Maintaining genome stability is a fundamental aspect of life. It is critical for proper gene expression and transmission. However, the genome is also highly dynamic, with alterations occurring in various forms during cell growth, division, and/or differentiation. One important aspect of these alterations is in gene dosage. For instance, gene duplications or amplifications have been known to occur aberrantly in somatic tissues and can cause diseases such as cancer (1). When gene duplication occurs in the germ line, it is thought to serve as a major driving force in genome evolution (2). Interestingly, amplifications of individual genes have also been known to occur as regulated developmental processes in many organisms, presumably to produce more gene products needed at specific stages of the life cycle (3–5). The mechanisms underlying these processes are not entirely clear and have been the subjects of extensive investigations for several decades. The articles by Nowacki et al. (6) and Heyse et al. (7) in PNAS reveal a surprising aspect of gene dosage variations in two species of ciliated protozoa, Stylonychia lemnae and Oxytricha trifallax, in which most genes exist as unlinked molecules or nanochromosomes in their somatic nuclei (8). It seems that changes in the copy number of these genes can be transmitted through sexual reproduction, setting up an unusual inheritance of somatic information, and RNA somehow serves as the messenger in this mother to daughter communication.
Ciliates live an interesting genetic life. This group of single-celled eukaryotes typically contains two types of nuclei, the micro- and macronucleus, which are the products of germ/soma differentiation. During the sexual process of conjugation, the micronucleus goes through meiosis, cross-fertilization, and mitosis to generate new micro- and macronucleus for the following vegetative life cycle, and the old macronucleus, being a somatic nucleus, degenerates and disappears at the end of this process. The differentiation of the new macronucleus involves an amazing array of regulated DNA rearrangement processes. Depending on the species, they can include internal DNA deletion, DNA fragmentation, telomere addition, inverted gene duplication, gene amplification, gene unscrambling, and endo-replication. These rearrangements are relatively modest in species such as Tetrahymena (9) but can be quite extensive in Oxytricha and Stylonychia, in which more than 90% of the genome is discarded and the retained DNA is fragmented into molecules averaging 2–3 kb in sizes, sufficient to contain just one or very few genes (8). This streamlined, highly polyploid somatic genome is responsible for essentially all transcriptional activities during the following vegetative life cycle. Lacking typical mitotic features such as centromeres and a spindle apparatus, the macronucleus divides amitotically. With essentially all genes being unlinked to each other, individual genes are presumably free to vary in dosage during vegetative growth and division, generating perhaps one of the most extreme cases of gene copy number control in biology.
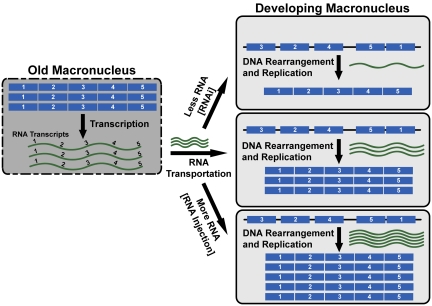
Recent studies on ciliate genomes have revealed the surprising involvement of RNA in DNA rearrangement. In Tetrahymena and Paramecium, internal DNA deletions occur at thousands of specific sites, and the process is guided by RNA in mechanisms highly related to RNAi (10–12). Injection of double-stranded RNA during conjugation actually induced the deletion of the corresponding genomic sequence (12). In Oxytricha, RNA is also found to guide the remarkable gene unscrambling process (Fig. 1). In this and other stichotrichous ciliates including Stylonychia, many genes in the germline genome are scrambled: they are interrupted by segments that need to be deleted, and the remaining segments are arranged out of order and often in the wrong orientation, which have to be reordered and assembled to create a functional gene every time a new somatic genome is formed during conjugation (13). The genetic information necessary for this gene unscrambling process apparently comes from the parental somatic genomes, which contain properly ordered genes already unscrambled in the previous conjugation. They make RNA copies that presumably travel to the developing new macronucleus to guide DNA rearrangements. Injection of RNA can indeed affect this process and creates artificially rearranged genes according to the mutated RNA templates injected (14). Studies of these rearrangement processes have now led to another surprise. The RNA encoded by the parental somatic nanochromosomes not only guides DNA rearrangements, but it also affects the copy numbers of these processed, gene-sized chromosomes in the offspring.
Fig. 1.
Parental RNA regulates somatic DNA copy numbers. During conjugation of Oxytricha and Stylonychia, the developing macronucleus (Right) rearranges its inherited genome to create functional genes. This rearrangement is guided by RNA (green wavy lines) made in the old macronucleus (Left), in which the DNA has already being rearranged in previous conjugation, and transported to the new macronucleus (Middle Right). Injection of extra RNA increased (Bottom Right) and RNAi treatments decreased (Top Right) the copy number of rearranged DNA, showing a role for RNA in regulating somatic DNA copy numbers.
