Abstract
Editing/proofreading by aminoacyl-tRNA synthetases is an important quality control step in the accurate translation of the genetic code that removes noncognate amino acids attached to tRNA. Defects in the process of editing result in disease conditions including neurodegeneration. While proofreading, the cognate amino acids larger by a methyl group are generally thought to be sterically rejected by the editing modules as envisaged by the “Double-Sieve Model.” Strikingly using solution based direct binding studies, NMR-heteronuclear single quantum coherence (HSQC) and isothermal titration calorimetry experiments, with an editing domain of threonyl-tRNA synthetase, we show that the cognate substrate can gain access and bind to the editing pocket. High-resolution crystal structural analyses reveal that functional positioning of substrates rather than steric exclusion is the key for the mechanism of discrimination. A strategically positioned “catalytic water” molecule is excluded to avoid hydrolysis of the cognate substrate using a “RNA mediated substrate-assisted catalysis mechanism” at the editing site. The mechanistic proof of the critical role of RNA in proofreading activity is a completely unique solution to the problem of cognate-noncognate selection mechanism.
Keywords: aminoacyl-tRNA synthetases, editing, double-sieve model, enzyme mechanism, X-ray crystallography
Proofreading mechanisms play a crucial role in the faithful flow of information as dictated by the genetic code. Proofreading occurs at two important steps in the cell, one during DNA replication and another during translation, and is essential for the transfer of genetic information (1). Such mechanisms require that the noncognate substrate is specifically recognized while rejecting the cognate substrate at the editing site. A high fidelity is ensured during aminoacylation of tRNA by editing/proofreading modules either free standing or covalently attached to aminoacyl-tRNA synthetases (aaRSs) (2, 3). The proofreading activity is crucial as editing defective aaRS leads to misincorporation of noncognate amino acids into proteins. Such statistical proteins may have altered function or misfold triggering cellular apoptosis thereby leading to various diseases including neurodegeneration (4, 5). Moreover, editing defective aaRS can lead to oxidative stress (6) and also heritable genetic changes that can be linked with genetic diseases (7).
AaRSs charge cognate amino acid on the corresponding tRNA. However, the inherent ability of smaller or isosteric amino acids to bind in a pocket designed for the cognate amino acid poses an obstacle in the correct aminoacylation of tRNA, e.g., Val (noncognate) and Ile (cognate) in the case of Isoleucyl-tRNA synthetase (IleRS) or Thr (noncognate) and Val (cognate) in the case of Valyl-tRNA synthetase (ValRS). This fundamental intermolecular recognition problem on an important and essential biological process was originally invoked by Linus Pauling (8). Alan Fersht proposed a simple and elegant “Double-Sieve Model” for aaRSs that face such a scenario, explaining the fidelity of aminoacylation reaction (9). The Double-Sieve Model posits that the catalytic site acts as a coarse sieve that attaches cognate as well as smaller or isosteric noncognate amino acids on tRNA while rejecting amino acids that are larger than cognate. The editing domain acts as a “fine” sieve and selectively binds only to the noncognate amino acid removing it from tRNA. Therefore, aaRSs use the small structural differences between similar substrates twice thus obtaining “hyperspecificity” (10). Hence, chemical discrimination in the case of isosteric and steric discrimination in the case of smaller noncognate amino acids are considered to be the major factors determining the fidelity of this process (10–12).
The first structural basis of the Double-Sieve mechanism for amino acids differing by a methyl group came with the structures of IleRS in complex with Ile and Val. The electron density for Ile was observed in catalytic domain only whereas the electron density for Val was observed in both catalytic and editing domain in respective complexes (13). The absence of Ile from the editing pocket of IleRS was considered to be the result of steric rejection. On similar lines, the discrimination of cognate substrate, differing by a methyl group, at the editing site has been reported on the basis of steric hindrance for different editing modules of aaRSs over the last few years (13–17) and also explained in biochemistry text books (9, 18).
We reasoned that the substrates for editing domains are either noncognate aminoacyl-adenylate (aa-AMP) for pretransfer editing or noncognate aminoacyl-tRNA (aa-tRNA) for posttransfer editing whereas free noncognate amino acids are end products. The cognate aa-tRNA/aa-AMP is not hydrolyzed to form free cognate amino acid at the editing site. Thus, the fact that cognate amino acid could not be observed in the editing pocket is not surprising as it is neither the substrate nor the product of the reaction.
