Abstract
Tissue-encysting coccidia, including Toxoplasma gondii and Sarcocystis neurona, are heterogamous parasites with sexual and asexual life stages in definitive and intermediate hosts, respectively. During its sexual life stage, T. gondii reproduces either by genetic out-crossing or via clonal amplification of a single strain through self-mating. Out-crossing has been experimentally verified as a potent mechanism capable of producing offspring possessing a range of adaptive and virulence potentials. In contrast, selfing and other life history traits, such as asexual expansion of tissue-cysts by oral transmission among intermediate hosts, have been proposed to explain the genetic basis for the clonal population structure of T. gondii. In this study, we investigated the contributing roles self-mating and sexual recombination play in nature to maintain clonal population structures and produce or expand parasite clones capable of causing disease epidemics for two tissue encysting parasites. We applied high-resolution genotyping against strains isolated from a T. gondii waterborne outbreak that caused symptomatic disease in 155 immune-competent people in Brazil and a S. neurona outbreak that resulted in a mass mortality event in Southern sea otters. In both cases, a single, genetically distinct clone was found infecting outbreak-exposed individuals. Furthermore, the T. gondii outbreak clone was one of several apparently recombinant progeny recovered from the local environment. Since oocysts or sporocysts were the infectious form implicated in each outbreak, the expansion of the epidemic clone can be explained by self-mating. The results also show that out-crossing preceded selfing to produce the virulent T. gondii clone. For the tissue encysting coccidia, self-mating exists as a key adaptation potentiating the epidemic expansion and transmission of newly emerged parasite clones that can profoundly shape parasite population genetic structures or cause devastating disease outbreaks.
Author Summary
The parasites Toxoplasma gondii and Sarcocystis neurona have lifecycles that include a sexual stage in a definitive host and an asexual stage in intermediate hosts. For T. gondii, laboratory studies have demonstrated that the sexual stage can serve the dual purpose of producing new, virulent genotypes through recombination and promoting expansion of single clones via self-mating. Self-mating and other life history traits of T. gondii, including transmission of asexual stages among intermediate hosts, are assumed to account for the clonal population genetic structure of this organism. However, the relative contributions of sexual recombination and self-mating verses other life history traits in causing disease outbreaks or in shaping Toxoplasma's population genetic structure have not been verified in nature, nor have these traits been extensively examined in related parasites. To address this knowledge gap, we conducted population genetic analyses on T. gondii and S. neurona strains isolated from naturally occurring outbreaks affecting humans and sea otters, respectively. Our results identify self-mating as a key trait potentiating disease outbreaks through the rapid amplification of a single clone into millions of infectious units. Selfing is likely a key adaptation for enhancing transmission of recently emerged, recombinant clones and reshaping population genetic structures among the tissue-cyst coccidia.
Introduction
Population genetic studies of pathogenic microbes have been paramount to our understanding of disease resulting from emerging and re-emerging infectious organisms [1]. Studies performed to determine the relative contributions of drift and recombination in the production of genetic diversity have identified that most pathogens have methods to alter, exchange and acquire genetic material that are intimately associated with pathogenicity [1], [2]. For viral pathogens, enhanced levels of drift, genomic reassortment [3], and incorporation of host genes [4] have all been linked to emergence of virulence. Likewise, horizontal gene transfer between bacterial species has facilitated assimilation of pathogenicity islands, plasmids, prophages, and other insertional elements essential for disease and drug resistance phenotypes [5]–[9]. For eukaryotic pathogens, meiotic sex serves an analogous purpose functioning to alter the genetic make-up, and therefore the biologic and virulence potential of strains [10]–[17]. A general paradigm describing disease epidemics for many pathogens is that genetic diversification, complemented by the acquisition of traits that enhance relative fitness and facilitate clonal expansion, leads to the emergence of novel, virulent genotypes. Just as the life history traits for generating genetic diversity vary widely among pathogen types, it is often the case that the mechanistic basis for subsequent clonal expansion of pathogenic strains is unique on a taxonomic level. Determining the mechanisms and contribution of these life history traits to disease is important for focusing prevention and treatment strategies to the most relevant pathogen strains and life cycle stages.
For the cyst-forming coccidia, which comprise a diverse group of parasites belonging to the phylum Apicomplexa, complex lifecycles that include both sexual and asexual stages have led to unusual population genetic structures for several species. For the widespread zoonotic pathogen, Toxoplasma gondii, the majority of strains infecting birds and mammals throughout North America and Europe are comprised of just three clonal lineages which exist as successful clones from a genetic out-cross [14], [18]. These three lineages have apparently emerged only recently due to an enhanced fitness that facilitated their ability to effectively outcompete other genotypes [13], [19]–[21]. Likewise, the veterinary pathogen Sarcocystis neurona possesses a surprisingly simple population genetic structure punctuated by the dominance of a few clonal lines in North America [22]–[25]. Similar clonal structures have been reported for other parasitic protozoa that possess sexual cycles [26] but identifying the precise genetic mechanisms that have led to the emergence of distinct clones among the different species in nature remains enigmatic.
In combination with population genetic data, the contributions of sexual out-crossing and clonal expansion as factors governing the emergence and eventual dominance of distinct, disease-producing clones have largely been inferred from laboratory studies of T. gondii among the cyst forming coccidia. Prior experiments demonstrated that a sexual cross between mouse-avirulent strains can produce genotypes representing a range of virulence in the mouse model, including some progeny several logs more virulent than the parents [14]. This study identified that natural out-crosses likely produce at least some virulent genotypes, which may subsequently have potential to emerge through clonal amplification to cause extensive disease [20], [21]. Clonal propagation is possible since T. gondii can effectively bypass the sexual stage in felid definitive hosts and cycle, presumably indefinitely, among intermediate hosts. This can occur horizontally via oral transmission through carnivory among intermediate hosts [19], [20] or vertically by transplacental transmission [27]–[30]. Toxoplasma gondii can also functionally bypass genetic diversification during the sexual stage by self-mating in the definitive host. Self-mating (also termed selfing, uni-parental mating, or self-fertilization) occurs when a single parasite clone can give rise to both male and female gametes capable of undergoing fertilization and producing viable offspring [31], [32]. In other words, no predetermined mating types are apparent and the end result is effectively clonal expansion via sex and meiosis.
Despite these important laboratory studies, the implications of these life-history traits and their relative effects on population genetic structures, especially in the context of virulence and disease outbreaks, have not been extensively studied in T. gondii or other cyst forming coccidia in a natural setting. Parasite life stages that are most important for causing mass-morbidity and mortality may be revealed through review of past, large-scale T. gondii-associated human outbreaks. For eleven reports of T. gondii-associated disease outbreaks in immune-competent people, eight events, including the four most devastating that caused disease or death in hundreds of individuals, were attributed to the oocyst form of the parasite, which is only produced during the sexual life cycle stage in the definitive feline host [20]. Furthermore, an outbreak of the related veterinary pathogen Sarcocystis neurona that resulted in the death of nearly 1.5% of the threatened Southern sea otter population over the course of a single month is thought to have resulted from exposure to infectious sporocysts originating in the definitive opossum host [33]. Circumstantial evidence, such as a complete lack [34] or much reduced [35]–[37] prevalence of T. gondii in certain island environments without cats, also gives weight to the importance of the definitive host stage in the parasite life cycle. Similarly, S. neurona has not been identified outside of its definitive host range in the Americas. The apparently profound importance of this stage in the lifecycle of not just T. gondii, but other related parasites, warrants further study to determine the influence it could impart to shaping parasite population genetic structures and which genetic mechanisms inherent to this life stage (i.e. selfing or out-crossing) are more likely to precede a disease outbreak in nature.
To determine the genetic basis governing the exposure, evolution, and emergence of virulent genotypes during natural outbreaks linked to sexual stages of these parasitic protozoa, we tested whether epidemic isolates exist as: 1. a diverse array of multiple, novel genotypes that are the products of an out-crossing event in the definitive host, or 2. epidemic clones of a single genotype derived via selfing in the definitive host. To distinguish between these two possibilities, high resolution genetic typing was used to characterize parasite strains associated with a T. gondii outbreak in humans [38] and a S. neurona outbreak in sea otters [33], both of which were associated with unusually high levels of morbidity and mortality. The population level genetic studies presented here argue that selfing in the definitive host plays a central role in the epidemic expansion of newly emerged, recombinant parasite strains, thus potentiating clonal outbreaks caused by tissue cyst-forming coccidia.
