Summary
Voltage-gated ion channels sense transmembrane voltage changes via a paddle-shaped motif that includes the C-terminal part of the third transmembrane segment (S3b) and the N-terminal part of the fourth segment (NTS4) that harbors positively charged, voltage-sensing residues. Here we find that residue triplets in S3b and NTS4 can be deleted individually, or even in some combinations, without compromising the channels’ basic voltage-gating capability. Thus, a high degree of complementarity between these S3b and NTS4 regions is not required for basic voltage gating per se. Remarkably, the voltage-gated Shaker K+ channel remains voltage gated after a 43-residue paddle sequence is replaced by a glycine triplet. Therefore, the paddle motif comprises a minimal core that suffices to confer voltage gating in the physiological voltage range, and a much larger, modulatory part. While arginines in NTS4 sense voltage, our study shows that the interposed hydrophobic residues help set the sensor’s characteristic chemical equilibrium between activated and deactivated states.
Introduction
Coordinated activity of voltage-gated ion channels generates action potentials, the electric signals used by nerve, muscle and endocrine cells. A fundamental question in the field has been how these electric signals are detected by the channel protein and how the resulting conformational changes are coupled to channel opening and closing. In the case of voltage-gated K+ (Kv) channels they are comprised of an ion conduction module surrounded by four voltage-sensing modules (Kubo et al., 1993; Lu et al., 2001; Jiang et al., 2003). Positively charged residues of the channel protein’s fourth transmembrane segment (S4) function as the main voltage-sensing residues, e.g., the four arginines (R1–R4) in the N-terminal part of S4 (NTS4) of the Shaker Kv channel (Noda et al., 1984; Catterall, 1988; Stühmer et al., 1989; Liman et al., 1991; Lopez et al., 1991; Papazian et al., 1991; Yang and Horn, 1995; Aggarwal and MacKinnon, 1996; Larsson et al., 1996; Mannuzzu et al., 1996; Seoh et al., 1996; Yang et al., 1996). Movement of these voltage-sensing residues results in transfer of as many as >12 elementary charges (or equivalent) across the transmembrane electric field (Schoppa et al., 1992; Aggarwal and MacKinnon, 1996; Seoh et al., 1996; Islas and Sigworth, 1999). Positively charged residues usually occupy every third position within S4, and are stabilized in the membrane plane by negatively charged protein residues or the phospho-head group of membrane phospholipids (Armstrong, 1981; Papazian et al., 1995; Seoh et al., 1996; Cuello et al., 2004; Freites et al., 2005; Ramu et al., 2006; Schmidt et al., 2006; Long et al., 2007; Xu et al., 2008; Milescu et al., 2009). The C-terminal part of S3 (S3b), NTS4, and their linker together form a helix-turn-helix motif termed the voltage-sensing paddle (Jiang et al., 2003; Long et al., 2007) (Fig. 1). Remarkably, the paddle from a given voltage-gated ion (or proton) channel or enzyme (Murata et al., 2005; Sasaki et al., 2006; Ramsey et al., 2006) can be transferred to another voltage-gated channel without loss of voltage-sensing function (Alabi et al., 2007; Bosmans et al., 2008).
Figure 1.
Sequence and structure of the voltage-sensing paddle. (A) Comparison of the paddle sequences of Shaker (upper) and Kv1.2-2.1 (chimeric) channels (lower) (Long et al., 2007). (B) Structure of the paddle of the Kv1.2-2.1 channel (PDB: 2R9R). The region of the paddle corresponding to the region of Shaker that we replaced by a glycine triplet in Fig. 5C is colored cyan (but recall that Shaker’s S3–S4 linker is much longer). The side chains of the residues that correspond to R1 – R4 in Shaker are shown as yellow sticks.
Membrane hyperpolarization drives NTS4 from the extracellular phase, via a short low-dielectric (hydrophobic) region, toward the intracellular phase (Yang and Horn, 1995; Aggarwal and MacKinnon, 1996; Larsson et al., 1996; Mannuzzu et al., 1996; Seoh et al., 1996; Yang et al., 1996; Starace et al., 1997; Glauner et al., 1999; Silverman et al., 2003; Ahern and Horn, 2004; Phillips et al., 2005; Ruta et al., 2005; Campos et al., 2007; Grabe et al., 2007; Pathak et al., 2007; Broomand and Elinder, 2008; Posson and Selvin, 2008; Tao et al., 2010). The accessibility pattern of different-length biotin-reagents tethered to substituted cysteines in the bacterial KvAP voltage-gated K+ channel (whose S3 – S4 linker is short) led Ruta et al. (2005) to conclude that S3b also undergoes substantial voltage-induced movement, consistent with the notion that S3b and NTS4 move together as a rigid body. On the other hand, the disulfide bond pattern of cysteine pairs substituted in S3b and NTS4 in the eukaryotic Shaker channel (whose S3 – S4 linker is long) led Broomand and Elinder (2008) to conclude that the two helices exhibit a large motion relative to each other. It is unclear whether these different conclusions reflect a difference in channel type and/or in experimental method. Additionally, it has been suggested that NTS4 alternates between different secondary structures during voltage gating (Long et al., 2007; Khalili-Araghi et al., 2010). If so, complementarity at the S3b– NTS4 interface should not be of such high degree as to prevent NTS4 from switching conformation.