The active sites of editing domains have evolved for acting on aa-tRNA/aa-AMP rather than to bind free amino acids when compared with the catalytic domains. Hence, simplification of editing pockets as pockets for smaller noncognate amino acid may not accurately depict the scenario as the editing pockets are large enough to accommodate aa-AMP or amino acid attached to terminal adenosine of tRNA (14–17). An extra methyl group in the case of a free amino acid adds significantly to its bulkiness when compared with an extra methyl group on aa-tRNA/aa-AMP, where either the tRNA or the AMP is expected to dominate the binding. Therefore, we hypothesized that steric hindrance that is sufficient to discriminate against bigger amino acid at the catalytic site may not be adequate to reject cognate aa-AMP/aa-tRNA in the editing pocket as the adenosine/tRNA is expected to make several identical contacts with the enzyme when compared to noncognate aa-AMP/aa-tRNA. All aa-tRNAs are scanned by the cis-editing domains either by acceptor arm flipping or by tRNA resampling mechanism (19), where the aminoacylated-tRNA is captured back by the aaRS competing with elongation factors to recheck the accuracy. Therefore, we set out to understand the mechanism of how the editing domain discriminates cognate and noncognate aminoacyl moiety differing by a methyl group using solution based direct binding assays and high-resolution structural data.
Results
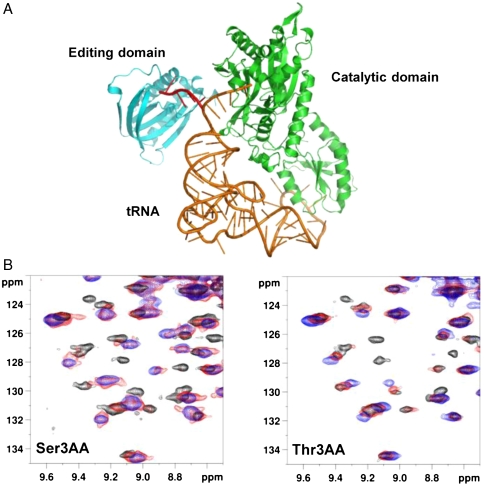
The editing domain of threonyl-tRNA synthetase (ThrRS) from Pyrococcus abyssi (Pab-NTD) was chosen for the study because of our ongoing studies on its editing mechanism (16, 20). ThrRS discriminates cognate threonine against noncognate smaller serine and isosteric valine. A conserved zinc ion present in the catalytic site completely prevents misactivation of isosteric valine (21) whereas smaller serine is mischarged on tRNAThr and is removed by its editing domain (15, 22, 23). Detailed mutational and structural analysis on Pab-NTD led us to elucidate the editing mechanism for the mischarged Ser-tRNAThr (16) (Fig. S1). Steric hindrance was proposed for the discrimination of Thr-tRNAThr based on structural analysis of Pab-NTD complexed with posttransfer substrate analog mimicking Ser-tRNAThr (16), as in the case of other editing domains (14, 15, 17). To capture the editing complexes, we have used nonhydrolysable posttransfer analogs mimicking Gly, Ser, and Thr charged on 3′ OH of tRNA, Gly3AA, Ser3AA, and Thr3AA respectively (Fig. 1A) and pretransfer analog, ThrAMS, mimicking Thr-AMP.
Fig. 1.
Binding of cognate (Thr3AA) and noncognate (Ser3AA) substrate analogs in the editing domain (Pab-NTD). (A): Ribbon representation of a model of archaeal ThrRS with tRNAThr to indicate that Ser3AA and Thr3AA mimic Ser-tRNAThr and Thr-tRNAThr respectively that flip from catalytic to editing domain of ThrRS. The flipping of 3′ end of tRNAThr to the editing domain is shown in red. (B): Superposition of 15N-1H HSQC excerpts obtained during titration of U -15N labeled Pab-NTD (200 μM) with increasing concentrations (0 μM: Black, 100 μM: Red, 500 μM: Blue) of Ser3AA and Thr3AA.
Binding Studies of Cognate and Noncognate Substrate Analogs in Editing Domain.
The binding of substrate analogs in U-15N labeled Pab-NTD was initially screened by two-dimensional 15N-1H HSQC experiments. As expected, titration of Ser3AA showed chemical shift perturbations for several resonances indicating its binding. However, surprisingly, titration of Thr3AA showed similar chemical shift perturbations for several resonances to that observed in the case of Ser3AA titration (Fig. 1B and Fig. S2A). The finding showed that the cognate substrate is not sterically excluded from binding to the editing pocket. The striking similarity in the pattern of chemical shift perturbations implies that the site of binding of Ser3AA and Thr3AA in the editing pocket of Pab-NTD is identical.