Results/Discussion
An outbreak linked to T. gondii oocyst ingestion was associated with a single parasite genotype
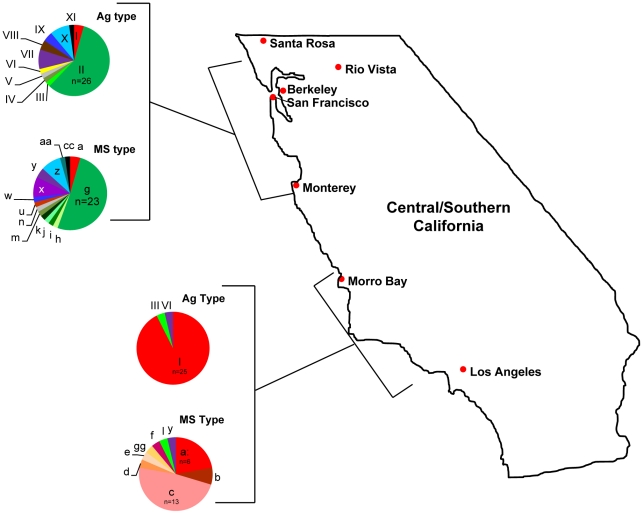
A microsatellite-based typing scheme using the markers B17, B18, TgMa, TUB2, W35 [39], and M95 [40] was applied to determine the molecular genotypes of T. gondii isolates associated with a human water-borne outbreak in Brazil. This outbreak, which occurred over a short time span in 2001, was linked to oocyst-contamination of a municipal water supply in the town of Santa Isabel do Ivai and resulted in infection and symptomatic disease in hundreds of people [38]. Initial genetic typing analyses performed on two T. gondii strains isolated from the water cistern implicated as the source of the outbreak [38], as well as isolates from chickens [41] and cats [42] from the immediate environment were limited to PCR-RFLP at a single locus, SAG2, leading to the conclusion that the outbreak strain was a canonical Type I strain (see below). Later, more extensive analysis by PCR-RFLP [43] and DNA sequencing on a limited set of markers [44] showed that the outbreak-associated strains from the water cistern were clonal and non-archetypal. The majority of people who seroconverted during the outbreak also possessed a serologic profile consistent with infection by the outbreak clone, and the outbreak genotype appeared to be highly prevalent in the surrounding environment immediately following the outbreak event, infecting 4/11 chickens (TgBrCk98, TgBrCk101–103) and 1 cat (TgCatBr85) [44] (Figure 1).
Figure 1. Genotype analysis of Toxoplasma gondii strains associated with an outbreak in Santa Isabel do Ivai, Brazil.
All T. gondii isolates were analyzed directly by sequencing at microsatellite (MS) loci and PCR-RFLP at the remaining loci except for Outbreak 1 and Outbreak 2 which were directly sequenced at all loci. Outbreak 1, Outbreak 2, TgCatBr85, and TgCkBr98–103 all possess one of two alleles at each locus, suggesting they are sibling progeny from a recent outcross. Outbreak 1 and Outbreak 2 were oocyst samples isolated from two separate water filters from water supplies implicated in the outbreak and possess identical genotypes indicative of a clonal outbreak. This suggests an outcross preceded the outbreak and was followed by a selfing event in the definitive host that enhanced the clonal expansion and transmission of the newly emerged, recombinant outbreak genotype. Shaded alleles indicate those which are identical to the Outbreak genotype. *Serotype, DNA sequence, and PCR RFLP data from Vaudaux et al. [44]; **Numbers indicate dinucleotide repeat count and letters indicate distinguishing SNPs surrounding the repeat region; MS: microsatellite; WC: water cistern; Ch: chicken; Lab: laboratory strain; na: not available.
To determine the extent of genetic relatedness among the outbreak-associated strains, high resolution MS typing and DNA sequencing using markers distributed on 11 of the 14 chromosomes was applied. This dataset distinguished the two water cistern, outbreak-associated strains at the genetic level from all others present in the environment, except for one chicken isolate (TgCkBr103) (Figure 1). Unfortunately, insufficient DNA remained from the cat isolate, TgCatBr85, which precluded testing whether it was genetically identical to the cistern isolates.
Utilizing the MS typing scheme confirmed the conclusion that the causal agent was a unique, emergent T. gondii strain with a potential for enhanced virulence. The additional typing provided in the current study refined the conclusions of previous studies in two key aspects. First, the much higher level of resolution provided by the markers used and the sequence level analysis imparts a higher level of confidence to the conclusion that the outbreak was in fact clonal. The possibility that the outbreak-associated clones are not genetically identical in lieu of additional typing cannot be excluded, but several facts strongly argue against this: 1. The 18 markers were distributed across all but three of the 14 chromosomes; 2. MS markers are prone to rapid evolution and therefore provide high resolution; 3. Strains from Brazil are genetically divergent from archetypal lines, as evidenced by the segregation of alleles amongst strains in Figure 1, and hence, less prone to linkage disequilibrium effects. Furthermore, only a single, oocyst-derived clonotype was isolated from independent filters collected from two different water-holding tanks providing additional evidence that these isolates resulted from self-mating rather than a genetic out-cross.
Second, this study refines previous work on the Santa Isabel outbreak by showing that the outbreak strain was actually rare in the surrounding environment, opposed to the high prevalence reported previously [44]. Moreover, close examination of the environmental isolates reveals that many of them, including those previously identified as the outbreak clone, and the outbreak clone itself, resemble recombinant progeny; only two allelic types are present that segregate independently across the loci examined (see TgCkBr98, 99, 100, 101, 102, 103, TgCatBr85 and Outbreak 1 and 2 in Figure 1). These data argue that prior to the outbreak, the epidemic clone was produced by a genetic out-cross and was subsequently expanded by self-mating. This confirms that the more extensive resolution provided by the current study was necessary to truly distinguish an epidemic clone in a region known to contain a diverse array of T. gondii genotypes, including many that are apparently siblings of this strain [44]. This result also speaks to the important role selfing in the definitive host can play; allowing a single, emergent genotype of low environmental prevalence to rapidly rise to dominance in the surrounding population by infecting several hundreds of hosts over a short time span.
Collectively these data support high-resolution genotyping schemes as important tools for detecting informative genetic signatures in this parasite species. Initial population genetic studies showed that T. gondii strain diversity was comprised of three main clonal groups: Type I, II, and III [18]. As a result of these early studies, many broader population genetic studies have since relied on typing at only one or just a few loci to classify strains as type I, II, or III. However, it is now apparent that strains from diverse geographic locales and host species are more often infected with strains bearing unique alleles or allelic combinations, so relying on a few markers is insufficient for robust conclusions [20]. The first quantitative analysis testing the accuracy of single locus typing found a very low predictive value for the loci analyzed to correctly identify strain genotype [45]. Indeed, results presented in the current study, when compared with results from more limited genetic studies of the same strains conducted previously [38], [41]–[44], provide a clear illustration of the value more extensive genetic typing can have in refining conclusions. This is especially relevant in outbreak investigations where variations in parasite genotype can be highly informative for explaining disease manifestation. High-resolution genetic typing appears to be critical for eliminating preconceived biases in epidemiologic investigations to ensure accurate discernment of disease-associated T. gondii strains and to recognize clonal outbreaks.
These results validate the utility of testing for epidemic clones from prospective and retrospective studies of T. gondii disease outbreaks [20]. In support of this, Dumar and colleagues applied a similar typing scheme to a T. gondii outbreak in Suriname and discovered that all five patients from whom they isolated parasites were infected with the same, previously undiscovered genotype [46]. Importantly, the outbreak in Suriname was another waterborne outbreak attributable to human exposure by infectious oocysts, further evidencing selfing in the definitive host as a key mechanism for allowing clonal expansion of virulent genotypes, ultimately resulting in disease epidemics.
Genetic typing of outbreak strains of the related pathogen, Sarcocystis neurona
Since parasite genetic material from past T. gondii outbreaks in humans is in limited supply for the majority of cases, we sought to further assess the role of self-mating in disease outbreaks by examining an epizootic of the related veterinary pathogen, Sarcocystis neurona, infecting the Southern sea otter (Enhydra lutris nereis) of California. As a threatened species, the Southern sea otter population is well monitored and accounted for by conservation groups, creating a unique opportunity to investigate infectious disease in a natural setting. Sea otters are also aberrant hosts for many terrestrial pathogens that can be washed to sea and their high susceptibility to many of these pathogens allows them to serve as a sentinel species for pathogens circulating in the adjacent terrestrial environment [44]. During April, 2004, the highest monthly mortality rate ever recorded in nearly 30 years of data collection occurred among Southern sea otters [33]. Over the course of approximately one month, at least 40 sea otters stranded dead or dying along an 18 kilometer stretch of coast within the 500–600 kilometer Southern sea otter range. Sixteen otters were in sufficient condition to allow for complete post-mortem analysis inclusive of PCR assessment and microscopic examination of tissues. Among these otters, the major cause of death for 15 of the 16 examined animals was S. neurona-associated brain and/or systemic disease [33].
