Abstract
Small proteins can fold remarkably rapidly, even in μs. What limits their rate of folding? The Engrailed homeodomain is a particularly well-characterized example, which folds ultrafast via an intermediate, I, of solved structure. It is a puzzle that the helix2-turn-helix3 motif of the 3-helix bundle forms in approximately 2 μs, but the final docking of preformed helix1 in I requires approximately 20 μs. Simulation and structural data suggest that nonnative interactions may slow down helix docking. Here we report the direct measurement of chain motions in I by using photoinduced electron transfer fluorescence-quenching correlation spectroscopy (PET-FCS). We use a mutant that traps I at physiological ionic strength but refolds at higher ionic strength. A single Trp in helix3 quenches the fluorescence of an extrinsic label on contact with it. We placed the label along the sequence to probe segmental chain motions. At high ionic strength, we found two relaxations for all probed positions on the 2- and 20-μs time scale, corresponding to the known folding processes, and a 200-ns phase attributable to loop closure kinetics in the unfolded state. At low ionic strength, we found only the 2-μs and 200-ns phase for labels in the helix2-turn-helix3 motif of I, because the native state is not significantly populated. But for labels in helix1 we observed an additional approximately 10-μs phase showing that it was moving slowly, with a rate constant similar to that for overall folding under native conditions. Folding was rate-limited by chain motions on a rough energy surface where nonnative interactions constrain motion.
Ultrafast folding proteins fold at close to the limits expected for the rates of chain diffusion on an energy landscape (1, 2). Measuring and understanding the underlying rate constants for these processes are fundamental to understanding the nature of protein-folding landscapes. Yet, because of the time regimes and difficulties in measuring these processes in proteins, data are scant (2–5). Drosphila melanogaster Engrailed homeodomain (EnHD) is a member of the homeodomain superfamily (6). EnHD is a 61-residue three-helix bundle comprising a helix-turn-helix motif, formed by helix 1 and helix 2 and a third helix docking onto the helix-turn-helix motif in perpendicular orientation. The protein folds in microseconds via the formation of a folding intermediate. Temperature jump (T-jump) experiments reveal biphasic kinetics on the 2-μs and 20-μs time scale corresponding to the formation of a folding intermediate and folding into the native state, respectively (7–9). A single-point mutation (L16A) destabilizes the native state, N, so that at physiological ionic strength it predominantly takes up the structure of the on-pathway folding intermediate, I, of the wild-type protein in equilibrium with a small fraction of more highly denatured state, U (9, 10). That intermediate of EnHD-L16A refolds at high salt concentrations, which shield repulsive positive charges in the protein. The simplified scheme of EnHD folding is: U↔I↔N.
Mutation L16A traps EnHD in the intermediate state along its folding pathway at physiological ionic strength, affording a rare, atomic-detailed structural characterization of a folding intermediate in solution by NMR spectroscopy (8). The solution structure of I has the helix2-turn-helix3 motif of the native protein to be intact, but helix1 is longer and in an ill-defined position without nuclear Overhauser enhancements (NOEs) with the motif (8) (Fig. 1A). The protein is poised to fold by a classical framework mechanism (11). Surprisingly, the 20-μs folding time constant observed in relaxation experiments (7, 9) for the final docking of helix1 on the motif is 10 times larger than that for formation of the fully folded motif in the intermediate. The wealth of previous experimental data on EnHD combined with the use of a new technique, here, namely photoinduced electron transfer fluorescence-quenching correlation spectroscopy (PET-FCS) (12), now allow a detailed analysis of rates of chain motions in a protein domain and show what is rate determining in the slow step of folding—the segmental motion of helix1.
Fig. 1.