In order to find out the difference in the binding affinities of Ser3AA and Thr3AA with Pab-NTD, we performed direct binding studies using isothermal titration calorimetry (ITC) experiments. Ser3AA showed a binding to Pab-NTD with a dissociation constant (Kd) of 3.4 μM whereas Thr3AA showed a weaker binding with a Kd in the range of 36.2 μM (Table 1 and Fig. S2B). It is difficult to assume that the discrimination of cognate substrate is based only on a 10-fold difference in affinity of cognate and noncognate substrates as this would mean hydrolysis of a considerable fraction of the pool of cognate Thr-tRNAThr. However, deacylation of Thr-tRNAThr by the editing domain of ThrRS is not observed (15, 16, 22, 23) suggesting the presence of an alternate strategy in the editing domain to discriminate the cognate substrate.
Table 1.
The thermodynamic parameters of binding of substrate analogs with Pab-NTD carried out at T = 303 K
| Ligand | ΔH Kcal/mole | −TΔS Kcal/mole | ΔG = ΔH - TΔS Kcal/mole | Kd μM |
| Ser3AA | -7.49 ± 0.20 | −0.01 | −7.50 | 3.4 ± 0.3 |
| Thr3AA | -0.86 ± 0.08 | −5.30 | −6.16 | 36.2 ± 10.0 |
| Gly3AA | -9.58 ± 0.62 | 3.33 | −6.25 | 33.3 ± 2.0 |
Structural Complexes of Editing Domain of ThrRS with Cognate and Noncognate Substrate Analogs.
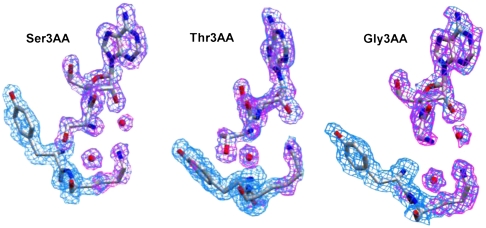
In order to know the exact mode of binding and the discrimination mechanism of Thr-tRNAThr, high-resolution crystal structures of Pab-NTD in complex with Ser3AA and Thr3AA were solved at 1.86 Å resolution. Ser3AA structure was rerefined using data collected from improved quality crystals that resulted in a higher resolution and a better redundant dataset in the same space group as Thr3AA to avoid crystal contact artifacts (Table S1). The electron density for the analogs and water molecules are extremely clear in the unbiased difference maps (Fig. 2). Different ratio of protein:ligand used in cocrystallization experiments show that Thr3AA can be captured in the editing pocket even at 1∶4 ratio as compared to 1∶2 for Ser3AA (Table S2). Multiple datasets for each complex were collected and the best datasets are presented here as the overall picture presented by all is identical as described below (Table S3).
Fig. 2.
Unbiased electron density of Ser3AA, Thr3AA and Gly3AA in the editing pocket of Pab-NTD. Unbiased electron density maps (Fo - Fc map at 2.0σ in pink and 2Fo - Fc map at 1.0σ in blue) for substrate analogs, W1, W2, Tyr120, and Lys121 side chain in respective complexes. No density corresponding to W1 was observed in both Fo - Fc and 2Fo - Fc maps in Pab-NTD-Thr3AA complex.