Voltage-induced motion of S4, through a physical coupler, then enables the channel gate, which is formed by the C-terminal end of S6 (CTS6), to move toward and from the central axis of the ion conduction pore and thereby close or open the pore (Liu et al., 1997; Doyle et al., 1998; Jiang et al., 2002; Kitaguchi et al., 2004). Mutagenesis-based functional studies have provided the initial clue as to which regions of the channel protein couple the motion of the voltage sensor to that of the channel gate (Lu et al., 2001; Lu et al., 2002; Tristani-Firouzi et al., 2002). Proper coupling requires two complementary sequences: 1) the S4–S5 linker and 2) CTS6 plus its immediate extension (XTS6)(Lu et al., 2002). This finding led to the following proposal regarding the crux of the coupling mechanism: as membrane hyperpolarization causes S4 to descend inwardly and rotate, the S4–S5 linker “pushes” onto CTS6-XTS6 to close the gate. This proposal has received structural support from crystallographic studies on Kv1.2 (Long et al., 2005). If the above coupling concept is valid, the goal of the voltage-sensing process is mainly to move the S4–S5 linker and thereby S6. Effective S4 movement must then both achieve this mechanical goal and transfer enough voltage-sensing residues across the electric field to engender the observed physiological voltage sensitivity.
The aim of the present study is to identify the essential characteristics of the paddle motif that confer voltage-sensing capability upon the Shaker channel.
Results
Deletion analysis of S3b and NTS4 segments
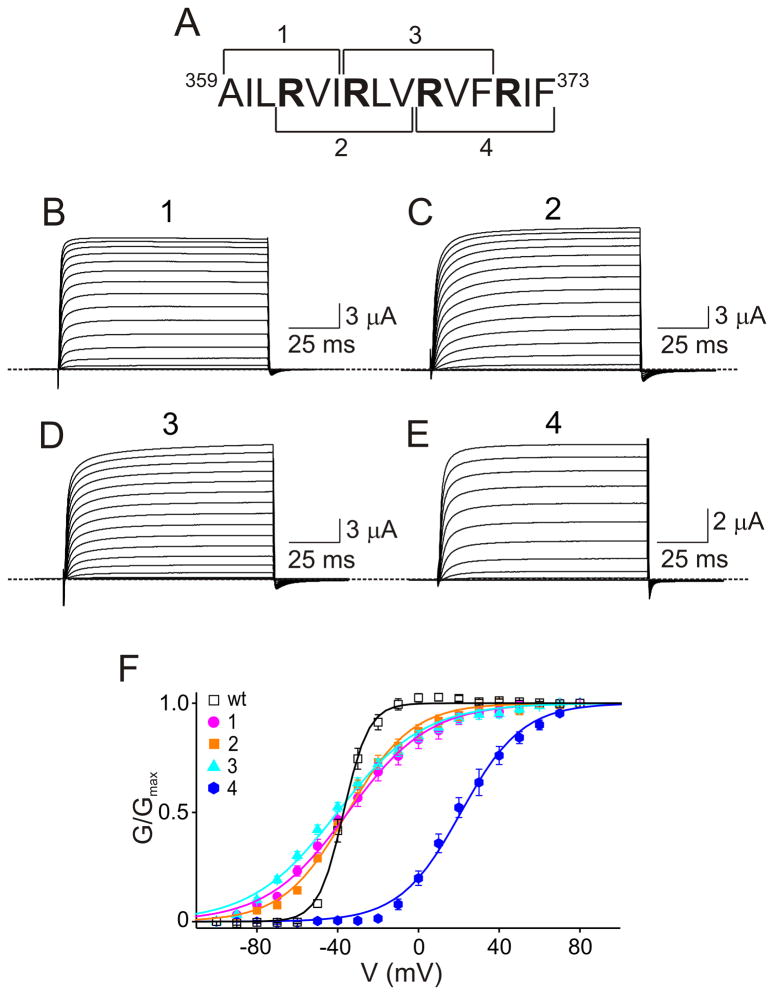
To examine the basic features of the paddle motif we performed, in the Shaker Kv channel, a systematic deletion analysis of S3b and NTS4. Remarkably, the channels remain voltage gated even after deletion of any one of the five residue triplets in the sequence encompassing S3b plus four trailing residues (Fig. 2A-G). Deleting the first triplet causes a marked rightward shift of the G-V curve whereas deleting any of the other four causes modest shifts (Fig. 2H). The channels also remain voltage gated after deletion of any of the adjacent four triplets in S4, each of which harbors one of the main voltage-sensing residues R1 – R4 (Aggarwal and MacKinnon, 1996; Seoh et al., 1996) (Fig. 3). All these results show that none of these triplets (or helical turns) in S3b and NTS4 is essential for the paddle to be a functional voltage sensor. They also imply that no high degree of complementarity between S3b and NTS4 is required for basic voltage sensing. We then systematically deleted pairs of adjoining triplets (i.e., 6 contiguous residues at a time) in NTS4 (Fig. 4A). These mutants, each missing one or two main voltage-sensing residues (R1; R1 and R2; R2 and R3; or R3 and R4 respectively), still remain voltage gated (Fig. 4B-E), albeit with reduced gain, i.e., a shallower G-V relation (Fig. 4F). The midpoint (V1/2) of the G-V curve of the mutant missing R3 and R4 is rightward shifted, a result consistent with the structural peculiarity that R3 and R4 are hydrogen bonded to certain negatively charged residues, interactions that help to stabilize the activated state (Long et al., 2007).