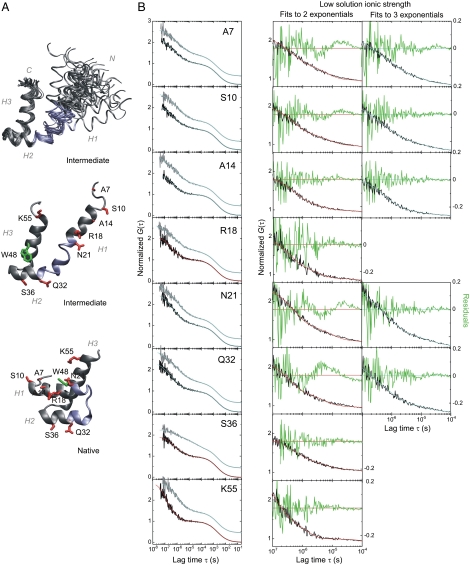
PET-FCS experiments of EnHD. (A) NMR solution structure of EnHD-L16A at low ionic strength (Upper and Middle) and of native EnHD (Lower). (Upper) Bundle of the 25 lowest-energy conformers of EnHD-L16A (PDB ID 1ZTR) in backbone representation with helices (H) 1, 2, and 3 indicated. (Middle) Lowest-energy structure of EnHD-L16A. Residue side chains modified to introduce the fluorophore AttoOxa11 are highlighted red. Trp48 is highlighted green. (Lower) Structure of native EnHD (PDB ID 2JWT) with the helix2-turn-helix3 motif in identical orientation as shown for EnHD-L16A. The chain segment between H1 and H2 that undergoes local refolding is highlighted blue in all three representations. (B) PET-FCS of EnHD-L16A. The left-hand column of panels shows ACFs recorded of each probe position at 2,000 mM (high) and 145 mM (low) solution ionic strength, in gray and black, respectively. Data are offset along the y-axis for reasons of clarity. The best data fits to a model for a single diffusing species with either three or two single-exponential relaxations are shown as cyan or red lines, respectively. The two columns of panels on the right side show close-ups of the sub-ms time domain of ACFs recorded at low solution ionic-strength and fitted to either two or three single-exponential relaxations (red or cyan, respectively). Residuals of data fits are shown in green (with y-axis on right). The three-exponential data fits from probe positions R18, S36, and K55 at low ionic strength did not yield physically meaningful results and were discarded (not shown).
Results
Design of a Fluorescence Reporter System for EnHD Chain Motions.
Oxazine fluorophores are quenched selectively by tryptophan (Trp) in proteins via photoinduced electron transfer (PET) from the Trp side chain to the fluorophore at van der Waals contact (13). By placing fluorophore and Trp at selected sites in a protein, we can transform conformational fluctuations into fluorescence fluctuations. Fluorescence correlation spectroscopy (FCS) analyzes fluorescence fluctuations at equilibrium arising from individual molecules diffusing through a confocal detection volume by Brownian motion (14, 15). By combining PET fluorescence quenching with FCS we can detect and analyze kinetics of protein chain motions and fast folding at equilibrium by correlation analysis (12, 16).
EnHD contains a single Trp at sequence position 48, which is largely buried in the native protein but solvent exposed in the denatured state and folding intermediate. We introduced the oxazine fluorophore AttoOxa11 via cysteine/maleimide chemistry individually at eight selected sites spanning the sequence of EnHD (Fig. 1A). The selection of sites for cysteine mutation was guided by protein structure analysis with the aim of minimizing perturbations. Measurements from far-UV circular dichroism (CD) spectroscopy suggested that the protein modifications did not perturb the folding equilibrium (Fig. S1), as did the kinetic analysis below. We could then detect and analyze the rate constants of side chain collisions between individual sites along the sequence of EnHD and Trp48 using PET-FCS, in order to derive a picture of the protein’s segmental chain motions during folding. We performed experiments at high and low solution ionic strength (2,000 mM and 145 mM, respectively) where previous studies have shown that the native state is either near-fully populated or the protein has insignificant native structure, respectively (8).
PET-FCS Experiments at High Solution Ionic Strength.