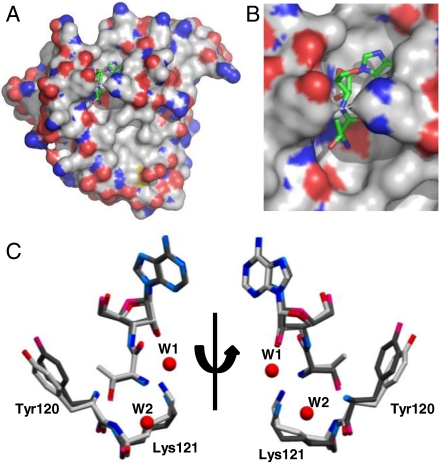
The Pab-NTD-Thr3AA complex presents a glimpse of the cognate substrate analog captured in an editing domain of the translational machinery (Fig. 3). The binding mode of adenosine as well as aminoacyl moiety of Thr3AA is similar to that of Ser3AA in Pab-NTD and is predominantly mediated through main chain atoms lining the editing pocket (Fig. S3 A and B). The side chains of invariant Val45, Ala82, Ala94, and Phe117 define the adenine pocket while Phe117 also provides a hydrophobic platform for binding the adenosine moiety. The α-amino groups as well as the side chain hydroxyls of Ser3AA and Thr3AA are recognized similarly in both the complexes by a conserved water molecule, W2, as well as main chain atoms of Pro80 and Lys121 respectively (Fig. S3 A and B). In Thr3AA complex, there is only a minor movement of main chain as well as side chain atoms of residues in the editing pocket. However, a subtle repositioning of the threonyl moiety of Thr3AA and a small rotation around its C-Cα axis, when compared with Ser3AA allows the extra methyl (γ-methyl) group in Thr3AA to be accommodated in the pocket without any steric hindrance. The γ-methyl group of threonyl moiety is in the range of van der Waals interaction distance (3.5–4.1 Å) with Tyr120 Cα as well as its side chain. Tyr120 side chain is comparatively moved away (0.54–1.39 Å) from the pocket in Thr3AA complex because of the extra methyl group interaction (Fig. 3C).
Fig. 3.
Cognate posttransfer analog captured in the editing domain of archaeal ThrRS. (A): Surface representation of Pab-NTD-Thr3AA complex. Thr3AA is shown in stick representation bound to the editing pocket of Pab-NTD. (B): A closer view of the bound Thr3AA in the editing pocket. (C): Superposition of the Pab-NTD-Ser3AA complex (darker shade) on Pab-NTD-Thr3AA complex (lighter shade) shows similar positioning of Ser3AA and Thr3AA as well as of residues in the editing pocket. A closer view of the superposition highlighting the movement of the side chains of Tyr120 and Lys121 is shown. Subtle change in the aminoacyl moiety of Ser3AA and Thr3AA can also be seen. W1 and W2 shown in the figure correspond to Pab-NTD-Ser3AA complex. W1 is absent in Pab-NTD-Thr3AA complex.
The most crucial repositioning is that of the invariant Lys121 side chain that comes closer to a distance of 3.39 Å and 3.80 Å with 2′ OH of Thr3AA in the two monomers present in the asymmetric unit (Fig. 3C). Such a movement of Lys121 side chain towards the ribose 2′ OH is not observed in both monomers of Ser3AA complexes where the distance between 2′ OH and Lys121 ϵ-amino group are 4.38 Å and 4.06 Å. As a result of this movement, the water molecule (W1) (Fig. S1) that is observed in Ser3AA complex is expelled out in Thr3AA complex (Fig. 2 and Table S4). W1 was earlier proposed to be essential for the removal of serine attached to tRNAThr and the Thr3AA complex provides direct evidence that in its absence the cleavage reaction does not proceed. The absence of catalytic water explains the discriminatory mechanism through a nonproductive mode of binding for Thr3AA in Pab-NTD. It is important to note that the extra methyl group of Thr-tRNAThr does not occupy the position of catalytic water or pose any steric hindrance to it. Thus, the Lys121 side chain that positions the catalytic water W1 along with 2′ OH of ribose in the case of Ser3AA complex is repositioned in Thr3AA complex and sterically excludes the catalytic water (Fig. S3C). The movements caused by the extra methyl group of threonine on Tyr120 are transmitted to Lys121 directly influencing the repositioning observed. The subtle repositioning presents an elegant mechanism by which nature discriminates a cognate aminoacyl-tRNA at the editing site.
Smaller Noncognate Substrates in the Editing Domain.
As smaller substrates are expected to bind in a pocket designed for bigger substrates, Gly3AA also showed clear binding with Pab-NTD with a Kd of 33.3 μM albeit with a 10-fold reduction when compared to Ser3AA (Table 1). We determined the Pab-NTD-Gly3AA complex at 2.4 Å (Fig. 2). The Cα of Gly3AA is positioned away when compared with that of Ser3AA suggesting the role of side chain in appropriate anchoring of substrate in the editing pocket (Fig. S3D). The Lys121 side chain is positioned as that observed in the case of Ser3AA along with the catalytic water (W1) coordinated by 2′ OH of the ribose, indicating that tRNAThr mischarged with glycine can be deacylated. However, it has been shown that the catalytic domain of ThrRS does not misactivate glycine to detectable levels (21) and Gly-tRNAThr may not be formed in the real scenario. Therefore, this study gives us a perspective of binding of smaller aminoacyl moiety in the editing pocket. More importantly, the Pab-NTD-Gly3AA complex further confirms the earlier conclusion that repositioning of Lys121 side chain to expel the catalytic water is due to the presence of extra methyl group in the case of Thr3AA complex.