Preliminary genetic analysis using only four polymorphic markers against parasite strains infecting a subset of these otters (n = 7) suggested they were genetically homogenous [25]. However, the limited polymorphism present in the markers used, and lack of information about the population genetic structure of S. neurona in California prevented a confident conclusion that they represented an epidemic clone. The present study developed and applied a battery of higher resolution, polymorphic microsatellite and gene-coding markers to type S. neurona strains. Additional samples were included, encompassing 12 S. neurona strains from otters that died during the outbreak, as well as additional strains from other geographic locations and/or time periods. The high number of sea otter deaths associated with this epizootic provided a unique opportunity to test whether self-mating, as identified in the human T. gondii outbreaks, could explain the genetic origin for the S. neurona strains that caused the outbreak. In addition, genetic data from the current study was combined with S. neurona typing data reported by Rejmanek et al. [23] to determine the population genetic structure of S. neurona in California spanning 15 years of study.
eBURST analysis reveals two main S. neurona clonal complexes in California
Sequence-level analysis of five surface antigen (Ag) genes (SnSAG1, 3, 4, 5, and 6) [25] and nine microsatellite (MS) markers (Sn2–Sn5, Sn7–Sn11) [23], [25] identified 12 Ag types and 33 MS types among 87 S. neurona-infected samples based on the allele combinations detected at each locus (Table 1; See Table S1 for complete strain and typing information). Seventy-four of the 87 samples were from mammals in California; other states represented include Georgia (n = 2), Illinois (n = 1), Missouri (n = 3), Washington (n = 5), and Wisconsin (n = 2). Combining Ag and MS alleles could distinguish 35 total genotypes, but for this study these typing schemes were analyzed independently because of the likelihood that these parts of the genome are under different selection pressures and subject to differing evolutionary processes [2]. The majority (56/87) of S. neurona strains were classified as either Ag type I or Ag type II (Figure 2A). Certain MS types were also over represented in the sample set, with MS types ‘a’, ‘c’, and ‘g’ accounting for 47/87 samples (Figure 2B). Importantly, 11/12 S. neurona strains from sea otters stranding during the mortality event in 2004 were an exact genetic clone at each marker analyzed (Ag type I, MS type ‘c’). The remaining outbreak sample (Ag type I, MS type ‘d’) differed from the other outbreak strains by only a single stepwise mutation at MS marker Sn4 (Table S1).
Table 1. Sarcocystis neurona genotyping data summary.
| Data Set | Host | Samples | Antigen types | MS types | total genotypes | eBurst complexes (MS) | eBurst singletons (MS) | Proportion complex 1 | Proportion complex 2 | Total proportion complex 1/2 |
| Overall | Sea otter | 57 | 9 | 20 | 20 | 5 | 2 | 0.47 | 0.32 | 0.79 |
| Harbor seal | 6 | 2 | 6 | 6 | nd | nd | 0 | 0.17 | 0.17 | |
| Raccoon | 2 | 1 | 2 | 2 | nd | nd | 0 | 0 | 0 | |
| Opossum | 13 | 6 | 7 | 9 | 1 | 5 | 0 | 0.31 | 0.31 | |
| Horse | 7 | 3 | 4 | 4 | nd | nd | 0 | 0.57 | 0.57 | |
| Porpoise | 1 | 1 | 1 | 1 | nd | nd | 0 | 1.00 | 1.00 | |
| Cat | 1 | 1 | 1 | 1 | nd | nd | 0 | 1.00 | 1.00 | |
| Total | 87 | 12 | 33 | 35 | 8 | 8 | 0.31 | 0.34 | 0.65 | |
| Monterey, CA (ATOS 1–400) | Sea otter | 30 | 9 | 12 | 12 | 4 | 2 | 0.07 | 0.63 | 0.70 |
| Opossum | 10 | 5 | 5 | 6 | 1 | 3 | 0 | 0.40 | 0.40 | |
| Horse | 4 | 2 | 2 | 2 | nd | nd | 0 | 0.75 | 0.75 | |
| Porpoise | 1 | 1 | 1 | 1 | nd | nd | 0 | 1 | 1.00 | |
| Total | 45 | 11 | 15 | 16 | 4 | 4 | 0.07 | 0.60 | 0.64 | |
| Morro Bay, CA (ATOS 800–1200) | Sea otter | 27 | 3 | 9 | 9 | 1 | 3 | 0.93 | 0 | 0.93 |
| Total | 27 | 3 | 9 | 9 | 1 | 3 | 0.93 | 0 | 0.93 |
MS: microsatellite.
nd: not done.
ATOS: The ‘As The Otter Swims’ number refers to each sea otter's stranding location, based upon defined and sequential 0.5 kilometer segments of the California coastline, starting with zero (0) just north of San Francisco and increasing numerically from north to south.
Figure 2. Sarocystis neurona genotyping results.
Distribution of the 12 Ag types (A) and 33 MS types (B) identified among all Sarcocystis neurona samples studied (n = 87). Ag type I and II accounted for the majority of all samples with 27 and 29 samples, respectively. The most numerous MS type identified was type g, accounting for 26 total samples. (C) Further analysis of MS types using the eBURST program on default settings for 9 loci (Sn2–Sn5, Sn7–Sn11), revealed that 64% of all isolates belonged to two clonal complexes. Clonal complex 1 (CC1) was comprised of MS types a, b, c, d, e, and gg and CC2 of types g, h, i, j, and k. All MS types in CC1 possessed Ag type I. MS types g, h, i, and j of CC2 possessed Ag type II, whereas MS type k possessed Ag type III. *MS type c was found in 11/12 examined S. neurona strains from sea otters that died during the 2004 epizootic.
Since this and all previous studies of S. neurona have found a high level of sequence homology among strains [22]–[25], [47], we chose to analyze strain relatedness with the eBURST algorithm [48], [49]. This program helps eliminate confounding effects that low sequence diversity and moderate levels of recombination can have on other methods of intra-specific sequence analysis, such as clustering, dendrograms, and phylogenetic trees, as demonstrated in [22]–[24], by only focusing on single clones and their most recent descendents [48]–[50]. We adapted the MS data for the nine markers that permit simultaneous comparison of all strains (Sn2–Sn5, Sn7–Sn11) to serve as a multi-locus typing scheme. This typing scheme, which is based on the number of repeats at each locus, was amenable to use with this program. Using the default settings, which group isolates based on the premise that they are single locus variants (SLVs), or share 8 out of 9 alleles, we identified 8 clonal complexes (CC1–8), only 3 of which contained more than two genotypes, and 8 singletons (genotypes differing by 2 or more alleles from all others) (Table 1; Figure 3). Intriguingly, just two clonal complexes, CC1 and CC2, accounted for almost 64% (56/87) of the strains analyzed in this study (Figure 2C). This result held true even when correcting for bias introduced by the outbreak event by removing these samples from the data set, as 44/75 samples (59%) still belonged to CC1 or CC2.
Figure 3. Modified eBURST analysis output.
Default eBURST settings were used to analyze Sarcocystis neurona sequence types based on MS markers Sn2–Sn5 and Sn7–Sn11. MS types identified are represented as small circles and designated by lowercase letters. Lines connect MS types that are identical at 8 out of 9 MS loci and are therefore considered part of a clonal complex (CC). eBURST identified 8 clonal complexes (designated CC1–CC8) and 8 singletons. Large colored ovals are overlain to indicate the Ag type (Ag types I–XII) that characterizes each MS type identified by eBURST. MS and Ag type color schemes refer to those described in Table S1. Results support an intermediate population structure with both clonal propagation and sexual recombination. All members of CC1 possess an identical Ag type (Ag type I). MS types x and bb were found in samples with different Ag types (VII and VIII). Ag types VII and VIII differ by a single di-nucleotide indel at Ag marker SnSAG3, likely representative of drift rather than recombination as a mechanism to account for allele differences in this case. In contrast, MS type k has a markedly different Ag type (III) compared to other members of the CC2, which all possess Ag type II. Ag types II and III have different alleles at all Ag loci examined, making a recombination event the most parsimonious explanation for the difference between MS type k and other members of CC2 rather than genetic drift.
All SLVs identified in this study differed by a single stepwise (i.e. a single di-nucleotide repeat) mutation, which supports the assumption that the eBURST groupings represent clonal complexes in which allelic variation is a result of mutation/drift and not recombination (Table S1) [50]. The only exceptions to this were SLVs ‘l’ and ‘o’, members of CC3, that differed by 3 di-nucleotide repeats at MS Sn11. These isolates were from a sea otter in California and a horse from Missouri so the greater number of stepwise mutations detected may be a result of extended geographic isolation, thus allowing time for more drift to occur (Table S1). A single mutation event that resulted in multiple stepwise mutations is also plausible.