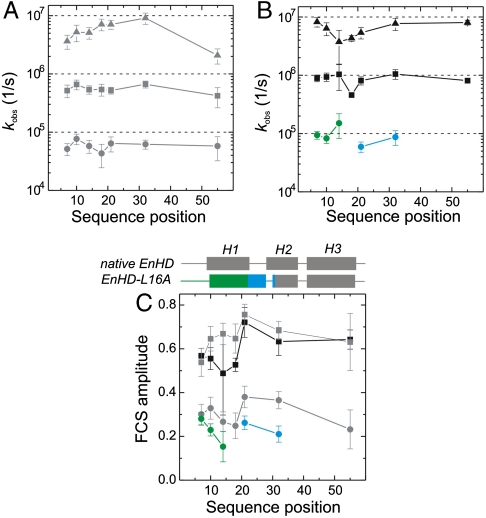
Autocorrelation functions (ACFs) recorded at 2,000 mM solution ionic strength exhibited complex kinetics for all eight probe positions, which arose from conformational fluctuations of EnHD-L16A during the approximately 2-ms passage through the confocal detection volume (Fig. 1B). Data recorded from a mutant where Trp48 was replaced by phenylalanine (EnHD-L16A-W48F, Fig. S2) showed a single decay on the ms time scale in the ACF that reported on translational diffusion, demonstrating that AttoOxa11/Trp48 fluorescence quenching was the exclusive origin of all additional sub-ms decays observed. The ACFs of EnHD-L16A were fitted to an FCS model containing the minimal number of exponential relaxations required to obtain randomly distributed residuals (Fig. 1B). All ACFs recorded at high ionic strength required a fitting model for a single diffusing species with three independent, single-exponential relaxations (Materials and Methods). The observed relaxations were essentially independent of probe position. The first two relaxations occurred at approximately 20 μs and approximately 2 μs (Fig. 2) and could be assigned to folding and formation of the intermediate with observed rate constants of kI-N = 55,000 ± 8,000 s-1 and kU-I = 550,000 ± 80,000 s-1, respectively, in agreement with quantities obtained from T-jump relaxation experiments of unlabeled wild-type EnHD (7) and mutant L16A (8, 9) (Table 2). The probe at position 36 had kinetics significantly slower than values derived from all other positions and previous T-jump experiments (Table 1). This finding indicated a mutation- and/or probe-induced perturbation at this position, and so the data from this mutant were not used for quantitative analysis. We observed an additional relaxation of approximately 200 ns for all probe positions. Such a kinetic phase was observed previously in infrared (IR) T-jump experiments but remained unassigned (9). The observed ns phase in PET-FCS is a signature of the unfolded state and reflects the rate constant of intrachain contact formation or loop closure, kU, in a coiled protein chain (16, 17). Taking into account differences in flexibility of interior compared with terminal chain segments (18), the observed loop closure kinetics resembled those determined for unfolded model peptides of similar size (19). Loop closure within the C-terminal sequence part, however, was surprisingly slow (kU(K55) = (2.1 ± 0.6) × 106 s-1) and might have resulted from some specific interactions introduced by the probe or local, transient formation of helical structure in the unfolded state of EnHD.
Fig. 2.
Sequence position-specific kinetics of protein chain motions. (A) Observed rate constants of folding (I-N, circles), of formation of the intermediate (U-I, squares), and of loop closure within the unfolded state (U, triangles) from PET-FCS data at high ionic strength (gray), plotted as function of sequence position. (B) Observed rate constants at low ionic strength (black) plotted as function of sequence position. Triangles and squares represent kinetics within U and of U-I, as shown in A. Green and blue circles reflect kinetics of helix1 motions within the intermediate and local refolding, respectively. (C) Observed amplitudes of FCS decays. Helical secondary structure in the sequence of native EnHD and its folding intermediate is illustrated on the top of the panel. Symbols and color code are the same as in A and B. All error bars are standard errors from data fits.
Table 2.