Figure 2.
Deletion analysis of Shaker’s S3b segment. (A) Sequence of S3b and four trailing residues. (B-G) Macroscopic currents of wild-type (B) and mutant (C-G) channels where the deleted residue triplets are indicated. The currents are elicited with the voltage protocol shown in B and with 100 mM Rb+ in the bath solution. The dotted lines indicate zero current levels. (H) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding midpoint voltages (V1/2 ; in mV) of −37 ± 0.2 (for the wild type shown in panel B), and −20 ± 0.3 (D), −26 ± 0.2 (E), −14 ± 0.3 (F), and −27 ± 0.6 (G) for the respective mutants, and apparent valences (Z) of 4.2 ± 0.3 (B), 2.8 ± 0.2 (D), 3.7 ± 0.4 (E), 2.5 ± 0.1 (F) and 2.3 ± 0.1 (G). All data shown are mean ± s.e.m. (n = 5–15). The G-V curve for the PYF deletion mutant was obtained by dividing the I-V curve by the driving force (the difference between membrane voltage and the K+ equilibrium potential).
Figure 3.
Residue triplet deletions in NTS4. (A) Sequence of NTS4. (B-E) Currents of mutant channels where the deleted residue triplets are indicated. The currents are elicited with the voltage protocol shown in Fig. 2B and with 100 mM Rb+ in the bath solution. (F) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding V1/2 values (in mV) of −2.0 ± 0.1 (B), 0 ± 0.3 (C), 15 ± 0.6 (D), and −8 ± 0.4 (E), and Z values of 1.3 ± 0.1 (B), 2.3 ± 0.2 (C), 1.7 ± 0.1 (D), and 2.2 ± 0.1 (E). The wild-type G-V curve (open symbols), taken from Fig. 2H, is re-plotted again for comparison. All data shown are as mean ± s.e.m. (n = 5–15).
Figure 4.
Hextuplet residue deletions in NTS4. (A) Sequence of NTS4 where the hextuplet residues deleted in individual constructs are indicated by brackets. (B-E) Currents of mutant channels, elicited with the protocol shown in Fig 2B, and with 100 mM Rb+ (B, C, E) or 20 mM Rb+ plus 80 mM Na+ (D) in the bath solution. (F) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding V1/2 (in mV) of −35 ±0.6 (B), −35 ± 0.2 (C), −40 ± 0.3 (D), and 21 ± 0.9 (E), and Z of 1.3 ± 0.2 (B), 1.6 ± 0.1 (C), 1.2 ± 0.1 (D), and 1.7 ± 0.1 (E). The wild-type G-V curve (open symbols), taken from Fig. 2H, is re-plotted for comparison. All data shown are mean ± s.e.m. (n = 4 – 15).
When we deleted the two triplets containing R1 and R2 in NTS4 together with (their structural neighbors) the two C-terminal triplets of S3b (i.e., a total of 12 residues; Fig. 5A), the construct remained voltage gated (Fig. 5B). Given that the S3–S4 linker in Shaker is not essential (Gonzalez et al., 2001), we substituted in the above construct a single glycine triplet for a further deletion comprising the S3 - S4 linker proper plus two S3b and four NTS4 residues (that is, in the final construct the glycine triplet bridges a 43-residue gap) (Fig. 5A). For visual reference, we render in cyan the sequence within the paddle of the Kv1.2-2.1 (chimera) structure (Long et al., 2007) that corresponds to the sequence we replaced in Shaker with a glycine triplet (Fig. 1, but recall that Shaker’s S3–S4 linker is much longer than the chimera’s). This Shaker mutant with minimal voltage sensor is still well gated by voltage (Fig. 5C). The midpoint of the G-V relation of both constructs (#1 and #2 in Fig. 5B, C) is only modestly rightward shifted but the slope (gain) is noticeably reduced (Fig. 5F). Additional deletion of R3 produces a mutant that resists closure with the strongest hyperpolarization tested (Fig. 5D), which suggests that voltage gating in the physiological range requires at least two of the four main voltage-sensing residues. Indeed, replacing R1–R3 with neutral residues (no deletion) also prevents the channels from closing under comparably strong hyperpolarization (Fig. 5E). These analyses indicate that the paddle in Shaker effectively comprises a minimal voltage-sensing core (containing 2 of the 4 main voltage-sensing residues) and a much larger, modulatory part of about 43 residues.
Figure 5.