Since recombination appeared to be rare between clonal complexes based on MS markers, we decided to overlay the results of the Ag typing analysis on the eBURST output (Figure 3). The results were consistent with previous claims of an intermediate population structure for S. neurona [22]–[25], [47] in that both clonal propagation and sexual recombination were supported. All members of CC1 and 29/30 members of CC2 possessed an identical Ag type (Ag types I and II, respectively). In contrast, all MS types in CC3 and CC8 possessed a distinct Ag type. There were also two cases (MS types ‘x’ and ‘bb’) where the same MS type was identified with two distinct Ag types (Ag types VII and VIII) and the reverse scenario also occurred where the same Ag type (VI) characterized three clonal complexes based on MS types (CC4, CC5, CC6), all of which could potentially indicate recombination events (Figure 3).
Overall, these data support a population structure that is highly clonal, though evidence for recombination is present as well. This intermediate population structure is similar to that described for T. gondii, though definitive conclusions will require a sample set less biased towards diseased animals [2]. It is worth noting here that the population structure of the organisms described in this study is, like all population genetic structures, only as resolved as the markers allow. For example, finer resolution can be achieved by applying the marker SnD2 from Rejmanek et al. [23] to SO4711, SO4786 and O7 to show that they are different strains. What this does not change, though, is that these strains are members of the same clonal complex and that resolution at this level is sufficient to identify an outbreak clone and to document geographic partitioning of strains along the California coastline (see below). This level of resolution is more robust to the possibility of strand slippage and evolution of new alleles during PCR that could make identical clones appear distinct with finer levels of resolution. An example of this may have occurred with SO4387, identified in this study as MS type ‘g,’ but by Rejmanek et al. [23] as MS type ‘i.’ These types differ by a single repeat at MS Sn9 (Table S1). It is also possible that this otter was co-infected with two closely related strains. Consistent identification of SLVs in many samples increases the confidence that they represent truly different strains. The outstanding potential these microsatellite markers have for more robust strain resolution, if interpreted cautiously, can facilitate addressing more specific questions, such as the identity and point source of an epidemic clone.
Temporal stability, geographic distribution, and host distribution of strains in California
The majority of strains (72/87; 83%) evaluated in this study were collected from two distinct 200 km stretches along the California coast or the adjacent terrestrial environment (Table S1; Figure 4). As such, we utilized this subset of the data to examine the temporal stability of strains and their geographic and host distribution in central California.
Figure 4. Geographic distribution of Sarcocystis neurona Ag and MS types in California.
All sea otter samples were collected in two distinct, ∼200 km stretches along the California coast: one in central California from just north of San Francisco Bay to just south of Monterey Bay, and one to the south from just north of Morro Bay to just north of Los Angeles. Nearly all (93%) of the 27 samples from the southern region belonged to eBURST defined clonal complex (CC) 1 and none were identified as CC2. In the north, 63% of 45 samples belong to CC2 and only two representatives of CC1 were found. Terrestrial isolates from California were from 10 opossums and 4 horses. These, along with one sample from a porpoise, were from the northern range and included as such. The majority of sea otter samples were from two small areas of coastline: one near Monterey Bay in the north and the other near Morro Bay in the south (see Table S1 for details).
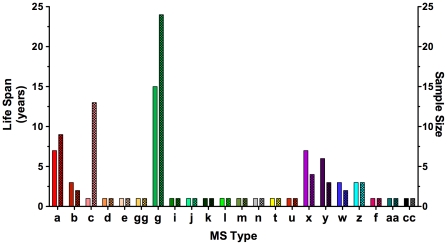
The total time period covered by the strains analyzed in this study is 15 years (1994–2009). Sample sizes were not evenly distributed across each year and some years (1996–1998) had no representative samples, so it is likely that genotype life spans are underestimated. Despite this, at least one clonal complex, CC2, appears to be very stable in nature over time, exhibiting a lifespan encompassing the entire length of this study. CC2 was sampled during 12 of the 13 years for which a sample was collected (Table S2). Within this complex, Ag type II, MS type ‘g’ had a lifespan of the full time period examined (15 years) and was the longest lived of any Ag or MS type (Figure 5; Table S2). The other clonal complexes present in California, CC2, CC3, CC6–CC8, appeared to be stable as well, with life spans ranging from 5–8 years (Table S2). Collectively these data provide supporting evidence for S. neurona's ability to propagate clonally. However, it will be important to test whether or not these allelic combinations appear more often than would be expected by chance to confirm clonal propagation as more sequencing data becomes available from strains collected from non-diseased animals and the position of the markers in the genome is identified [51]. Interestingly, the genotype associated with the outbreak, Ag type I, MS type ‘c’, was only found during 2004 (Figure 5). These samples were all associated with otters dying during the epizootic in April, 2004, except for two samples that were obtained from sick otters in the same area four months after the event ended (Table S1). The implications these observations may have for strain virulence are discussed below.
Figure 5. Sarocystis neurona MS type lifespan and sample size in California.
Lifespans for microsatellite (MS) types (solid bars) were defined as the time period from identification the first representative sample to the last during the 15 years (1994–2009) encompassed in this study. Sample sizes are indicated by checkered bars. MS type ‘g,’ a member of eBURST defined clonal complex 2 (CC2), was the longest lived and most prevalent MS type, having a representative sample in all of the years for which samples were available. There were no S. neurona samples available for testing during 1996–1998. MS type ‘c,’ the genotype implicated in the sea otter epizootic, was only found in 2004 during the month of the outbreak and four months thereafter in the same region.
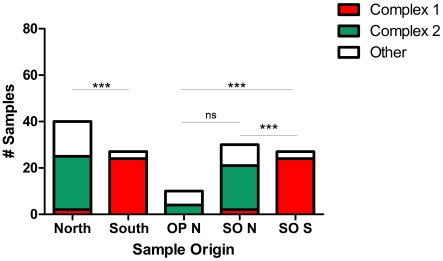
On visual inspection, it appeared that the genetic composition of S. neurona strains from the Monterey Bay area was distinct from the southern strains obtained in or near Morro Bay (Figure 4; Table 1). We further tested this hypothesis by conducting χ2 analysis on the proportion of the majority clonal complexes (CC1 and CC2) that comprised each population. There was a highly significant difference between northern and southern strains (Figure 6). Significance remained when analysis was restricted to sea otter samples, in order to eliminate any confounding effects due to host species, because all southern strains were from sea otters (Figure 6). This conclusion is consistent with data reported previously on S. neurona strains from coastal California [24], [25], but contrasts with the conclusions of Rejmanek et al. [23].
Figure 6. Geographic partitioning and host associations of Sarcocystis neurona strains.
Distinct S. neurona populations as defined by the proportion of the population belonging to the dominant eBURST defined clonal complexes (CC) 1 or 2 were found infecting animals in the northern and southern ranges examined in California (see Figure 4). This difference remained significant by Chi-Square analysis when only sea otter samples were compared. When samples from sea otters from the northern range were compared to opossum samples from the adjacent terrestrial environment, no significant difference was found. There were no samples from terrestrial mammals in the southern range. OP N: opossum samples from the northern range; SO N: sea otter samples from the northern range; SO S: sea otter samples from the southern range; ns: not significant; ***p<0.00001.
We also sought to identify a potential terrestrial source for S. neurona strains present in the marine environment. Experimental evidence for the model organism, T. gondii, supports a route of infection for sea otters through ingestion of S. neurona sporocysts that were washed to the ocean in contaminated fresh water and then concentrated in the otters' filter-feeding invertebrate prey [52]–[54]. Implicating opossums as the ultimate terrestrial source of infection is supported by comparing the prevalence of the majority clonal complexes (CC1 and CC2) in sea otters and opossums in the northern, Monterey Bay area study site (the only locale from which opossum samples were obtained). Strain prevalence differences between these groups were not statistically different, suggesting that monitoring strain types in coastal dwelling opossums will be predictive of genotypes infecting adjacent marine dwelling otters (Figure 6). Observational data from the outbreak noting an abundance of razor clams and evidence of sea otter movement into the area for feeding (i.e. accumulation of broken shells on the shore) just prior to the event, further support this model of land-to-sea parasite transfer [33]. Sea otters very rarely consume known intermediate hosts of S. neurona [55], leaving the ingestion of sporocysts as the most biologically plausible route for sea otter infection regardless of the land-to-sea transport mechanism, and strongly supporting the conclusion that this outbreak originated from a selfing event in the opossum host.