Average relaxation times for EnHD-L16A from PET-FCS, and IR-monitored T-jump kinetics for overall folding of L16A and fragment 16–59
| Method, conditions | τI-N(10-5 s) | τU-I(10-6 s) | τU(10-7 s) |
| FCS, high salt * | 1.8 ± 0.1 | 1.8 ± 0.3 | 2.2 ± 0.5 |
| IR, T-jump high salt † | 1.8 ± 0.6 | 3.2 ± 0.6 | 3.2 ± 0.5 |
| IR, T-jump high salt 16–59† | - | 3.6 ± 0.4 | 2.5 ± 0.3 |
| FCS, low salt * | - | 1.3 ± 0.2 | 1.8 ± 0.2 |
| IR, T-jump low salt † | - | 1.9 ± 0.3 | 2.5 ± 0.4 |
| IR, T-jump low salt 16–59 † | - | 2.5 ± 0.4 | 0.83 ± 0.03 |
*Kinetic parameters from fitted PET-FCS data of EnHD-L16A recorded at 25 °C averaged over all positions except 36. τI-N is the relaxation time for docking of helix1, τU-I for the relaxation of the folding intermediate and τU for motions in the denatured state.
†Values from ref. 9 for folding of EnHD-L16A and the fragment 16–59, which contains the helix-turn-helix motif but lacks Helix1.
Table 1.
Time constants for relaxations in the folding intermediate of EnHD-L16A *
| Sequence position | aI-N | τI-N(10-5 s) | aU-I | τU-I(10-6 s) | aU | τU(10-7 s) |
|
aI |
τI(10-5 s) |
|||||
| 7 (N term) | 0.30 ± 0.05 | 1.9 ± 0.5 | 0.54 ± 0.06 | 1.9 ± 0.5 | 0.40 ± 0.07 | 2.8 ± 0.7 |
| 0.28 ± 0.03 | 1.1 ± 0.2 | 0.57 ± 0.03 | 1.1 ± 0.2 | 0.45 ± 0.04 | 1.2 ± 0.2 | |
| 7† | 0.18 ± 0.02 | 2.3 ± 0.5 | 0.43 ± 0.03 | 2.1 ± 0.3 | 0.42 ± 0.03 | 1.8 ± 0.3 |
| 10 (H1) | 0.33 ± 0.05 | 1.3 ± 0.3 | 0.65 ± 0.06 | 1.5 ± 0.3 | 0.33 ± 0.06 | 1.9 ± 0.6 |
| 0.23 ± 0.03 | 1.2 ± 0.2 | 0.55 ± 0.05 | 1.1 ± 0.2 | 0.38 ± 0.05 | 1.6 ± 0.4 | |
| 14(H1) | 0.27 ± 0.05 | 1.7 ± 0.4 | 0.67 ± 0.05 | 1.9 ± 0.3 | 0.42 ± 0.05 | 2.0 ± 0.4 |
| 0.15 ± 0.07 | 0.6 ± 0.3 | 0.49 ± 0.19 | 1.0 ± 0.5 | 0.30 ± 0.22 | 2.7 ± 1.5 | |
| 18(H1) | 0.25 ± 0.06 | 2.3 ± 1.0 | 0.65 ± 0.07 | 1.9 ± 0.4 | 0.56 ± 0.06 | 1.4 ± 0.3 |
| n.o. | n.o. | 0.53 ± 0.03 | 2.2 ± 0.5 | 0.57 ± 0.03 | 2.3 ± 0.3 | |
| 21(H1) | 0.38 ± 0.05 | 1.6 ± 0.3 | 0.67 ± 0.05 | 1.9 ± 0.3 | 0.51 ± 0.04 | 1.4 ± 0.3 |
| 0.26 ± 0.03 | 1.7 ± 0.4 | 0.72 ± 0.07 | 1.2 ± 0.2 | 0.55 ± 0.07 | 1.9 ± 0.4 | |
| 32(H2) | 0.36 ± 0.04 | 1.6 ± 0.3 | 0.68 ± 0.04 | 1.5 ± 0.2 | 0.45 ± 0.04 | 1.1 ± 0.2 |
| 0.21 ± 0.04 | 1.2 ± 0.3 | 0.63 ± 0.06 | 1.0 ± 0.2 | 0.52 ± 0.06 | 1.3 ± 0.3 | |
| 36(H2) | 0.31 ± 0.07 | 5.9 ± 1.7 | 0.18 ± 0.06 | 9 ± 6 | 0.32 ± 0.05 | 10 ± 2 |
| n.o. | n.o. | 0.32 ± 0.02 | 6.3 ± 0.9 | 0.63 ± 0.03 | 5.9 ± 0.6 | |
| 55(H3) | 0.23 ± 0.09 | 1.7 ± 0.8 | 0.63 ± 0.13 | 2.3 ± 0.9 | 0.56 ± 0.16 | 4.8 ± 1.4 |
| n.o. | n.o. | 0.64 ± 0.04 | 1.2 ± 0.1 | 1.26 ± 0.06 | 1.3 ± 0.1 |
*Kinetic parameters from fitted PET-FCS data of EnHD-L16A recorded at 25 °C at high (bold, upper of each pair of cells) and low (lower of each pair of cells) ionic strength. The relaxation time constant τ equals 1/k. a denotes the amplitude of the observed relaxation.