The invariant Lys121 in Pab-NTD plays a crucial role in the recognition of the substrate as all its anchoring points; i.e., α-amino group, main chain carbonyl, and side chain ϵ-amino group are utilized to interact with the side chain hydroxyl, α-amino group, and 2′ OH of the analog respectively, either directly or by water-mediated interaction (Fig. S3E and Table S5). The side chain ϵ-amino group also makes a van der Waals interaction with the Cα of the analog. These interactions are conserved in all the complexes suggesting a decisive role played by Lys121 in recognition of the substrate and positioning or expelling the catalytic water accordingly.
Furthermore, we also determined the structure of Pab-NTD with cognate pretransfer analog, Thr-AMS. The complex shows that Thr-AMS is bound with its threonyl moiety positioned out of the editing pocket (Fig. S4). Similar conformation for Ser-AMP analog, SerAMS, was observed earlier suggesting lack of pretransfer editing in this editing module (16) (Fig. S4).
Discussion
The study opens up an interesting question on the fundamental mechanism of discrimination of cognate amino acids at the proofreading step. The editing domains have been proposed to discriminate noncognate amino acids that are isosteric or smaller in size by chemical or steric discrimination mechanisms, respectively. For example, in the case of ValRS, that discriminates Val against isosteric Thr, the editing domain uses chemical discrimination as the cognate and noncognate amino acids differ in the polarity of the side chains. Similar strategy is also used by PheRS to discriminate Phe against Tyr and by AlaRS to discriminate Ala against Ser. However, when the chemistry of the side chains cannot be exploited for discrimination then the size of the side chain comes into picture as in the case of IleRS, LeuRS, and ThrRS. Generally, cognate amino acids larger by a methyl group are thought to be sterically rejected from binding during proofreading (3, 9, 11–17). Our results show that steric rejection of cognate substrate by editing domains may not be sufficient, at least in a few cases. We show that the penalty of binding Thr-tRNA in a pocket designed for Ser-tRNA at the editing site is around 50-fold less than that observed for Thr in a pocket designed for Ser at the catalytic site in Seryl-tRNA synthetase (24). It goes in accordance with the current view that proteins, particularly their side chains, are more dynamic in nature rather than being considered having a rigid position. The solution based binding studies in conjunction with the high-resolution structural information presented here reaffirms such a subtle, but critical, mobility of residues lining the active site. While the binding affinities at the editing site are lower by 10-fold for Thr3AA/Gly3AA when compared to Ser3AA, underlining the selection mechanism, it does not completely rule out the binding of cognate substrate. Moreover, Ser3AA and Thr3AA are anchored in a near identical conformation in the editing pocket as opposed to different nonproductive conformations proposed for the binding of bigger noncognate amino acids in the catalytic domain (24). Therefore, as originally envisaged in the double-sieve model, the structural differences between Ser and Thr have been used by the proofreading module to present a productive vs nonproductive mode of binding and thus avoiding the removal of the cognate amino acid from the tRNA.
In this context, the case of CP1 domain, responsible for proofreading in various Class I aaRSs, is interesting as it is used to discriminate Val/Leu/Ile differing by a methyl group. Moreover, nature has designed changes in this editing pocket to reject a particular amino acid in one context and recognize it in another; e.g., Ile is recognized in editing domain of Leucyl-tRNA synthetase (LeuRS), whereas it is rejected at the editing site of IleRS. The structural complexes of CP1 domains with pre and posttransfer substrate analogs show that the hydrophobic side chains of substrate analogs are anchored by weak van der Waals interactions only and thereby adopt different conformations within the editing pocket (14, 17) (Fig. S5). Although, each structure independently explains the cognate discrimination mechanism, our analysis of all the known substrate complexes together suggests that the editing pocket in both Class I and Class II aaRSs may bind the cognate substrates (Fig. S5). It would be interesting to have a holistic view of the discrimination mechanism in all proofreading modules and could any other editing domain share a similar mechanistic mode of discrimination for cognate substrate like Pab-NTD remains to be explored.