Parasite genotypes and virulence
Disease is a complex manifestation of the interplay between intrinsic pathogen factors (i.e. pathogen genotype) and numerous external factors, including dose, host immune status, and environmental conditions such as weather that can influence transmission. Delineating the relative contribution of each of these factors to a given disease outbreak is a difficult process, as is illustrated by the outbreaks described in this study. It is plausible that the S. neurona strain associated with the 2004 epizootic is intrinsically more virulent than other strains since it was only identified during the time period surrounding the outbreak and may have been too virulent for continued propagation. Also, the majority of otters infected died within 24–48 hours of stranding and had high IgM titers [33]. The rapid rise and subsequent fall of a virulent strain type is a phenomenon noted in many outbreaks of a diverse array of pathogens from viruses (e.g. Influenza virus [3]) to bacteria (e.g. Leptospira interrogans [56]) to fungi (e.g. Coccidioides immitis [57]). However, this phenomenon may also be attributable to sampling biases [2] or environmental factors [57] making the assumption that the virulent genotype is not adaptive inaccurate. Equally in the case of the sea otter outbreak, numerous external factors, including concurrent infection with other pathogens and domoic acid poisoning, abundant food source with potential for contamination with sporocysts, and a large rainstorm preceding the event that could have increased sporocyst deposition, may have played a contributing role in conferring this S. neurona strain with a virulent phenotype [33].
Similarly, the T. gondii strain implicated in the 2001 Brazil outbreak appeared to rise in prevalence during the outbreak but then decline over time in the local environment [44]. This was also a unique, newly identified genotype that caused symptomatic disease in 155 immune-competent individuals—an unusual phenomenon for this normally asymptomatic parasite. Importantly, though, ∼270 other individuals with access to the same water cistern seroconverted during this time with no overt signs of disease [44], invoking a role for environmental and host factors in this outbreak.
A striking character of both these outbreak events is the key role self-mating in the definitive host served as a catalyst allowing virulent pathogen genotypes to rapidly reach high levels under the right conditions to precipitate a disease epidemic.
Self-mating potentiated the emergence of the S. neurona and T. gondii epidemic clones
Epidemic clonality associated with sporocyst or oocyst ingestion strongly suggests that self-mating in the definitive host was the key event leading to these outbreaks. Selfing in the definitive host has been confirmed experimentally for T. gondii [31], [32] but only indirectly assumed for S. neurona [58]. Prior to this study, rigorous genetic characterization of selfing events in nature were lacking and the question as to whether a productive sexual out-cross or a selfing event precedes an outbreak linked to oocysts or sporocysts had not previously been tested.
Early population genetic studies using limited, poorly resolved markers identified a paucity of mixed strain T. gondii or S. neurona infections in nature and these data have previously been interpreted to suggest that most definitive host infections would be by a single strain and therefore out-crossing would be rare in nature [59]. However, more recent studies using unbiased, multi-locus typing schemes have consistently identified mixed strain infections among natural intermediate hosts suggesting that prey species of definitive hosts are more frequently harboring mixed strain infections than previously envisaged [60]–[72]. Hence, the lack of mixed strain infections identified in earlier studies may simply reflect the techniques used, such as bioassay or limited genetic typing, that were biased toward certain strains and likely missed multiple infections and the true diversity of genotypes present.
As more high resolution, multilocus genetic markers are being applied against previously characterized strains of T. gondii, an increasing number are being re-classified as recombinants, defined as products of sexual out-crossing events, including strains previously linked to outbreaks [20]. Given the virulent nature of the two outbreaks examined here, and the evidence that out-crossing between two avirulent, haploid parents can produce progeny with enhanced virulence [14], we originally hypothesized that out-crossing might explain the genetic origin and expansion of the outbreak strains, rather than self-mating. Intriguingly, close examination of the environmental isolates surrounding the T. gondii outbreak supported this hypothesis because the epidemic clone was one of many progeny produced by a local genetic out-cross. However, the available evidence indicated that, while out-crossing certainly preceded the outbreak, it was the subsequent selfing event that was responsible for the epidemic expansion and transmission of the virulent clone that caused the outbreak. Certainly this dataset argues that sex and self-mating combined to produce the T. gondii clonal outbreak. Further typing of additional outbreaks is warranted to examine whether or not an out-cross is independently sufficient to cause an epidemic attributable to multiple, recombinant progeny.
This two-step process of local epidemic expansion via a sexual out-cross followed by clonal propagation of a few progeny with enhanced adaptations or virulence is reminiscent of the process envisioned on a larger scale for the pandemic rise of the archetypal T. gondii clones (Types I, II, and III), also found to be the progeny of an out-cross [13], [14]. Documenting this process in real time at a local level has provided key insight into mechanisms that account for clonal propagation in nature. It was previously proposed based on laboratory studies that clonal dominance of archetypal T. gondii strains was attributable to an enhanced ability for oral transmission through carnivory, a hypothesis which certainly warrants further investigation in natural settings [19]. However, recent studies have since shown that this trait does not operate as originally proposed [73], [74]. These findings raised the possibility that other life history traits may likewise be important in perpetuating clones.
In this light, it is worth noting that all aspects of the parasite lifecycle that promote clonal propagation, namely selfing, oral transmission through carnivory, and transplacental transmission, contribute in part to clonality in the population structure. However, when considering their relative roles, the advantage in fecundity the sexual stage can impart during a selfing event to a single parasite genotype, as documented in this study, provides strong evidence this mechanism is likely the major contributor to localized or regional clonal dominance of certain strains. The basic reproductive number (R0), or number of secondary infections a single infected individual will cause, is many orders of magnitude greater in the definitive host (which releases millions of environmentally stable, infectious propagules capable of waterborne or aerosolized transmission [75]) compared to an intermediate host (in which the infectious units produced can only be passed to those directly feeding on tissues). Oocysts or sporocysts can also successfully infect intermediate hosts at much lower doses (even a single oocyst) than tissue cysts [76], [77]. Oocyst deposition therefore exists as a potent mechanism for causing widespread epidemics and establishes a plausible rationale for explaining how selective sweeps can occur among these heterogamous pathogens. Determining what factors govern whether these sweeps occur on a local, and presumably more frequent, epidemic level or reach pandemic proportions are important subjects for future research.
Our results also confirm that fecal contamination of food and water sources represents a major threat to human and animal health, hence targeting the definitive host or the oocyst stage of these parasites is an excellent first-step strategy to disrupt transmission. This conclusion is further supported by studies showing the importance of the definitive host stage for maintaining continued transmission of this parasite in island communities [34]–[37] and how local vaccination of definitive feline hosts can significantly reduce T. gondii infection rates [78].
The scope of explanatory power for this selfing model can also be extended to other highly clonal, cyst forming parasites, including the clonal outbreak linked to S. neurona and likely other pathogenic Sarcocystis spp. and Neospora spp. This finding is significant since many aspects of the T. gondii life cycle have previously been proposed to be unique to this species among the tissue encysting coccidia, including its broad host range inclusive of nearly all warm-blooded vertebrates and its ability to be transmitted through carnivory among intermediate hosts [19]–[21,] (but also see: [79], [80]–[83]). Notably, selfing has also been demonstrated in more distantly related Apicomplexan parasites, including Eimeria spp. and Plasmodium spp. [31]. In addition, the processes of homothalism and same-sex mating identified in fungi serve the analogous purpose of clonal propagation via a mechanism more generally thought to serve in genetic recombination and out-crossing [84]. This suggests that selfing, as a genetic mechanism of clonal propagation, has potential to play a pivotal and previously under-recognized role for a diverse array of eukaryotic pathogens in the expansion of genotypes that cause disease epidemics and/or emerge as highly successful clonotypes to rapidly alter population genetic structures.
Materials and Methods
Ethics statement
Work in California was conducted under United States Fish and Wildlife Service (USFWS) permit MA 491 672724-9 issued to United States Geological Survey Biological Resource Discipline (USGS492 BRD). Harbor seal carcasses were gathered and samples processed as part of Northwest Marine Mammal Stranding Network activities authorized under Marine Mammal Protection Act (MMPA) Stranding Agreements (SA), and Section 109(h) (16 U.S.C. 1379(h)). Additional specimens were acquired under MMPA Section 120, and the National Marine Fisheries Service (NMFS) MMPA Research Permit 782–1702.