†Values measured in low ionic-strength solution containing 15% (w/v) sucrose as viscogen. Sucrose did not induce any detectable structural perturbation in EnHD-L16A as judged by far-UV CD spectroscopy (Fig. S3B). n.o.: not observed—fits to two exponentials.
PET-FCS Experiments at Low Solution Ionic Strength.
ACFs recorded at 145 mM ionic strength, where the native state is insignificantly populated and the protein predominantly the intermediate, differed from those recorded at high ionic strength (Fig. 1B). ACFs fitted well to a model for a single diffusing species with either three, as at high salt, or now only two independent single-exponential relaxations, depending on the probed position. The kinetic parameters of all probe positions extracted from data fits are shown in Fig. 2 B and C and are summarized in Table 1. While probes at position 14, 18, 36, and 55 fitted well to a model for a single diffusing species with two single-exponential relaxations, data from positions 7, 10, 21, and 32 required three single-exponential decays. The probe at position 14 fitted equally well to the two- or three-exponential model. An approximately 2-μs (k = 800,000 ± 200,000 s-1) and approximately 200-ns relaxation (k = (6.2 ± 1.8) × 106 s-1) were present in all constructs, similar to that observed at high ionic strength, and were essentially independent of probe position (Fig. 1B). The approximately 2-μs relaxation was in good agreement with kinetic quantities for formation of the intermediate determined previously at identical low ionic-strength solution conditions by T-jump spectroscopy (7, 9) (Table 2). The additional third kinetic phase from probes at position 7, 10, 21, and 32 was of varying amplitude and appeared between 5 and 15 μs (Figs. 1B and 2 B and C). This kinetic phase cannot result from overall folding because N was insignificantly populated at those solvent conditions (7, 8). Far-UV CD spectroscopy showed that protein modifications did not restabilize the native state either (Fig. S1). The observed third relaxation occurred for residues in helix1, which is in an ill-defined orientation relative to the folded helix2-turn-helix3 motif in the intermediate as revealed by NMR spectroscopy (8) (Fig. 1A). The amplitude of this relaxation increased as the probe was moved toward the N terminus, coinciding with the level of uncertainty with which the orientation of helix1 could be assigned by NMR spectroscopy due to the lack of NOEs (Figs 1 and 2C). The corresponding observed rate constants, kI = 60,000–150,000 s-1, can thus be assigned to chain motions within the intermediate. kI was only slightly faster than the rate constant for overall folding that involves docking of helix1 onto the helix2-turn-helix3 motif (9) (Fig. 2 A and B and Table 1).
All three rate constants were affected by solvent viscosity: A 60% increase of viscosity, caused by the viscogen sucrose, lowered the rate constants by approximately 50% (Table 1 and Fig. S3A), suggesting that the detected chain motions are diffusive.