Simultaneous deletions in S3b and NTS4. (A) Sequences of S3b through NTS4. The lines under the upper sequence indicate the pair of hextuplet residues simultaneously deleted from S3b and NTS4 (construct #1); the line under the lower sequence indicates the 43 residue sequence replaced by a glycine triplet (construct #2). (B) Currents of channel construct #1 described in panel A. (C) Currents of channel construct #2 in panel A. (D) Currents of mutant channels similar to those in panel C, except that R3 is also deleted. (E) Currents of mutant channels containing three mutations (R1M, R2Q and R3N). Currents in panels B and C were elicited with the protocol shown in Fig 2B, whereas for panels D and E, the test pulses ranged from −100 mV to 60 mV (D) or to 30 mV (E). They were recorded in the presence of 20 mM Rb+ (B, C) or 100 mM Rb+ (D, E) in the bath solution. (F) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage. The curves through the data for the mutant shown in panels B and C are Boltzmann fits, yielding V1/2 (in mV) of −23 ± 0.3 (B) and −18 ± 0.3 (C), and Z of 1.7 ± 0.1 (B) and 2.1 ± 0.1 (C). The wild-type G-V curve (open symbols), taken from Fig. 2H, is re-plotted for comparison. All data shown are mean ± s.e.m. (n = 5–30).
Role of hydrophobic residues in determining characteristics of the voltage sensor
The NTS4 segment exhibits three striking characteristics (Fig. 6A). First, it harbors four arginines that are the main voltage-sensing residues (Aggarwal and MacKinnon, 1996; Seoh et al., 1996). Second, these positively charged residues are generally located at every third position. Third, the two residues intercalated between the positively charged ones are mainly hydrophobic.
Figure 6.
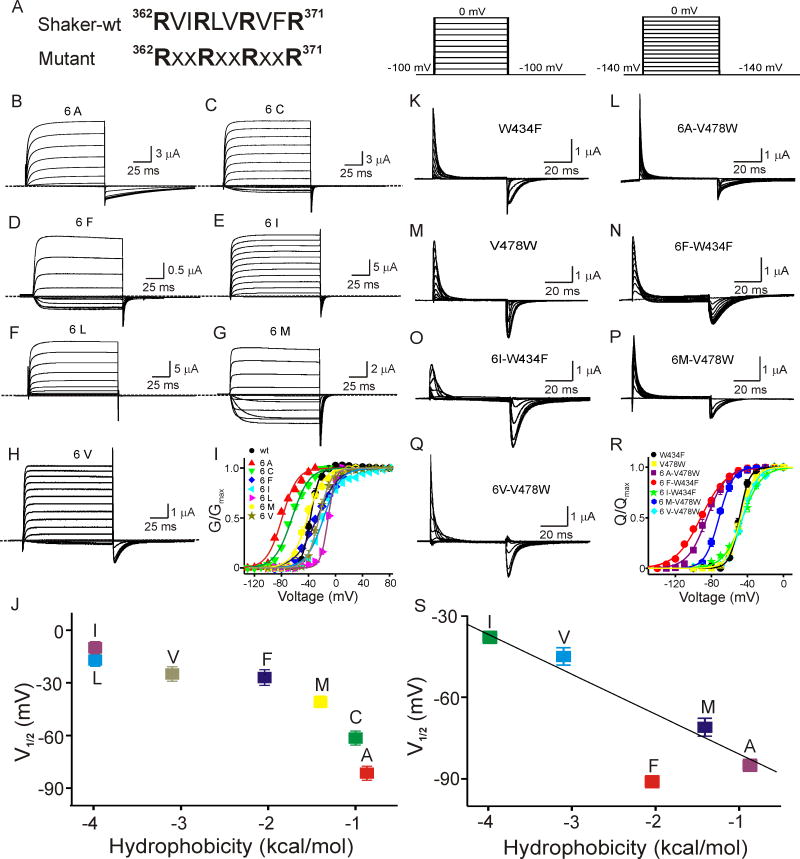
Effect of hydrophobicity of non-charged intercalated residues in NTS4 on voltage dependence of ionic and gating currents. (A) Wild-type (upper) and mutant (lower) NTS4 sequences, where all six residues indicated by X are identical in a given mutant but vary between constructs. (B-H) Currents of mutant channels, recorded with the protocol shown in Fig 2B and with 100 mM Rb+ in the bath solution, except for panels B and C where the membrane voltage was stepped from the −130 mV holding potential to 10 mV (B) or 30 mV (C) in 10-mV increments. (I) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding V1/2 (in mV) of −80 ± 0.1 (B), −64.0 ± 0.8 (C), −27 ± 0.7 (D), −17 ± 0.8 (E), −10 ± 1.0 (F), −41 ± 1.0 (G) and −25 ± 0.9 (H), and Z of 2.3 ± 0.2 (B), 2.0 ± 0.1 (C), 1.6 ± 0.1 (D), 2.1 ± 0.1 (E), 4.7 ± 0.4 (F), 2.1 ± 0.2 (G) and 3.0 ± 0.4 (H). The wild-type G-V curve (black dots), taken from Fig. 2H, is re-plotted for comparison. All data shown are mean ± s.e.m. (n = 7–15). (J) The midpoint of mutant G-V curves is plotted against hydrophobicity of residue type X. (K-Q) Gating currents of non-conducting W434F and V478W mutant channels (Perozo et al., 1993; Hackos et al., 2002), with and without additional replacement of six residues by a single type of hydrophobic residue. Current was elicited with the protocol shown above the respective column. (R) Normalized gating current (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding V1/2 (in mV) of −48 ± 0.4 (K), −47 ± 0.6 (L), −39 ± 2 (M), −45 ± 1 (N), −85 ± 1 (O), −91 ± 2 (P) and −71 ± 2 (Q), and Z of 4.7 ± 0.6 (K), 3.5 ± 0.2 (L), 4.1 ± 0.3 (M), 3.2 ± 0.4 (N), 2.9 ± 0.6 (O), 1.8 ± 0.5 (P) and 3.3 ± 0.3 (Q). All data shown are mean ± s.e.m. (n = 6–14). (S) The midpoint of each mutant Q-V curve is plotted against the hydrophobicity of the substituted residues. The line is a linear fit to the upper four data points. (see also Fig. S1)
To investigate the role of these hydrophobic residues we simultaneously replaced all six residues between the four arginines with a single residue type, one type at a time, testing all twenty naturally occurring residues (Fig. 6A). Only those constructs expressed current where the sixfold substitution was with one of seven relatively hydrophobic residue types, namely, Ala, Cys, Ile, Leu, Met, Phe, or Val (Fig. 6B-H). The G-V midpoint tends further rightwards with increasing residue hydrophobicity (Fig. 6I, J). The position of this midpoint may, in principle, be determined mainly by gating transitions involving the bulk of voltage-sensor movement, or by transitions further downstream, or both. For