Furthermore, the possibility of Pab-NTD being an exception in the mode of cognate amino acid discrimination is not ruled out, because of its proposed evolutionary link with D-aminoacyl-tRNA deacylase (DTD) (16, 20, 25). The unique mode of discrimination might suggest that the primordial editing domains utilized similar means of discrimination of cognate and noncognate substrate that has been preserved in archaeal ThrRS. Another possibility is that the editing domain of ThrRSs have been evolved also for rejecting valine, as it is isosteric to threonine, in addition to serine and hence the module binds threonine. However, considering the fact that valine is not activated by the catalytic domain to detectable levels and that both serine and threonine hydroxyl groups are involved in a network of hydrogen bonding interactions in the editing site, it is unlikely to be the scenario. In any case, aaRSs have been the paradigm for studying enzyme specificity and the strategy of cognate discrimination in Pab-NTD presents a unique mode of discriminating substrates. It is worth noting that nature is finding it difficult to evolve a design mechanism for a pocket to sterically exclude Thr from Ser completely, particularly when it is part of a larger molecule, like tRNA, and rather rely on an intricate mode of discrimination. This paradox apparently arose due to large editing pockets to accommodate its substrates where steric hindrance is insufficient to totally discriminate substrates differing by a methyl group. The mechanistic analysis underscores a unique mode for cognate substrate discrimination while proofreading by aaRS for maintaining a high fidelity during translation of the genetic code.
Our mechanistic study shows the crucial role played by the tRNA in substrate-assisted catalysis by positioning the catalytic water along with protein side chain (Lys121). Such an active role played by the tRNA molecule, which is the substrate itself, seems to be essential for providing the discriminatory role by excluding the catalytic water. It is interesting to note that tRNA-mediated proofreading of noncognate substrate has been implicated in PheRS (26) and CP1 domain of LeuRS (27). Moreover, a thorough analysis of all the structural complexes of proofreading modules provides strong evidence that such a mechanism may exist in other editing modules as well. Thus, the mechanistic proof of the critical role of RNA, through substrate-assisted catalysis, to the problem of cognate-noncognate substrate selection is a completely unique solution for proofreading during translation.
Overall, the study shows that cognate substrate can be accommodated in the editing pocket thus questioning the paradigm that it is rejected by the editing domains. The study also implies that these mechanisms are designed in such a tipping point that a mutation in the editing modules even far away from the editing site could lead to deleterious consequences especially in the light of a recent study that showed that proofreading modules of aaRS recheck the fidelity of the aminoacylation process by capturing the elongation factor bound aminoacylated-tRNA before it reaches the ribosomes (19). Because cognate aa-tRNAs are normally enriched in the cellular pool, in conjunction with our study, it would imply that a small mutational perturbation in the editing domain may result in hydrolysis of the cognate amino acid from tRNA thus depleting the correctly charged tRNA pool which results in disease conditions (28). Generally, editing defects are defined on the basis of efficiency of deacylation of mischarged tRNA. However, another important aspect of editing is the ability to not to act on correctly charged tRNA. Any mutation that leads to deacylation of the cognate aa-tRNA or even worse if a cross reaction occurs with other species of aa-tRNA, it would deplete its cellular pool resulting in disease conditions.
Materials and Methods
Binding Studies.
The nonhydrolyzable analog Thr3AA was bought from Jena Biosciences while Ser3AA, Gly3AA, and ThrAMS were purchased from RNA-Tech. The binding of substrate analogs with Pab-NTD was checked using HSQC titrations and chemical shift perturbations in U-15N Pab-NTD upon titration of analogs were monitored. ITC was used to determine the thermodynamic parameters of these bimolecular interactions. The experimental details are mentioned in the SI Text.
Crystallization, Structure Determination, and Refinement.
Pab-NTD was purified and the complexes were crystallized in similar conditions as mentioned earlier (16). The X-ray diffraction datasets were collected on the in-house X-ray facility. The structures were solved by molecular replacement using the Pab-NTD coordinates (PDB ID: 1Y2Q). Detailed descriptions of the structure determination of the complexes are mentioned in the SI Text.
Accession Codes.
The coordinates of Ser3AA, Thr3AA, Gly3AA and ThrAMS complex with Pab-NTD have been deposited in the Protein Data Bank with access codes 3PD2, 3PD3, 3PD4, and 3PD5 respectively.