Discussion
Proteins fold by chain diffusion on an energy surface (2). Conventional laboratory kinetics usually measures the overall rate determining steps in structure formation. Here, we measured, using PET-FCS, relaxation times at equilibrium of chain motions in EnHD, a protein of very well characterized folding pathway. The complete assignment of these relaxations to physical processes would not be possible on the basis of the FCS-PET data alone. But combining our previously published data from experiment and simulation with the new results from fluorescence fluctuation analysis on a series of mutants and structural perturbation by salt, we can assign the processes with confidence. Also, by making experiments on many mutants, we have built-in controls that should detect artefacts.
Previously, using laser T-jump methods on the folding of EnHD, we found two major events: the formation of the helix2-turn-helix3 motif, with 1/k of approximately 2 μs and the docking of helix1 at approximately 20 μs, detectable from overall changes in Trp fluorescence and backbone IR absorption on folding. Experiments on the folding intermediate isolated the rate constant for formation of the helix2-turn-helix3 motif (9). The 2- and 20-μs time constants determined here from substitutions at seven positions were in excellent agreement with those previously measured by T-jump monitored IR absorption on unmodified protein (Table 2), allowing their assignment to the overall folding processes and also showing that the probes did not perturb the kinetics, apart from an eighth mutation at position 36. The very fast relaxation of 200–300 ns corresponds to chain motions in unfolded states (16).
Whereas conventional T-jump experiments tend to detect the major rate determining processes associated with folding, PET-FCS monitors all intrachain processes that lead to contact between the dye and quencher. PET-FCS will detect relaxations in the unfolded state where the chain segments are free to diffuse, the relaxations arising from processes that lead to unfolding, and any quenching chain motions in folded or partly folded states. There were key observations from the effects of perturbation of folding equilibria by salt concentration on PET-FCS. (i) At high ionic strength where EnHD-L16A is predominantly folded there were in all cases three relaxations that corresponded to fluorescence quenching in the unfolded state via the slow undocking of helix1 in 20 μs, followed by the unfolding of the helix2-turn-helix3-motif in 2 μs and intrachain collisions in the unfolded state in 200–300 ns. (ii) At low ionic strength EnHD-L16A has helix1 undocked and just the helix2-turn-helix3-motif as well-defined tertiary fold. For residues in the native-like parts of the helix2-turn-helix3-motif, just the 2-μs and 200–300-ns relaxations were observed, as expected. But (iii) residues in helix1, the N terminus, and the plastic region between helix1 and helix2 had, at low ionic strength, an approximately 10-μs relaxation. That process must clearly arise from segmental motions that allow quenching in the folding intermediate from collisions of the probe at various positions with Trp48.
The 10-μs time constant for that motion is very similar to that for the docking of helix1 under conditions where the protein is folded. The time constant for docking appears, therefore, to be that for the segmental diffusion of helix1 over the energy surface. It is evident from NMR that some nonnative interactions have to be broken for helix1 to be able to dock onto the preformed helix2-turn-helix3 motif in order to fold (8). The C-terminal end of helix1 and the N-terminal end of helix2 are elongated and shortened, respectively, in the intermediate compared to the native structure. Both structural elements contain nonnative contacts and have to refold in the intermediate in order to form a turn in native EnHD (8) (Figs. 1A and 2C). This local refolding event rationalizes the appearance of additional kinetics at sequence positions 21 and 32 in the folding intermediate, which frame the refolding segment, showing that these sites are mobile. We note that we avoided modifications within the corresponding segment because they introduce substantial energetic perturbations (7). Further, the comparatively slow time constant of chain motions of helix1 within the intermediate, initiating folding under native conditions, can be explained by breaking of the nonnative interactions. This interpretation is also supported by very recent computer simulations (20).
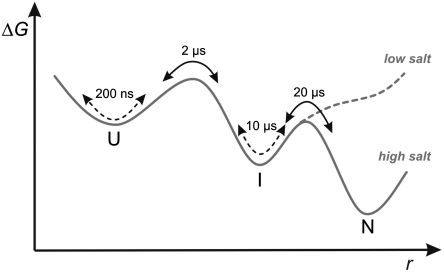
We can now fill in the kinetic data in refined detail to add to the structural information found from NMR, ϕ-analysis, and atomistic simulation. The elementary events during folding of EnHD are illustrated in a simplified free-energy profile in Fig. 3. Folding starts by diffusive chain motions on the 200-ns time scale within a largely unfolded protein, followed by cooperative formation of the folding intermediate in approximately 2 μs comprising the helix2-turn-helix3 motif and helix1 in ill-defined position. Preformed helix1 in the intermediate moves slowly at approximately 10 μs because it involves breaking of some nonnative interactions and docks onto the helix2-turn-helix3 motif in the rate determining step. The time scales of these processes are most suitable for benchmarking computer simulation.