example, mutating three uncharged residues in the S4 region distal to R3 right-shifts the G-V curve mostly by altering a downstream transition (Smith-Maxwell et al., 1998; see also Lopez et al., 1991). To determine whether our systematic hydrophobic mutations affect the behavior of the voltage sensor proper rather than downstream transitions, we examined the mutations’ effect on the voltage dependence of gating currents, non-linear capacitive currents directly related to voltage sensor movement (Armstrong and Bezanilla, 1973). Five of the seven mutant channels express sufficient gating current to allow examination (Fig. 6K-Q). The midpoint position of the mutants’ gating charge-voltage (Q-V) relation turns increasingly positive as hydrophobicity of the intercalated residues increases (Fig. 6R, S). Except for the mutant containing the aromatic residue phenylalanine, midpoint shift and hydrophobicity (Radzicka and Wolfenden, 1988) appear to be strongly correlated (Fig. 6S). These results imply that upon membrane hyperpolarization the voltage-sensing NTS4 moves from (on average) a relatively hydrophilic environment towards a more hydrophobic one.
Periodicity of positively charged residues in S4
We also tested whether the striking periodicity of S4 — two hydrophobic residues between each pair of positively charged residues — is important for forming a functional voltage-gated channel. Among the mutants described above, the one with sixfold valine replacement not only expresses well but also exhibits a G-V (or Q-V) relation more resembling that of wild type (Fig. 6I, R). We therefore chose valine to test variable spacing between the first four arginines (R1–R4). We made Shaker mutants where the three (identical) intercalations between the first four arginines consist of a number (from zero to five) of valine residues. Of the six constructs only the one with two spacer (valine) residues between each arginine pair expressed detectable current (Fig. S1). The general alternating pattern of one positively charged residue and two hydrophobic residues is therefore important.
Functional replacement of Shaker’s voltage sensor by its counterpart from CNGA1
The above results show that no high degree of complementarity between NTS4 containing the first four arginines and S3b is required for voltage sensing, but that the alternating pattern of one positively charged and two neutral residues is important. These findings predict the possibility, in principle, of replacing the paddle (hairpin) motif of Shaker with that from a non-voltage-gated channel, provided the latter’s S4 sequence exhibits the requisite alternating sequence. In this regard, cyclic nucleotide-gated (CNG) channels are suitable donor candidates. Although they are gated by ligands not voltage, their S4 sequence has the same residue pattern as that of canonical voltage-gated channels.
A previous study has shown that EAG Kv channels remain voltage gated following replacement of their S4 segment by its counterpart from an olfactory CNG channel (Tang and Papazian, 1997). Building on this study, we used the Shaker channel and the retinal CNGA1 channel to test the above prediction. The S4 of CNGA1 contains four arginine residues that correspond to the first four arginines in Shaker (Fig. 7A). However, CNGA1’s S3b has a sequence quite unlike that of Shaker, and the length and sequence of its S3–S4 linker are also very different. Despite these disparities, the chimeric Shaker construct containing the paddle sequence from ligand-gated CNGA1 remains fully voltage gated, albeit with a markedly right-shifted G-V curve (Fig. 7B-D). Such shift is not surprising, given that the neutral residues separating the conserved arginines differ, because these intercalated residues have a very strong influence on the G-V curve’s midpoint (Fig. 6I). Regardless of the quantitative differences in voltage-gating parameters between wild-type Shaker channels and our chimeric construct, the observation that the chimeric channels are fully voltage gated fulfills our prediction. This result and those described earlier help to explain the apparent high degree of interchangeability of paddle sequences within the super-family of voltage-gated proteins (including CNG channels), on condition that the general alternating residue pattern of S4 is preserved (Alabi et al., 2007; Bosmans et al., 2008).
Figure 7.