Sarcocystis neurona and Toxoplasma gondii DNA and genetic typing markers
Parasite DNA was obtained either from infected host tissues or parasite isolates maintained in tissue culture as described previously [25]. Samples were analyzed using a typing scheme that included the surface antigen markers: SnSAG1, SnSAG3, SnSAG4, SnSAG5, SnSAG6 [25] and 9 microsatellite markers Sn2–Sn5 and Sn7–Sn11 originally described by Asmundsson and Rosenthal [85] but applied as modified in Wendte et al. [25] and Rejmanek et al. [23]. Three additional microsatellite markers were designed by the following method: Publically available Sarcocystis neurona expressed sequence tags (ESTs) were downloaded from the NCBI dbEST database (http://www.ncbi.nlm.nih.gov/dbEST) and the S. neurona Gene Index (maintained by the Computational Biology and Functional Genomics Laboratory at the Dana Farber Cancer Institute, http://compbio.dfci.harvard.edu/tgi/) databases. The downloaded ESTs were assembled into contigs using the SeqMan (Lasergene) application. Contig sequences were then processed with the MISA microsatellite identification program (http://pgrc.ipk-gatersleben.de/misa/) with the following repeat parameters: definition (unit size-minimum repeats): 2-12, 3-7, 4-5, 5-4, 6-3, 7-3, 8-2, 9-2, 10-2, 11-2, 12-2, 13-2, 14-2, 15-2; interruptions (maximum difference between 2 simple sequence repeats): 25.
Approximately 50 microsatellites of sufficient length and/or complexity were identified. Three (Sn1520, Sn1863 and Sn515) of these markers were not previously published and possessed sufficient non-redundant flanking sequence to allow for nested primer design and produced robust size-polymorphic PCR amplification products. Primers were validated as described [25] and found to be specific and sensitive for S. neurona DNA in tissues (data not shown). The primers designed are as follows: Sn1520 Fext- GGGGCAGAACCATCGTAGTA, Rext- GTGAAGCATTTCCCCTACGA, Fint- GGCGGTAGTCACTTGCTGA, Rint- GTGGGAGAAGACGGTCGTTA; Sn1863 Fext- CATGGCGTGCGTTAACTAAA, Rext- CGTACAAACACACGCTCCAC, Fint- CCATTCATCGACAGCGACTA, Rint- TGAGACAGCCGTCAAACACT; Sn515 Fext- CTTCTAGCGGCTGTTTCTCC, Rext- TCTGTGTGGGTGTGGAAGTC, Fint- GACCCCCTCTCTGCTTCTCT, Rint- ACGCAAATGCGAACATATCA. Representative sequences for each allele at each locus were placed in GenBank under the following accession numbers: Sn1520: HM851251, HM851252, HM851253, HM851254, HM851255; Sn1863: HM851256, HM851257, HM851258, HM851259; Sn515: HM851249, HM851250. PCR, DNA sequencing and analysis were conducted as described previously, except, to control for bias in scoring results, random sample IDs were assigned to samples before sequencing so that sequence analysis for some loci was blinded [25].
For this study, S. neurona DNA from 15 sea otters and 4 harbor seals was analyzed. Additionally, samples from 21 sea otters, 2 harbor seals, 3 horses, and 2 raccoons previously described by Wendte et al. [25] at the SnSAG antigen loci and MS Sn9, were further typed in this study at the remaining 10 MS loci. Finally, S. neurona DNA from 21 sea otters, 1 porpoise, 4 horses, 13 opossums, and 1 cat that was previously typed by Rejmanek et al. [23] at SnSAG3, SnSAG4, and MS markers Sn2–Sn5 and Sn7–Sn11 were combined with the data in this study for a total sample set that included 87 samples from 57 sea otters, 6 harbor seals, 2 raccoons, 13 opossums, 7 horses, 1 porpoise, and 1 cat. In all, 75 of the 87 samples were from California. Other states represented include Georgia (n = 2 samples), Illinois (n = 1), Missouri (n = 3), Washington (n = 4), and Wisconsin (n = 2). Some overlap existed between the samples typed in this study and those reported by Rejmanek et al.: samples SO4387, SO4413, H1, H2, and H3 in this study are reported as SO1, SO2, Horse 1, Horse 2, and Horse 3 in Rejmanek et al. [23], respectively. Complete information about the sample origins is found in Table S1.
Toxoplasma gondii isolates from a water cistern (n = 2), chickens (n = 11), and one cat associated with a human waterborne toxoplasmosis outbreak [44], as well as laboratory strain CEP were typed at microsatellite loci B17, B18, TgMA, TUB2, W35 [39] and M95 [40]. Markers were PCR amplified and sequenced to assign alleles as for S. neurona markers [25]. Representatives of each microsatellite allele at each locus were placed in Genbank under accession numbers: B17: HM851260–67; TgMA: HM851268–73; W35: HM851274–77; M95: HM851278–81.
Genotyping and eBURST analysis
Because different parts of the genome are likely under different selective pressures, all S. neurona samples were categorized by an antigen (Ag) type designated by roman numerals and a microsatellite (MS) type indicated by a lowercase letter designation. Ag types were defined by the presence/absence of mutually exclusive antigen genes (SnSAG1, SnSAG5, or SnSAG6) and the inheritance pattern of alleles at SnSAG3 and SnSAG4 [23], [25]. MS types were assigned on the basis of allele combinations defined by the number of di- or tri- nucleotide repeats at each locus (Sn2–Sn5 and Sn7–Sn11, Sn1520, Sn1863). Sn515 was a complex repeat in which each isolate possessed one of two alleles. Samples from the study by Rejmanek et al. [23] were not typed at the SnSAG1-5-6 loci, but were placed into Ag groups based on the allelic profile at SnSAG3 and SnSAG4 and by the Ag group their MS type was associated with in samples typed at all markers. For example, based on the alleles at SnSAG3 and SnSAG4, sample SO4 (Table S1) could be placed either in Ag type II or V, but its MS type was only found associated with Ag type II in samples where all markers were typed, making this the most likely, though not definitive, Ag type designation. The S. neurona strains assessed by Rejmanek et al. [23] were also not typed at MS markers Sn1520, Sn1863, and Sn515. Presumptively classifying these samples into MS types based on alleles at Sn2–Sn5 and Sn7–Sn11 is likely accurate, though, since these three markers did not provide additional resolution to MS types for the 46 additional S. neurona strains described in this study.
The alleles present at MS markers Sn2–Sn5 and Sn7–Sn11 were used for creation of a multi-locus sequence typing scheme by which all isolates could be compared. The numerical designation of alleles allowed the detection of which MS types formed clonal complexes using the eBURST program [48]. Default settings were used which grouped MS types on the basis of sharing alleles at 8 of the 9 markers analyzed.
To assess T. gondii isolates for clonality, MS alleles were combined with previously published DNA sequence analysis at three genetic loci, PCR-RFLP or DNA sequencing at 10 loci, and serologic analysis as described by Vaudaux et al. [44].
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 and χ2 values were considered significant at P = 0.05.
Supporting Information
Sarcocystis neurona genotypes and sample source information.
(0.02 MB PDF)
Sarcocystis neurona genotype presence over time in California.
(0.01 MB PDF)
Acknowledgments
Thanks to all members of the Grigg lab for helpful discussions and to Robin Miller for identifying the MISA program for microsatellite identification. Thank you to Pat Conrad and Dan Rejmanek, for kindly providing S. neurona strains SN1 and SN3 and primers for marker Sn4, and to J. P. Dubey for S. neurona strains isolated from two raccoons. Specimens were provided by various Northwest Marine Mammal Stranding Network participants including staff, interns, and volunteers from Washington Department of Fish and Wildlife, Cascadia Research Collective, and the Portland State University Mammal Stranding Network. MEG is a scholar of the Canadian Institute for Advanced Research (CIFAR) Program for Integrated Microbial Biodiversity.
Footnotes
The authors have declared that no competing interests exist.
