Abstract
We exploit the unusual genome organization of the ciliate cell to analyze the control of specific gene amplification during a nuclear differentiation process. Ciliates contain two types of nuclei within one cell, the macronucleus and the micronucleus; and after sexual reproduction a new macronucleus is formed from a micronuclear derivative. During macronuclear differentiation, most extensive DNA reorganization, elimination, and fragmentation processes occur, resulting in a macronucleus containing short DNA molecules (nanochromosomes) representing individual genetic units and each being present in high copy number. It is believed that these processes are controlled by small nuclear RNAs but also by a template derived from the old macronucleus. We first describe the exact copy numbers of selected nanochromosomes in the macronucleus, and define the timing during nuclear differentiation at which copy number is determined. This led to the suggestion that DNA processing and copy number control may be closely related mechanisms. Degradation of an RNA template derived from the macronucleus leads to significant decrease in copy number, whereas injection of additional template molecules results in an increase in copy number and enhanced expression of the corresponding gene. These observations can be incorporated into a mechanistic model about an RNA-dependent epigenetic regulation of gene copy number during nuclear differentiation. This highlights that RNA, in addition to its well-known biological functions, can also be involved in the control of gene amplification.
Keywords: epigenetics, copy number, genome organization, nuclear differentiation, ciliates
Selective amplification of genes is frequently observed in differentiating eukaryotic cells. This amplification may also occur spontaneously under selective conditions or during transformation of cells. Gene amplification generally results in an increase in the number of transcripts and gene products in a gene dosage-dependent manner (1). Examples are amplification of rRNA genes in Xenopus oocytes (2–4), amplification of chorion genes after polytene chromosome formation in ovarian follicle cells of diptera (5), or amplification of the DHFR locus observed under methotrexate selection (6). Various mechanisms for gene amplification have been reported: a rolling circle replication resulting in many extrachromosomal copies, as described for the rDNA genes in Xenopus oocytes (7); amplification by initial double strand breaks, followed by sister chromatid fusion and repeated breakage–fusion–bridge cycles, as in the case of the DHFR locus in CHO cells (4, 8); or selective activations of replication origins within the same S-phase, leading to an onion-skin–like structure observed during amplification of the chorion genes in diptera (9). In general, initiation and regulation of gene amplification processes are still poorly understood, but a correlation with chromatin structure, transcriptional activity, or, in the case of rDNA in Tetrahymena, the function of a noncoding RNA in origin recognition has been described (1, 4, 10).
In this study, we exploit the unique ciliate model system to analyze the regulation of gene amplification during a nuclear differentiation process. These eukaryotic unicellular organisms contain two types of nuclei, the large DNA-rich macronucleus and the diploid micronuclei. Whereas the macronucleus is transcriptionally highly active, the micronuclei are transcriptionally almost inert during vegetative growth, and are required during sexual reproduction (conjugation) (11). After conjugation, a new macronucleus arises from a micronuclear derivative, and the old macronucleus degenerates (12). During this differentiation process, extensive DNA reorganization, DNA fragmentation, and DNA elimination events occur in the developing macronucleus (macronuclear anlage), which are most extreme in spirotrichous ciliates, such as Oxytricha, Stylonychia, or Euplotes. In these organisms, up to 90% of micronucleus-specific DNA sequences are eliminated, resulting in a macronucleus with greatly reduced kinetic complexity (13) and fragmented DNA molecules (nanochromosomes). More precisely: the micronuclear genome is rich in repetitive DNA sequences, from which ∼5–10% are transposon-like elements. The macronuclear precursor DNA sequences [macronucleus-destined sequences (MDS)] in the micronucleus are interrupted by short noncoding DNA sequences [internal eliminated sequences (IES)] and are not associated with telomeric repeats (11, 12). In stichotrichous ciliates, such as Oxytricha and Stylonychia, these MDSs are not always arranged linearly but, in ∼30%, appear in scrambled disorder (14–16).