Replacement of Shaker’s paddle sequence by that from CNGA1. (A) Sequence alignment of S3b through NTS4, for wild-type Shaker and for CNGA1. The chimeric Shaker construct (whose sequence is not shown) contained the CNGA1 sequence shown in panel A. (B and C) Currents of wild-type and chimeric Shaker channels, the latter recorded with a protocol in which the test pulses ranged from 0 mV to 160 mV in 10-mV increments and with 100 mM Rb+ in the bath solution. (D) Normalized tail currents (mean ± s.e.m.) plotted against membrane voltage, where the curves through the data are Boltzmann fits, yielding V1/2 (in mV) of −37 ± 0.2 (B) and 106 ± 0.5 (C), and Z of 4.2 ± 0.3 (B) and 2.9 ± 0.2 (C) (mean ± s.e.m.; n = 15 for B and 8 for C). The wild-type data, taken from Fig. 2, are re-plotted for comparison.
Discussion
We report a number of key findings. First, the Q-V curve undergoes a positive shift with increasing hydrophobicity of the pairs of residues that alternate with the four main voltage-sensing arginines, which suggests that the three turns of the NTS4 helix encounter (on average) a more hydrophobic environment in the deactivated state (Fig. 6). Second, as the number (0–5) of hydrophobic residues separating the arginines in NTS4 is varied, only the construct with two spacer residues between each pair of arginines expresses functional voltage-gated channels (Fig. S1). Third, residue triplets within S3b and NTS4 can be deleted individually (or even in some combinations) without compromising the channels’ basic voltage-gating capability (Figs. 2–5). It follows that no high degree of complementarity between the S3b and NTS4 regions is required for basic voltage gating. This feature is important because it allows S4 to adopt different (e.g., secondary) conformations as it moves along the gating sequence. It also follows that, at any given time during voltage gating, only a small subset of residues in the voltage-sensing part of the paddle may engage in specific and essential interactions with its surroundings. Fourth, replacement of a 43-residue paddle sequence with a glycine triplet leaves the resulting channels (that lack R1 and R2) still well gated by voltage (Fig. 5). Given that the (truncated) S3b and NTS4 sequences are now joined by a mere glycine triplet, any sizeable translational motion of S4 should result in movement of S3. Fifth, if R3 is removed as well (or if R1 – R3 in the wild-type are all replaced by neutral mutations) the channels resist closure under strongest hyperpolarization tested (Fig. 5). Finally, a 47-residue sequence in Shaker’s paddle can be functionally replaced by the corresponding sequence from the ligand-gated (not voltage-gated) CNGA1 channel (Fig.7), even though these two sequences bear little resemblance other than the periodic placement of R1–R4 in their NTS4. We next discuss these findings in the context of the Kv1.2-2.1 chimera crystal structure (Long et al., 2007).
In the Kv1.2-2.1 chimera in an activated state (Long et al., 2007), NTS4 is exposed to the extracellular phase. The residues that correspond to Shaker’s R1 and R2 are positioned to interact with phospholipid head groups and/or water molecules, whereas R3 and R4 form ionized hydrogen bonds with certain negatively charged residues (the external negative cluster). Thus, in the activated state NTS4 is generally exposed to a relatively hydrophilic environment. K5 and R6 form ionized hydrogen bonds with certain other negatively charged residues (the internal negative cluster). Intriguingly, whereas the R1–R3 region adopts an α-helical conformation, the R4–R6 region adopts a 310 helical conformation so that the side chains of K5 and R6 point in the “same” angular direction with respect to the helix axis, interacting with the internal negative cluster.
Previous functional studies have strongly suggested that hyperpolarization tilts and twists NTS4, driving it inwards (Yang and Horn, 1995; Aggarwal and MacKinnon, 1996; Larsson et al., 1996; Mannuzzu et al., 1996; Seoh et al., 1996; Yang et al., 1996; Glauner et al., 1999; Silverman et al., 2003; Phillips et al., 2005; Ruta et al., 2005; Campos et al., 2007; Grabe et al., 2007; Pathak et al., 2007; Broomand and Elinder, 2008; Posson and Selvin, 2008; Tao et al., 2010). This inference and the structure of Kv1.2-2.1 (Long et al., 2007) predict the following. First, NTS4 in the deactivated state will be located in an (on average) more hydrophobic environment (formed by protein residues and/or lipid molecules) than in the activated state. Second, two of the four voltage-sensing arginines may need to be preserved to interact effectively with the internal negative cluster, and thus help maintain the voltage sensor in a stable deactivated conformation at physiological hyperpolarization. Third, the typical residue periodicity of NTS4 is important in allowing NTS4 to change conformation (α helix to 310 helix) on its way inwards (Long et al., 2007; Khalili-Araghi et al., 2010), so two arginines can adopt the “same” angular orientation to interact most efficiently with the internal negative cluster and help stabilize the 310 helical structure. Simultaneously, the intercalated hydrophobic residues, facing the other side, interact with their (relatively hydrophobic) surroundings to further stabilize the 3 10-helical conformation. Thus, the alternating positively charged and hydrophobic residues work in concert, together creating the curious, characteristic residue periodicity of S4. Fourth, NTS4 is not expected to be constrained by surrounding structural elements to such an extent that it cannot alternate between (e.g., two helical) conformations or twist.