The work was supported by the Intramural Research Program of the NIH and NIAID AI001018 and the National Science Foundation 0525750 (MEG); by the Howard Hughes Medical Institute Research Scholars Program and Morris Animal Foundation Wildlife Training Fellowship D10ZO-416 (JMW); by NOAA Fisheries Prescott grant program and WDFW (DML). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li W, Raoult D, Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009;33:892–916. doi: 10.1111/j.1574-6976.2009.00182.x. [DOI] [PubMed] [Google Scholar]
- 2.Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 3.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 4.Powers C, DeFilippis V, Malouli D, Fruh K. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol. 2008;325:333–359. doi: 10.1007/978-3-540-77349-8_19. [DOI] [PubMed] [Google Scholar]
- 5.Aires-de-Sousa M, Correia B, de Lencastre H. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J Clin Microbiol. 2008;46:2912–2917. doi: 10.1128/JCM.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amorim ML, Faria NA, Oliveira DC, Vasconcelos C, Cabeda JC, et al. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45:2881–2888. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci U S A. 2005;102:1968–1973. doi: 10.1073/pnas.0406993102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 10.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrnes EJ, III, Li W, Lewit Y, Ma H, Voelz K, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 13.Boyle JP, Rajasekar B, Saeij JP, Ajioka JW, Berriman M, et al. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc Natl Acad Sci U S A. 2006;103:10514–10519. doi: 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science. 2001;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- 15.Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 16.Jenni L, Marti S, Schweizer J, Betschart B, Le Page RW, et al. Hybrid formation between African trypanosomes during cyclical transmission. Nature. 1986;322:173–175. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- 17.Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annu Rev Microbiol. 2009;63:195–221. doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- 19.Su C, Evans D, Cole RH, Kissinger JC, Ajioka JW, et al. Recent expansion of Toxoplasma through enhanced oral transmission. Science. 2003;299:414–416. doi: 10.1126/science.1078035. [DOI] [PubMed] [Google Scholar]
- 20.Grigg ME, Sundar N. Sexual recombination punctuated by outbreaks and clonal expansions predicts Toxoplasma gondii population genetics. Int J Parasitol. 2009;39:925–933. doi: 10.1016/j.ijpara.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol. 2008;62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- 22.Asmundsson IM, Dubey JP, Rosenthal BM. A genetically diverse but distinct North American population of Sarcocystis neurona includes an overrepresented clone described by 12 microsatellite alleles. Infect Genet Evol. 2006;6:352–360. doi: 10.1016/j.meegid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Rejmanek D, Miller MA, Grigg ME, Crosbie PR, Conrad PA. Molecular characterization of Sarcocystis neurona strains from opossums (Didelphis virginiana) and intermediate hosts from Central California. Vet Parasitol. 2010;170:20–29. doi: 10.1016/j.vetpar.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundar N, Asmundsson IM, Thomas NJ, Samuel MD, Dubey JP, et al. Modest genetic differentiation among North American populations of Sarcocystis neurona may reflect expansion in its geographic range. Vet Parasitol. 2008;152:8–15. doi: 10.1016/j.vetpar.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, et al. Limited genetic diversity among Sarcocystis neurona strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Vet Parasitol. 2010;169:37–44. doi: 10.1016/j.vetpar.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 27.Dubey JP. History of the discovery of the life cycle of Toxoplasma gondii. Int J Parasitol. 2009;39:877–882. doi: 10.1016/j.ijpara.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Hide G, Morley EK, Hughes JM, Gerwash O, Elmahaishi MS, et al. Evidence for high levels of vertical transmission in Toxoplasma gondii. Parasitology. 2009;136:1877–1885. doi: 10.1017/S0031182009990941. [DOI] [PubMed] [Google Scholar]
- 29.Innes EA, Bartley PM, Buxton D, Katzer F. Ovine toxoplasmosis. Parasitology. 2009;136:1887–1894. doi: 10.1017/S0031182009991636. [DOI] [PubMed] [Google Scholar]
- 30.Miller M, Conrad P, James ER, Packham A, Toy-Choutka S, et al. Transplacental toxoplasmosis in a wild southern sea otter (Enhydra lutris nereis). Vet Parasitol. 2008;153:12–18. doi: 10.1016/j.vetpar.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Cornelissen AW, Overdulve JP. Sex determination and sex differentiation in coccidia: gametogony and oocyst production after monoclonal infection of cats with free-living and intermediate host stages of Isospora (Toxoplasma) gondii. Parasitology. 1985;90(Pt 1):35–44. doi: 10.1017/s003118200004899x. [DOI] [PubMed] [Google Scholar]
- 32.Pfefferkorn ER, Pfefferkorn LC, Colby ED. Development of gametes and oocysts in cats fed cysts derived from cloned trophozoites of Toxoplasma gondii. J Parasitol. 1977;63:158–159. [PubMed] [Google Scholar]
- 33.Miller MA, Conrad PA, Harris M, Hatfield B, Langlois G, et al. A protozoal-associated epizootic impacting marine wildlife: mass-mortality of southern sea otters (Enhydra lutris nereis) due to Sarcocystis neurona infection. Vet Parasitol. 2010;172:183–194. doi: 10.1016/j.vetpar.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace GD, Marshall L, Marshall M. Cats, rats, and toxoplasmosis on a small Pacific island. Am J Epidemiol. 1972;95:475–482. doi: 10.1093/oxfordjournals.aje.a121414. [DOI] [PubMed] [Google Scholar]
- 35.Dubey JP, Rollor EA, Smith K, Kwok OC, Thulliez P. Low seroprevalence of Toxoplasma gondii in feral pigs from a remote island lacking cats. J Parasitol. 1997;83:839–841. [PubMed] [Google Scholar]
- 36.Munday BL. Serological evidence of Toxoplasma infection in isolated groups of sheep. Res Vet Sci. 1972;13:100–102. [PubMed] [Google Scholar]
- 37.Wallace GD. Serologic and epidemiologic observations on toxoplasmosis on three Pacific atolls. Am J Epidemiol. 1969;90:103–111. doi: 10.1093/oxfordjournals.aje.a121054. [DOI] [PubMed] [Google Scholar]
- 38.de Moura L, Bahia-Oliveira LM, Wada MY, Jones JL, Tuboi SH, et al. Waterborne toxoplasmosis, Brazil, from field to gene. Emerg Infect Dis. 2006;12:326–329. doi: 10.3201/eid1202.041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajzenberg D, Dumetre A, Darde ML. Multiplex PCR for typing strains of Toxoplasma gondii. J Clin Microbiol. 2005;43:1940–1943. doi: 10.1128/JCM.43.4.1940-1943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackston CR, Dubey JP, Dotson E, Su C, Thulliez P, et al. High-resolution typing of Toxoplasma gondii using microsatellite loci. J Parasitol. 2001;87:1472–1475. doi: 10.1645/0022-3395(2001)087[1472:HRTOTG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 41.Dubey JP, Navarro IT, Graham DH, Dahl E, Freire RL, et al. Characterization of Toxoplasma gondii isolates from free range chickens from Parana, Brazil. Vet Parasitol. 2003;117:229–234. doi: 10.1016/j.vetpar.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Dubey JP, Navarro IT, Sreekumar C, Dahl E, Freire RL, et al. Toxoplasma gondii infections in cats from Parana, Brazil: seroprevalence, tissue distribution, and biologic and genetic characterization of isolates. J Parasitol. 2004;90:721–726. doi: 10.1645/GE-382R. [DOI] [PubMed] [Google Scholar]
- 43.Dubey JP, Velmurugan GV, Chockalingam A, Pena HF, de Oliveira LN, et al. Genetic diversity of Toxoplasma gondii isolates from chickens from Brazil. Vet Parasitol. 2008;157:299–305. doi: 10.1016/j.vetpar.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaudaux JD, Muccioli C, James ER, Silveira C, Magargal SL, et al. Identification of an atypical strain of Toxoplasma gondii as the cause of a waterborne outbreak of toxoplasmosis in Santa Isabel do Ivai, Brazil. J Infect Dis. 2010;202:1226–1233. doi: 10.1086/656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann T, Graham DH, Dahl ER, Bahia-Oliveira LM, Gennari SM, et al. Variation in the structure of Toxoplasma gondii and the roles of selfing, drift, and epistatic selection in maintaining linkage disequilibria. Infect Genet Evol. 2004;4:107–114. doi: 10.1016/j.meegid.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, et al. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin Infect Dis. 2007;45:e88–95. doi: 10.1086/521246. [DOI] [PubMed] [Google Scholar]