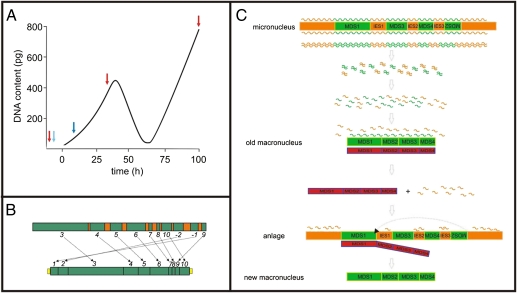
The macronuclear differentiation in spirotrichous ciliates can be divided into three distinct phases (Fig. 1A). Immediately after fusion of the two haploid micronuclei and mitotic division of the diploid zygote nucleus, the developing anlage enters a first DNA amplification stage, resulting in the formation of polytene chromosomes, which then become degraded and up to 90% of their DNA is eliminated. Second rounds of DNA amplification lead to the mature macronucleus (11–13). During this differentiation process, a large number of events take place at the DNA level. Early during the formation of the polytene chromosomes, transposon-like elements are excised, followed by religation of the DNA; and before DNA elimination, all IESs are removed. Also, MDSs of scrambled genes have to be reordered by the end of the polytene chromosome stage (15). Bulk repetitive DNA sequences as well as unique micronucleus-specific sequences become eliminated during polytene chromosome degradation, and macronucleus-destined DNA is fragmented into small DNA molecules, the nanochromosomes, ranging in size between ∼0.4 and ∼40 kbp. In general, each of these nanochromosomes encodes only one ORF; in rare cases, up to three ORFs can be found on one nanochromosome (17). Telomeric DNA sequences are added de novo to the macronuclear nanochromosomes (18). Finally, these nanochromosomes are amplified during a second DNA amplification stage. The average copy number of nanochromosomes varies among different ciliate species. For example, it has been estimated for Stylonychia that between 15,000 and 20,000 different nanochromosomes are present in the macronucleus, with an average copy number of 15,000 (11). Furthermore, as already visible on agarose gels, the copy number of individual nanochromosomes differs substantially (11, 12, 19), and by using quantitative Southern blot analyses, rough copy numbers have been determined for a few nanochromosomes (20–22). Therefore, differential amplification of nanochromosomes has to be tightly regulated during macronuclear differentiation.
Fig. 1.
Nuclear differentiation processes in stichotrichous ciliates. (A) Schematic diagram of the DNA content in the differentiating macronucleus of Stylonychia lemnae. Following mitotic division of the zygote nucleus, the macronuclear anlage enters a first DNA amplification stage, leading to the formation of polytene chromosomes. They become degraded and greater than 90% of their DNA is eliminated, resulting in a DNA poor stage. A second DNA amplification phase then leads to the mature macronucleus. Blue arrows indicate time-points at which nucleic acid injections into conjugating cells (light blue) and early exconjugant cells (dark blue) were performed. Red arrows indicate nuclear types (micronucleus, developing anlagen, or mature macronucleus) out of which DNA was isolated and copy numbers were determined. (B) Schematic diagram of the micronuclear and macronuclear version of actin I gene. In the micronucleus, the macronucleus-destined sequences (MDSs, green) are interrupted by internal eliminated sequences (IESs, orange) and occur in a scrambled disorder. During nuclear differentiation, IESs become excised and MDSs are rearranged into the correct order (as indicated by arrows), resulting in the macronuclear nanochromosome carrying telomeric repeats (yellow). (C) Model (2JLP model) of the epigenetic control of macronuclear differentiation in stichotrichous ciliates [afterJuranek and Lipps (12)]. The complete micronuclear genome becomes bidirectionally transcribed, and the resulting dsRNAs are processed into scanRNAs. The scan RNAs invade the old macronucleus where those scan RNAs homologous to the nanochromosomes (green) are retained and degraded (23). Simultaneously, templates (red) are generated in the old macronucleus comprising the sequence of whole nanochromosomes. scanRNAs homologous to micronucleus-specific DNA sequences (orange) and templates then travel into the anlage. Here the scanRNAs mark micronucleus-specific sequences and recruit chromatin-modifying proteins, resulting in excision and degradation of micronucleus-specific DNA. At the same time, the template is involved in correction of imprecise IES excision and in reordering of MDSs.
Although the morphological events of macronuclear differentiation are well described, we are only beginning to understand the molecular mechanisms involved in the regulation of macronuclear differentiation. There is very good evidence in some ciliates that small RNAs (scanRNAs) mark the sequences to be excised and eliminated. These RNAs recruit chromatin-modifying enzymes, and eventually DNA sequences to be removed during differentiation are organized into heterochromatin (23, 24). Although this pathway may result in the removal of specific DNA sequences, it cannot account for reordering of the MDSs of scrambled genes, proof-reading of imprecise excision processes (15), de novo addition of telomeric sequences, or differential amplification of nanochromosomes to specific copy numbers. A model explaining IES excision and reordering of MDSs was proposed by Prescott et al. (25). According to this model, a template (either DNA or RNA) derived from the old macronucleus guides the excision of IESs and subsequent rearrangement of MDSs from scrambled genes by recombination processes. The present view of the regulation of macronuclear differentiation in stichotrichous ciliates involves scanRNAs required for the heterochromatization of DNA sequences to be removed, whereas a template derived from the old macronucleus simultaneously directs MDS reordering and corrects imprecise excision of IESs (Fig. 1C) (12). In fact, direct evidence for the existence and relevance of a template for MDS reordering was recently provided in Oxytricha. When artificial template molecules (either DNA or RNA) were injected into conjugating cells, MDS reordering according to these templates was observed (26).
