 1A and patient 1B were identical to the Mexican strain (Figure 4). A limitation to the study is that specimens from both patient 1A and 1B could not undergo complete viral genome sequencing, because no live virus could be cultured.
Pandemics have several characteristics including multiple waves and a shift to a higher death rate among younger populations (17). In addition to our two cases, severe pH1N1 infection was also observed in a younger population in Mexico, the US and other parts of the world (6,7,18). Furthermore, our epidemiological investigation, along with the wide global spread, confirms the extensive and efficient person-to-person transmission of influenza pH1N1. Noteworthy, although pH1N1 is sensitive to oseltamivir and resistant to adamantanes, recently widespread circulation of oseltamivir-resistant, adamantanesensitive human H1N1 has been observed (19,20). With concomitant circulation of these strains, including in Alberta (19), gene reassortment may result in a new strain resistant to all anti-influenza medications, eliminating a cornerstone in the current strategy for pandemic control and containment. Resistance has been reported in children receiving oseltamivir therapy for H1N1 infection, without evidence of reassortment (21). Irrespective, if a death occurs in a patient with confirmed pH1N1 who was treated with oseltamivir, assessment of resistance would be warranted.
Acknowledgments
The authors thank all public health, laboratory and hospital staff throughout Canada for their ongoing efforts in caring for patients in this epidemic investigation.
Footnotes
CONFLICTS OF INTEREST: No potential conflicts of interest relevant to the present article were reported by any of the authors.
REFERENCES
- 1.Centers for Disease Control and Prevention Swine influenza A (H1N1) infection in two children – southern California. Mar–Apr. 2009. < http://www.cdc.gov/mmwr/preview/mmwrhtml/mm58d0421a1.htm?s_cid=mm58d0421a1_e> (Accessed on June 29, 2010).
- 2.Public Health Agency of Canada < http://www.phac-aspc.gc.ca/alert-alerte/h1n1/surveillance-archive/20090426-eng.php> (Accessed on June 29, 2010).
- 3.World Health Organization < http://www.euroflu.org/cgi-files/bulletin_v2.cgi> (Accessed on June 29, 2010).
- 4.Dawood FS, Jain S, Finelli L, et al. Emergence of novel swine origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Zarychanski R, Pinto R, et al. Critically ill patients with 2009 influenza A (H1N1) infection in Canada. JAMA. 2009;302:1872–9. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 6.Jain S, Kamimoto L, Bramley A, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April – June 2009. N Engl J Med. 2009;361:1935–44. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- 7.Steven A, Pettila V, Seppelt I, et al. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361:1925–34. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 8.Health Canada Early detection of severe or emerging respiratory infections through severe respiratory illness (SRI) surveillance. < http://www.phac-aspc.gc.ca/sars-sras/pdf/hc-ri-enhanced-surveillance-pop_e.pdf> (Accessed on June 29, 2010).
- 9.Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeBlanc J, Li Y, Bastien N, Forward K, Davidson R, Hatchette T. Switching gears for a pandemic: Validation of a duplex RT-PCR for simultaneous detection and confirmation of pandemic influenza A (H1N1) J Clin Microbiol. 2009;47:3805–13. doi: 10.1128/JCM.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Zhang J, Huang K, et al. Systemic infection of avian influenza A virus H5N1 subtype in humans. Hum Pathol. 2009;40:735–9. doi: 10.1016/j.humpath.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galrinho A, Tavares J, Caria R, Veiga C. Myocarditis due to influenza virus complicated by intravascular coagulopathy. Rev Port Cardiol. 2000;19:835–8. [PubMed] [Google Scholar]
- 13.Ikonen N, Strengell M, Kinnunen L, et al. High frequency of cross-reacting antibodies against 2009 pandemic influenza A (H1N1) virus among the elderly in Finland. Euro Surveill. 2010;15:1–8. [PubMed] [Google Scholar]
- 14.Chiu YT. Taiwanese scientists find genetic link to SARS. Nature Med. 2003;9:1335. doi: 10.1038/nm1103-1335b. [DOI] [PubMed] [Google Scholar]
- 15.Xiong P, Zeng X, Song MS, et al. Lack of association between HLA-A, -B and -DRB1 alleles and the development of SARS: A cohort of 95 SARS-recovered individuals in a population of Guangdong, southern China. Int J Immunogenet. 2008;35:69–74. doi: 10.1111/j.1744-313X.2007.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci USA. 2009;106:8611–6. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller MA, Viboud C, Balinska M, Simonsen L. The signature features of influenza pandemics – implications for policy. N Engl J Med. 2009;360:2595–8. doi: 10.1056/NEJMp0903906. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention . Update: Novel influenza A (H1N1) virus infections – Worldwide. May 6, 2009. < www.cdc.gov/mmwr/preview/mmwrhtml/mm5817a1.htm> (Accessed on June 29, 2010). [Google Scholar]
- 19.Pabbaraju K, Ho KC, Wong S, et al. Adamantane resistance in circulating human influenza A viruses from Alberta, Canada (1970–2007) Antiviral Res. 2008;79:81–6. doi: 10.1016/j.antiviral.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 2008;4:e100–3. doi: 10.1371/journal.ppat.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephenson I, Democratis J, Lackenby A, et al. Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin Infect Dis. 2009;48:389–96. doi: 10.1086/596311. [DOI] [PubMed] [Google Scholar]