- 47.Elsheikha HM, Schott HC, 2nd, Mansfield LS. Genetic variation among isolates of Sarcocystis neurona, the agent of protozoal myeloencephalitis, as revealed by amplified fragment length polymorphism markers. Infect Immun. 2006;74:3448–3454. doi: 10.1128/IAI.01215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. Displaying the relatedness among isolates of bacterial species — the eBURST approach. FEMS Microbiol Lett. 2004;241:129–134. doi: 10.1016/j.femsle.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 50.Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arkush KD, Miller MA, Leutenegger CM, Gardner IA, Packham AE, et al. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis). Int J Parasitol. 2003;33:1087–1097. doi: 10.1016/s0020-7519(03)00181-4. [DOI] [PubMed] [Google Scholar]
- 53.Lindsay DS, Collins MV, Mitchell SM, Wetch CN, Rosypal AC, et al. Survival of Toxoplasma gondii oocysts in Eastern oysters (Crassostrea virginica). J Parasitol. 2004;90:1054–1057. doi: 10.1645/GE-296R. [DOI] [PubMed] [Google Scholar]
- 54.Miller MA, Miller WA, Conrad PA, James ER, Melli AC, et al. Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: new linkages between terrestrial mammals, runoff and toxoplasmosis of sea otters. Int J Parasitol. 2008;38:1319–1328. doi: 10.1016/j.ijpara.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Ebert EE. A Food Habits Study of Southern Sea Otter Enhydra Lutris Nereis. California Fish and Game. 1968;54:33–37. [Google Scholar]
- 56.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. doi: 10.1371/journal.pntd.0000056. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisher MC, Koenig GL, White TJ, Taylor JW. Pathogenic clones versus environmentally driven population increase: analysis of an epidemic of the human fungal pathogen Coccidioides immitis. . J Clin Microbiol. 2000;38:807–813. doi: 10.1128/jcm.38.2.807-813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butcher M, Lakritz J, Halaney A, Branson K, Gupta GD, et al. Experimental inoculation of domestic cats (Felis domesticus) with Sarcocystis neurona or S. neurona-like merozoites. Vet Parasitol. 2002;107:1–14. doi: 10.1016/s0304-4017(02)00107-3. [DOI] [PubMed] [Google Scholar]
- 59.Sibley LD. Recent origins among ancient parasites. Vet Parasitol. 2003;115:185–198. doi: 10.1016/s0304-4017(03)00206-1. [DOI] [PubMed] [Google Scholar]
- 60.Boughattas S, Ben-Abdallah R, Siala E, Souissi O, Aoun K, et al. Direct genotypic characterization of Toxoplasma gondii strains associated with congenital toxoplasmosis in Tunisia (North Africa). Am J Trop Med Hyg. 2010;82:1041–1046. doi: 10.4269/ajtmh.2010.09-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aspinall TV, Guy EC, Roberts KE, Joynson DH, Hyde JE, et al. Molecular evidence for multiple Toxoplasma gondii infections in individual patients in England and Wales: public health implications. Int J Parasitol. 2003;33:97–103. doi: 10.1016/s0020-7519(02)00230-8. [DOI] [PubMed] [Google Scholar]
- 62.Dubey JP, Lopez-Torres HY, Sundar N, Velmurugan GV, Ajzenberg D, et al. Mouse-virulent Toxoplasma gondii isolated from feral cats on Mona Island, Puerto Rico. J Parasitol. 2007;93:1365–1369. doi: 10.1645/GE-1409.1. [DOI] [PubMed] [Google Scholar]
- 63.Dubey JP, Moura L, Majumdar D, Sundar N, Velmurugan GV, et al. Isolation and characterization of viable Toxoplasma gondii isolates revealed possible high frequency of mixed infection in feral cats (Felis domesticus) from St Kitts, West Indies. Parasitology. 2009;136:589–594. doi: 10.1017/S0031182009006015. [DOI] [PubMed] [Google Scholar]
- 64.Dubey JP, Su C, Cortes JA, Sundar N, Gomez-Marin JE, et al. Prevalence of Toxoplasma gondii in cats from Colombia, South America and genetic characterization of T. gondii isolates. Vet Parasitol. 2006;141:42–47. doi: 10.1016/j.vetpar.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 65.Dubey JP, Sundar N, Pineda N, Kyvsgaard NC, Luna LA, et al. Biologic and genetic characteristics of Toxoplasma gondii isolates in free-range chickens from Nicaragua, Central America. Vet Parasitol. 2006;142:47–53. doi: 10.1016/j.vetpar.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 66.Dubey JP, Vianna MC, Sousa S, Canada N, Meireles S, et al. Characterization of Toxoplasma gondii isolates in free-range chickens from Portugal. J Parasitol. 2006;92:184–186. doi: 10.1645/GE-652R.1. [DOI] [PubMed] [Google Scholar]
- 67.Dubey JR, Bhaiyat MI, de Allie C, Macpherson CN, Sharma RN, et al. Isolation, tissue distribution, and molecular characterization of Toxoplasma gondii from chickens in Grenada, West Indies. J Parasitol. 2005;91:557–560. doi: 10.1645/GE-463R. [DOI] [PubMed] [Google Scholar]
- 68.Elbez-Rubinstein A, Ajzenberg D, Darde ML, Cohen R, Dumetre A, et al. Congenital toxoplasmosis and reinfection during pregnancy: case report, strain characterization, experimental model of reinfection, and review. J Infect Dis. 2009;199:280–285. doi: 10.1086/595793. [DOI] [PubMed] [Google Scholar]
- 69.Lindstrom I, Sundar N, Lindh J, Kironde F, Kabasa JD, et al. Isolation and genotyping of Toxoplasma gondii from Ugandan chickens reveals frequent multiple infections. Parasitology. 2008;135:39–45. doi: 10.1017/S0031182007003654. [DOI] [PubMed] [Google Scholar]
- 70.Ragozo AM, Pena HF, Yai LE, Su C, Gennari SM. Genetic diversity among Toxoplasma gondii isolates of small ruminants from Brazil: novel genotypes revealed. Vet Parasitol. 2010;170:307–312. doi: 10.1016/j.vetpar.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 71.Sundar N, Cole RA, Thomas NJ, Majumdar D, Dubey JP, et al. Genetic diversity among sea otter isolates of Toxoplasma gondii. Vet Parasitol. 2008;151:125–132. doi: 10.1016/j.vetpar.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 72.Parameswaran N, Thompson RC, Sundar N, Pan S, Johnson M, et al. Non-archetypal Type II-like and atypical strains of Toxoplasma gondii infecting marsupials of Australia. Int J Parasitol. 2010;40:635–640. doi: 10.1016/j.ijpara.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fux B, Nawas J, Khan A, Gill DB, Su C, et al. Toxoplasma gondii strains defective in oral transmission are also defective in developmental stage differentiation. Infect Immun. 2007;75:2580–2590. doi: 10.1128/IAI.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan A, Fux B, Su C, Dubey JP, Darde ML, et al. Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc Natl Acad Sci U S A. 2007;104:14872–14877. doi: 10.1073/pnas.0702356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dubey JP. Oocyst shedding by cats fed isolated bradyzoites and comparison of infectivity of bradyzoites of the VEG strain Toxoplasma gondii to cats and mice. J Parasitol. 2001;87:215–219. doi: 10.1645/0022-3395(2001)087[0215:OSBCFI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 76.Dubey JP. Comparative infectivity of oocysts and bradyzoites of Toxoplasma gondii for intermediate (mice) and definitive (cats) hosts. Vet Parasitol. 2006;140:69–75. doi: 10.1016/j.vetpar.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 77.Dubey JP, Lunney JK, Shen SK, Kwok OC, Ashford DA, et al. Infectivity of low numbers of Toxoplasma gondii oocysts to pigs. J Parasitol. 1996;82:438–443. [PubMed] [Google Scholar]
- 78.Mateus-Pinilla NE, Dubey JP, Choromanski L, Weigel RM. A field trial of the effectiveness of a feline Toxoplasma gondii vaccine in reducing T. gondii exposure for swine. J Parasitol. 1999;85:855–860. [PubMed] [Google Scholar]
- 79.Dubey JP, Lindsay DS, Saville WJ, Reed SM, Granstrom DE, et al. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM). Vet Parasitol. 2001;95:89–131. doi: 10.1016/s0304-4017(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 80.Mansfield LS, Mehler S, Nelsonc K, Elsheikha HM, Murphy AJ, et al. Brown-headed cowbirds (Molothrus ater) harbor Sarcocystis neurona and act as intermediate hosts. Vet Parasitol. 2008;153:24–43. doi: 10.1016/j.vetpar.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 81.Miller MA, Barr BC, Nordhausen R, James ER, Magargal SL, et al. Ultrastructural and molecular confirmation of the development of Sarcocystis neurona tissue cysts in the central nervous system of southern sea otters (Enhydra lutris nereis). Int J Parasitol. 2009;39:1363–1372. doi: 10.1016/j.ijpara.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Costa KS, Santos SL, Uzeda RS, Pinheiro AM, Almeida MA, et al. Chickens (Gallus domesticus) are natural intermediate hosts of Neospora caninum. Int J Parasitol. 2008;38:157–159. doi: 10.1016/j.ijpara.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Gondim LS, Abe-Sandes K, Uzeda RS, Silva MS, Santos SL, et al. Toxoplasma gondii and Neospora caninum in sparrows (Passer domesticus) in the Northeast of Brazil. Vet Parasitol. 2010;168:121–124. doi: 10.1016/j.vetpar.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 84.Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe. 2010;8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asmundsson IM, Rosenthal BM. Isolation and characterization of microsatellite markers from Sarcocystis neurona, a causative agent of equine protozoal myeloencephalitis. Mol Ecol Notes. 2006;6:8–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sarcocystis neurona genotypes and sample source information.
(0.02 MB PDF)
Sarcocystis neurona genotype presence over time in California.
(0.01 MB PDF)