Fig. 3.
Simplified free-energy profile of EnHD folding. A two-dimensional projection of the free-energy surface is shown along an arbitrary reaction coordinate, r. Kinetics accessible to conventional relaxation experiments are illustrated as black arrows. Chain motions within populated states detected by PET-FCS are illustrated as broken black arrows. At low solution ionic strength EnHD-L16A is trapped in the denatured state ensemble, comprising U and I.
Materials and Methods
Protein Synthesis and Modification.
EnHD was overexpressed in Escherichia coli C41 (DE3) cells and purified as described elsewhere (10). Single-point cysteine mutants of EnHD-L16A were modified using the maleimide derivative of the fluorophore AttoOxa11 (Atto-Tec GmbH). An eight-fold molar excess of fluorophore yielded quantitative modification. Labeling reactions were carried out for 2 h at 25 °C in 50 mM 3-(morpholino)propane-sulfonic acid (MOPS), pH 7.0, containing 6 M GdmCl, and a 10-fold molar excess of tris(2-carboxyethyl)phosphine (TCEP) to prevent oxidation of cysteine side chains. Labeled protein was purified to homogeneity using size-exclusion chromatography.
Sample Preparation and Bulk Measurements.
All measurements were performed at 25 °C in filtered solutions (0.22 μm syringe filters) containing 50 mM sodium acetate, pH 5.7, with the solution ionic strength adjusted to 145 mM (low salt) or 2,000 mM (high salt) using sodium chloride. Far-UV CD spectra of 10-μM protein samples were recorded on a thermostatted Jasco J-815 spectropolarimeter.
Solution viscosities were measured using an automated, thermostatted falling-ball micro-viscometer (Anton Paar).
PET-FCS.
PET-FCS experiments were performed on a home-built, confocal fluorescence microscope setup. The setup consisted of a Nikon Eclipse TE2000-U microscope body equipped with a high numerical aperture objective lens (Nikon, Plan Apo VC 60x/1.40 oil) and a He-Ne laser at 633 nm (Melles Griot) as excitation source. The excitation power was adjusted to 400 μW before entering the back aperture of the lens, low enough to prevent photo-physical artefacts in the ACFs. The temperature was adjusted to 25 °C using a custom-built objective-type heater. The fluorescence signal of the sample was collected by the same objective lens and optically and spatially filtered using a dichroic mirror (Semrock, BrightLine FF 494/540/650), a 150-μm pinhole, and a long-pass cut-off filter (Semrock, RazorEdge 647 nm). A 50% cubic, nonpolarizing beam splitter (ThorLabs) was used to partition fluorescence photons between two fiber-coupled avalanche photo-diode detectors (Perkin Elmer, SPCM-AQR-15-FC) to overcome detector dead time and after-pulsing effects in ns experiments. ACFs were recorded in the cross-correlation mode using a digital hardware correlator device (Flex02-01D, Correlator.com). Sample/glass surface interactions in LabTek sample chambers (Nunc) were suppressed by surface passivation using poly-L-lysine hydrobromide (Sigma-Aldrich), and 0.3 mg/ml BSA and 0.05% Tween-20 as additives in buffered solutions containing 1 nM labeled protein. The accumulated measurement time for each dataset was 30 min.