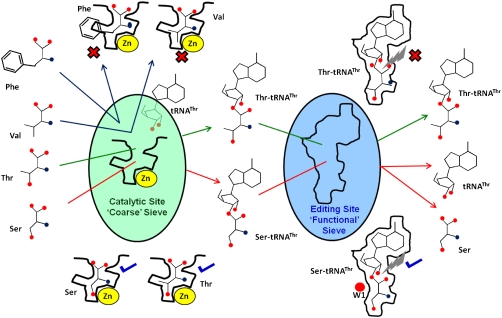
Fig. 4.
Double-sieve model with reference to Archaeal ThrRS. The catalytic site acts as a coarse sieve and discriminates against larger Phe based on size and isosteric Val as it lacks β-hydroxyl group. Cognate Thr as well as noncognate smaller Ser is (mis)charged on tRNAThr to form Thr-tRNAThr or Ser-tRNAThr, respectively. The Editing domain acts as a fine “functional” sieve hydrolyzing only Ser-tRNAThr into Ser and tRNAThr and not cognate Thr-tRNAThr.
Supplementary Material
Acknowledgments.
T.H. thanks University Grant Commission (UGC), India for Senior Research Fellowship. R.S. acknowledges funding from Swarnajayanti Fellowship of DST, India. The authors declare no competing financial interests.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org.
See Commentary on page 21949.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014299107/-/DCSupplemental.
References
- 1.Jakubowski H, Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev. 1992;56:412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 4.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 5.Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008;17:1643–1652. doi: 10.1110/ps.037242.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacher JM, Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci USA. 2006;104:1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauling L. In: The probability of errors in the process of synthesis of protein molecules, In Festschrift für Pr. Arthur Stoll., editor. Basel: Birkhauser Verlag; 1957. pp. 597–602. [Google Scholar]
- 9.Fersht AR. Enzyme structure and mechanism. San Francisco: Freeman; 1977. [Google Scholar]
- 10.Fersht AR, Kaethner MM. Enzyme hyperspecificity. Rejection of threonine by Valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 11.Fersht AR, Dingwall C. Evidence for the double-sieve editing mechanism in protein synthesis. Steric exclusion of isoleucine by valyl-tRNA synthetases. Biochemistry. 1979;18:2627–2631. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- 12.Fersht AR. Sieves in sequence. Science. 1998;280:541. doi: 10.1126/science.280.5363.541. [DOI] [PubMed] [Google Scholar]
- 13.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 14.Lincecum TL, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 15.Dock-Bregeon A-C, et al. Achieving error-free translation: the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16:375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Hussain T, et al. Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. EMBO J. 2006;25:4152–4162. doi: 10.1038/sj.emboj.7601278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukunaga R, Yokoyama S. Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J Mol Biol. 2006;359:901–912. doi: 10.1016/j.jmb.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: W.H. Freeman and Co.; 2002. [Google Scholar]
- 19.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwivedi S, Kruparani SP, Sankaranarayanan R. A D-amino acid editing module coupled to the translational apparatus in archaea. Nature Struct Mol Biol. 2005;12:556–557. doi: 10.1038/nsmb943. [DOI] [PubMed] [Google Scholar]
- 21.Sankaranarayanan R, et al. Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nat Struct Biol. 2000;7:461–465. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- 22.Beebe K, Merriman E, Pouplana LR, Schimmel P. A domain for editing by an archaebacterial tRNA synthetase. Proc Natl Acad Sci USA. 2004;101:5958–5963. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korencic D, et al. A freestanding proofreading domain is required for protein synthesis quality control in archaea. Proc Natl Acad Sci USA. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilokapic S, et al. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006;25:2498–2509. doi: 10.1038/sj.emboj.7601129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferri-Fioni M-L, et al. Structure of crystalline D-Tyr-tRNATyr deacylase. J Biol Chem. 2001;276:47285–47290. doi: 10.1074/jbc.M106550200. [DOI] [PubMed] [Google Scholar]
- 26.Ling J, Roy H, Ibba M. Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci USA. 2007;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagiwara Y, Field MJ, Nureki O, Tateno M. Editing mechanism of aminoacyl-tRNA synthetases operates by a hybrid ribozyme/protein catalyst. J Am Chem Soc. 2010;132:2751–2758. doi: 10.1021/ja9095208. [DOI] [PubMed] [Google Scholar]
- 28.Ling J, et al. Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc Natl Acad Sci USA. 2007;104:15299–15304. doi: 10.1073/pnas.0704441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.