Our present findings corroborate these predictions. First, increasing the hydrophobicity of the neutral residues in NTS4 favors the deactivated state of the voltage sensor (Fig. 6). Second, we cannot delete or replace more than two of the four arginines without losing the ability to close the channels with physiological hyperpolarization (Fig. 5). Third, alternation of one positively charged and two hydrophobic residues in NTS4 is important (Fig. S1). Fourth, NTS4 is not greatly constrained by surrounding structural elements, as systematic deletion of residue triplets (or in some combinations) from either NTS4 or S3b does not eliminate the channel’s basic voltage-gating capability (Figs. 2–5).
Whereas positively charged arginines in NTS4 function as the main voltage-sensing residues, the interposed hydrophobic residue pairs help to keep the voltage sensor poised between activated and deactivated states. [As an empirical energetic metric, the apparent free energy associated with membrane insertion of an isolated S4 is modestly positive (Hessa et al., 2005).] Consequently, the voltage sensor can readily acquire sufficient energy from the electric field to overcome the chemical energy of the system, thereby raising the probability of its reaching the deactivated state. In general, only the more hydrophobic aliphatic residues (I, L or V) allow the Q-V and G-V curve to fall in the physiological voltage range (Fig. 6), a fact that explains why such residues are found at these positions in NTS4, or why experimental replacement with other residue types usually causes, if anything, only a leftward shift of the Q-V curve.
In summary, by substituting a glycine triplet for a 43-residue sequence in the Shaker paddle motif, we have engineered a well-gated Shaker construct containing a miniature voltage sensor in each subunit. This finding indicates that the eliminated sequence is not needed for the strict purpose of basic voltage gating. In fact, the paddle of a non-voltage-gated CNG channel can be transplanted into the Shaker channel and be functional, even though its sequence bears little resemblance to that of Shaker, except for the four arginines that sense voltage in Shaker. Thus, the paddle motif effectively consists of two parts: a minimal core essential for voltage gating per se, and a comparatively much larger, modulatory part. The core, which contains two of the four main voltage-sensing arginines, suffices to produce voltage gating in the physiological voltage range. In the construct with a miniature voltage sensor, these two arginines, besides sensing voltage, presumably also interact with the external and internal clusters of negatively charged residues (Long et al., 2007) to help stabilize the conformation of NTS4 in an activated or a deactivated state, respectively. The modulatory part confers additional important features upon the voltage sensor, namely such specific gating characteristics as midpoint voltage, gain, speed, number of voltage-sensing residues, extent of S4 movement, and number of gating states, as well as pharmacological profile (Swartz and MacKinnon, 1997; Li-Smerin and Swartz, 2000). While the hydrophobic residue pairs that separate the voltage-sensing arginines help keep the voltage sensor poised between the activated and deactivated states, hyperpolarization apparently provides the necessary additional energy for the voltage-sensing part of S4 to tilt, twist, and/or move inwardly from a (water-exposed) rather hydrophilic environment to a (protein- and lipid-lined and on average) more hydrophobic region, a motion that is then propagated through the S4–S5 linker to S6, closing the channel gate.
Experimental Procedures
The cDNA of Shaker(-IR) (Hoshi et al., 1990) was cloned in the pGEMHess vector (Liman et al., 1992). The mutant channel cDNAs were produced through PCR-based mutagenesis and confirmed with DNA sequencing. The cRNAs were synthesized with T7 polymerase using the corresponding linearized cDNAs as templates. Channel currents were recorded from oocytes (previously injected with appropriate cRNA) (Spassova and Lu, 1998) using a two-electrode voltage clamp amplifier (Warner OC-725C; Harvard Apparatus), filtered at 1 kHz and sampled at 10 kHz using an analog-to digital converter (Digidata 1322A; MDS Analytical Technologies) which was interfaced with a personal computer. pClamp8 software (MDS Analytical Technologies) was used for amplifier control and data acquisition. To elicit currents the voltage across the oocyte membrane was stepped from the −100 mV holding potential to various test voltages in 10-mV increments and back to −100 mV. The resistance of electrodes filled with 3 M KCl was ~0.2 MΩ. Unless specified otherwise, the bath solution contained (in mM): a mixture of 20 RbCl and 80 NaCl (for ionic or gating current measurements) or 100 RbCl (to further enhance and slow down the tail current), 0.3 CaCl2, 1 MgCl2 and 10 HEPES; pH was adjusted to 7.6 with RbOH. Ionic currents of Shaker constructs were corrected for background current using templates obtained in the presence of 1 μM Agitoxin 2 [Kd 1 nM (Garcia et al., 1994)]. Gating currents were isolated with the P/4 protocol (Armstrong and Bezanilla, 1973). Data analysis and curve fitting were performed with OriginPro 8 (OriginLab Corp.). The figures were made using OriginPro 8, PyMOL 1.0 (DeLano Scientific), and CorelDRAW X14 (Corel Corp.).