To show this point, Nowacki et al. (6) inject RNA (transcribed in vitro) into conjugating cells of Oxytricha at a time before macronuclear differentiation begins and analyze the progeny produced (Fig. 1). The effects are significant: injection of RNA from either or both strands increased corresponding gene copy numbers in the macronucleus of the offspring by about threefold for telomere binding protein β-gene and 5- to 12-fold for ActinI gene. The endogenous RNA that guides DNA rearrangements can also be reduced by RNAi treatments as previously shown. When this was performed for two other genes, telomere binding protein α-gene and DNA polymerase α-gene, DNA copy numbers in the offspring were reduced by 1.8- and 7.7-fold, respectively. Importantly, these changes in copy number seemed heritable through sexual reproductions. Two of three injected clones produced progeny with elevated somatic gene copy numbers on conjugation, showing the epigenetic inheritance of this somatic property. In the accompanying paper, Heyse et al. (7) report a similar phenomenon in Stylonychia, although the effects on copy numbers are lower (within twofold) after RNAi treatments or template RNA injections.
Epigenetic inheritance of gene dosages might have an interesting biological role.
It is not clear how the RNA actually affects DNA copy number in this process. A possible explanation, as offered by Heyse et al. (7), is that it might act through gene rearrangement. Higher amounts of template RNA could promote or speed up the rearrangement process, thus producing successfully rearranged genes sooner and conferring an advantage in the next endoreplication phase to produce more DNA copies. In this scenario, the main molecular action of the RNA is in guiding DNA rearrangements, and its influence on DNA copy number would be a secondary effect. However, in the absence of more data, other possibilities remain open. Transacting RNA has long been known to regulate DNA copy number of ColE1 plasmids in Escherichia coli (15). Nanochromosomes of these ciliates are small free molecules much like plasmids. It is interesting that their copy numbers also seem to be modulated by transacting RNA, although the mechanism remains to be deciphered.
This epigenetic inheritance of gene dosages might have an interesting biological role. The special genomic structure of these ciliates offers remarkable potentials for genome-wide gene dosage variations within a somatic lifespan. Indeed, copy numbers of different nanochromosomes in a culture have been known to differ by many folds, and the copy number of a given gene can also change with the age of the culture (16–18). The study by Heyse et al. (7) further confirms this variation in several genes. It is conceivable that, through cell division and DNA assortment, individual gene copy numbers can vary as a result of selection and adaptation in the open environment of these free-living ciliates, thus producing a somatic genome with the most advantageous gene dosage combination for the niche. When a new macronucleus is formed through conjugation, such a desirable dosage composition is recreated using the process described here, thereby facilitating the immediate success of their progeny. Thus, using the special genetic tools that they have, these ciliates may have found a way to ensure the proper upbringing of their offspring in the ever-changing environment in which they live.
Acknowledgments
I wish to thank Mr. Yu-sheng Lin for his help in preparing the references and figure. The work in this laboratory on DNA rearrangements is supported by grants from the National Science Council of Taiwan and intramural funding of Academia Sinica.
Footnotes
The author declares no conflict of interest.
See companion articles on pages 22134 and 22140.
References
- 1.Tanaka H, Yao MC. Palindromic gene amplification—an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- 2.Korbel JO, et al. The current excitement about copy-number variation: How it relates to gene duplications and protein families. Curr Opin Struct Biol. 2008;18:366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- 4.Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oögenesis. Proc Natl Acad Sci USA. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spradling AC. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27:193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- 6.Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci USA. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyse G, Jönsson F, Chang W-J, Lipps HJ. RNA-dependent control of gene amplification. Proc Natl Acad Sci USA. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: An RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- 10.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 12.Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- 13.Greslin AF, Prescott DM, Oka Y, Loukin SH, Chappell JC. Reordering of nine exons is necessary to form a functional actin gene in Oxytricha nova. Proc Natl Acad Sci USA. 1989;86:6264–6268. doi: 10.1073/pnas.86.16.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eguchi Y, Tomizawa J. Complex formed by complementary RNA stem-loops and its stabilization by a protein: Function of CoIE1 Rom protein. Cell. 1990;60:199–209. doi: 10.1016/0092-8674(90)90736-x. [DOI] [PubMed] [Google Scholar]
- 16.Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- 17.Harper DS, Song K, Jahn CL. Overamplification of macronuclear linear DNA molecules during prolonged vegetative growth of Oxytricha nova. Gene. 1991;99:55–61. doi: 10.1016/0378-1119(91)90033-8. [DOI] [PubMed] [Google Scholar]
- 18.Steinbruck G. Overamplification of genes in macronuclei of hypotrichous ciliates. Chromosoma. 1983;88:156–163. [Google Scholar]