ACFs, G(τ), were fitted to a model for a diffusion in two dimensions of a single species with either two or three additional single-exponential relaxations (21, 22):
where τ is the lag time, N is the average number of molecules in the detection focus, τD is the translational diffusion time constant, and an and τn are observed amplitude and time constant, respectively, of the nth relaxation. For a two- and a three-exponential model, n is 2 and 3. A two-dimensional model for translational diffusion is of sufficient accuracy for detection foci where the horizontal dimension (x, y) is much smaller than the lateral (z) one, which was the case in the used setup. All ACFs shown in Fig. 1 are normalized to N for reasons of clarity.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011666107/-/DCSupplemental.
References
- 1.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 3.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Bieri O, Kiefhaber T. Elementary steps in protein folding. Biol Chem. 1999;380:923–929. doi: 10.1515/BC.1999.114. [DOI] [PubMed] [Google Scholar]
- 5.Cellmer T, Henry ER, Hofrichter J, Eaton WA. Measuring internal friction of an ultrafast-folding protein. Proc Natl Acad Sci USA. 2008;105:18320–18325. doi: 10.1073/pnas.0806154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: A framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 7.Mayor U, et al. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 8.Religa TL, Markson JS, Mayor U, Freund SM, Fersht AR. Solution structure of a protein denatured state and folding intermediate. Nature. 2005;437:1053–1056. doi: 10.1038/nature04054. [DOI] [PubMed] [Google Scholar]
- 9.Religa TL, et al. The helix-turn-helix motif as an ultrafast independently folding domain: The pathway of folding of Engrailed homeodomain. Proc Natl Acad Sci USA. 2007;104:9272–9277. doi: 10.1073/pnas.0703434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayor U, Grossmann JG, Foster NW, Freund SM, Fersht AR. The denatured state of Engrailed homeodomain under denaturing and native conditions. J Mol Biol. 2003;333:977–991. doi: 10.1016/j.jmb.2003.08.062. [DOI] [PubMed] [Google Scholar]
- 11.Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 12.Neuweiler H, Doose S, Sauer M. A microscopic view of miniprotein folding: Enhanced folding efficiency through formation of an intermediate. Proc Natl Acad Sci USA. 2005;102:16650–16655. doi: 10.1073/pnas.0507351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuweiler H, et al. Detection of individual p53-autoantibodies by using quenched peptide-based molecular probes. Angew Chem Int Ed Engl. 2002;41:4769–4773. doi: 10.1002/anie.200290044. [DOI] [PubMed] [Google Scholar]
- 14.Magde D, Elson E, Webb WW. Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- 15.Rigler R, Mets U, Widengren J, Kask P. Fluorescence correlation spectroscopy with high count rate and low background: Analysis of translational diffusion. Eur Biophys J Biophy. 1993;22:169–175. [Google Scholar]
- 16.Neuweiler H, Johnson CM, Fersht AR. Direct observation of ultrafast folding and denatured state dynamics in single protein molecules. Proc Natl Acad Sci USA. 2009;106:18569–18574. doi: 10.1073/pnas.0910860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuweiler H, Lollmann M, Doose S, Sauer M. Dynamics of unfolded polypeptide chains in crowded environment studied by fluorescence correlation spectroscopy. J Mol Biol. 2007;365:856–869. doi: 10.1016/j.jmb.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Fierz B, Kiefhaber T. End-to-end vs interior loop formation kinetics in unfolded polypeptide chains. J Am Chem Soc. 2007;129:672–679. doi: 10.1021/ja0666396. [DOI] [PubMed] [Google Scholar]
- 19.Krieger F, Fierz B, Bieri O, Drewello M, Kiefhaber T. Dynamics of unfolded polypeptide chains as model for the earliest steps in protein folding. J Mol Biol. 2003;332:265–274. doi: 10.1016/s0022-2836(03)00892-1. [DOI] [PubMed] [Google Scholar]
- 20.McCully ME, Beck DAC, Fersht AR, Daggett V. Refolding the Engrailed homeodomain: Structural basis for the accumulation of a folding intermediate. Biophys J. 2010;99:1628–1636. doi: 10.1016/j.bpj.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 22.Krichevsky O, Bonnet G. Fluorescence correlation spectroscopy: The technique and its applications. Rep Prog Phys. 2002;65:251–297. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.