Supplementary Material
Acknowledgments
We thank P. De Weer for critical review and discussion of our manuscript, and K. Baek for help with figure preparation. Molecular models were prepared with PyMOL 1.0 (DeLano Scientific). This study was supported by grant GM55560 from the National Institutes of Health to Z. Lu. Z. Lu is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- Ahern CA, Horn R. Specificity of charge-carrying residues in the voltage sensor of potassium channels. J Gen Physiol. 2004;123:205–216. doi: 10.1085/jgp.200308993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM. Sodium channels and gating currents. Physiol Rev. 1981;61:644–683. doi: 10.1152/physrev.1981.61.3.644. [DOI] [PubMed] [Google Scholar]
- Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomand A, Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci U S A. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Structure and function of voltage-sensitive ion channels. Science. 1988;242:50–61. doi: 10.1126/science.2459775. [DOI] [PubMed] [Google Scholar]
- Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais CJ, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Freites JA, Tobias DJ, von HG, White SH. Interface connections of a transmembrane voltage sensor. Proc Natl Acad Sci U S A. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia ML, Garcia-Calvo M, Hidalgo P, Lee A, MacKinnon R. Purification and characterization of three inhibitors of voltage-dependent K+ channels from Leiurus quinquestriatus var. hebraeus venom. Biochemistry. 1994;33:6834–6839. doi: 10.1021/bi00188a012. [DOI] [PubMed] [Google Scholar]
- Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Rosenman E, Bezanilla F, Alvarez O, Latorre R. Periodic perturbations in Shaker K+ channel gating kinetics by deletions in the S3-S4 linker. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9617–9623. doi: 10.1073/pnas.171306298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe M, Lai HC, Jain M, Jan YN, Jan LY. Structure prediction for the down state of a potassium channel voltage sensor. Nature. 2007;445:550–553. doi: 10.1038/nature05494. [DOI] [PubMed] [Google Scholar]
- Hackos DH, Chang TH, Swartz KJ. Scanning the intracellular S6 activation gate in the shaker K+ channel. J Gen Physiol. 2002;119:521–532. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, White SH, von HG. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Islas LD, Sigworth FJ. Voltage sensitivity and gating charge in Shaker and Shab family potassium channels. J Gen Physiol. 1999;114:723–742. doi: 10.1085/jgp.114.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Khalili-Araghi F, Jogini V, Yarov-Yarovoy V, Tajkhorshid E, Roux B, Schulten K. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys J. 2010;98:2189–2198. doi: 10.1016/j.bpj.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaguchi T, Sukhareva M, Swartz KJ. Stabilizing the closed S6 gate in the Shaker Kv channel through modification of a hydrophobic seal. J Gen Physiol. 2004;124:319–332. doi: 10.1085/jgp.200409098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Baldwin TJ, Jan YN, Jan LY. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- Li-Smerin Y, Swartz KJ. Localization and molecular determinants of the Hanatoxin receptors on the voltage-sensing domains of a K+ channel. J Gen Physiol. 2000;115:673–684. doi: 10.1085/jgp.115.6.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Hess P, Weaver F, Koren G. Voltage-sensing residues in the S4 region of a mammalian K+ channel. Nature. 1991;353:752–756. doi: 10.1038/353752a0. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Liu Y, Holmgren M, Jurman ME, Yellen G. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 1997;19:175–184. doi: 10.1016/s0896-6273(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, MacKinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Lopez GA, Jan YN, Jan LY. Hydrophobic substitution mutations in the S4 sequence alter voltage-dependent gating in Shaker K+ channels. Neuron. 1991;7:327–336. doi: 10.1016/0896-6273(91)90271-z. [DOI] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. Ion conduction pore is conserved among potassium channels. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- Milescu M, Bosmans F, Lee S, Alabi AA, Kim JI, Swartz KJ. Interactions between lipids and voltage sensor paddles detected with tarantula toxins. Nat Struct Mol Biol. 2009;16:1080–1085. doi: 10.1038/nsmb.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- Noda M, Shimizu S, Tanabe T, Takai T, Kayano T, Ikeda T, Takahashi H, Nakayama H, Kanaoka Y, Minamino N. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature. 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- Papazian DM, Shao XM, Seoh SA, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 1991;349:305–310. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Perozo E, MacKinnon R, Bezanilla F, Stefani E. Gating currents from a nonconducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 1993;11:353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- Phillips LR, Milescu M, Li-Smerin Y, Mindell JA, Kim JI, Swartz KJ. Voltage-sensor activation with a tarantula toxin as cargo. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzicka A, Wolfenden R. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- Schoppa NE, McCormack K, Tanouye MA, Sigworth FJ. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Silverman WR, Roux B, Papazian DM. Structural basis of two-stage voltage-dependent activation in K+ channels. Proc Natl Acad Sci U S A. 2003;100:2935–2940. doi: 10.1073/pnas.0636603100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Maxwell CJ, Ledwell JL, Aldrich RW. Uncharged S4 residues and cooperativity in voltage-dependent potassium channel activation. J Gen Physiol. 1998;111:421–439. doi: 10.1085/jgp.111.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassova M, Lu Z. Coupled ion movement underlies rectification in an inward-rectifier K+ channel. J Gen Physiol. 1998;112:211–221. doi: 10.1085/jgp.112.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace DM, Stefani E, Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- Stühmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- Swartz KJ, MacKinnon R. Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron. 1997;18:675–682. doi: 10.1016/s0896-6273(00)80307-4. [DOI] [PubMed] [Google Scholar]
- Tang CY, Papazian DM. Transfer of voltage independence from a rat olfactory channel to the Drosophila ether-a-go-go K+ channel. J Gen Physiol. 1997;109:301–311. doi: 10.1085/jgp.109.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristani-Firouzi M, Chen J, Sanguinetti MC. Interactions between S4-S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J Biol Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.