In this report, we investigate copy number regulation of nanochromosomes during macronuclear differentiation, and provide experimental evidence that this process too seems to be regulated by an RNA-template derived from the old macronucleus and, as such, is epigenetically inherited.
Results
Copy Number of Macronuclear Nanochromosomes in Stichotrichous Ciliate Stylonychia lemnae.
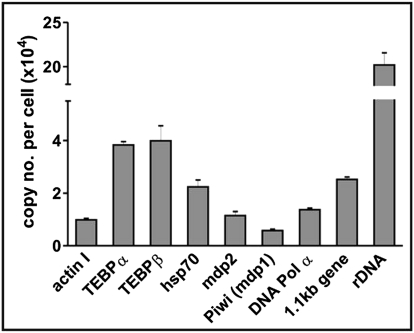
We first used quantitative real-time PCR (qRT-PCR) analyses to determine the copy numbers of nine selected nanochromosomes in the macronucleus of Stylonychia lemnae (Fig. 2). Our results showed comparable ranges of nanochromosomal copy numbers to earlier estimations of ∼15,000 copies per macronucleus on average. Fig. 2 illustrates that the copy numbers of the nanochromosomes examined can vary by at least 30-fold. The least abundant nanochromosome that we determined carried the Piwi (mdp1) gene (∼6,000 copies), whereas the most abundant was the nanochromosome encoding rRNA (∼200,000 copies). Interestingly, despite major copy number variations, the nanochromosomes encoding the telomere-end binding proteins TEBPα and TEBPβ were present at a very similar copy number. Both TEBPs are prominent proteins and required in equimolar amounts in the ciliate cell for telomere protection and maintenance (27–29).
Fig. 2.
Gene copy numbers in the macronucleus of Stylonychia. Copy numbers were determined by qRT-PCR as described in Material and Methods. Each column represents the copy number of a specific nanochromosome in the mature macronucleus of one cell. For each gene, three individual qRT-PCR experiments were performed and triplicates were analyzed. n = 9. Error bars represent SD.
We determined the copy number of the same set of nanochromosomes after an additional 3 mo vegetative growth, and no considerable changes in copy number could be observed. This demonstrates that the variations in copy numbers between different nanochromosomes did not result from unequal DNA amplification during vegetative growth but that nanochromosomes are stably retained, even though the ciliate macronucleus undergoes amitosis, a process whereby two daughter cells receive unequal amounts of DNA after binary fission. Our results also agree with an earlier observation that recombinant nanochromosomes could be faithfully retained and stably inherited in the spirotrich Euplotes crassus (30).
Although qRT-PCR has widely been used to assign gene copy numbers and gene expression levels, it has not previously been applied to determine copy numbers in the nuclei of ciliates. As a precaution we therefore addressed the reproducibility of this method by testing more than one set of primers from different regions of the same nanochromosomes (for hsp70, tebpβ, rDNA, and the 1.1kb gene, a macronuclear nanochromosome encoding a gene of unknown function). We found that the copy numbers for a given nanochromosome did not differ substantially (on average, <12%). This suggests that qRT-PCR yields reliable and reproducible results when quantifying gene copy numbers in ciliate cells.
In summary, macronuclear nanochromosomes occur at a specific copy number that is stable during vegetative growth, and nanochromosomes encoding proteins required in equimolar amounts in the cell show the same copy number as demonstrated for tebpα/β.
Timing of DNA Amplification During Macronuclear Differentiation.
To understand and analyze the control of differential gene amplification, we assigned the time-point at which the determination of copy numbers takes place during macronuclear differentiation. Therefore the copy numbers of four macronuclear-destined sequences previously analyzed in macronuclei (actin I, mdp2, tebpα, and tebpβ) were determined in micronuclei and in the developing macronucleus during polytene chromosome formation (Fig. S1). To avoid amplification of any contaminating macronuclear DNA in the DNA isolated from micronuclei or the polytene chromosome stage, micronucleus-specific primer combinations were used in these analyses (Table S1 and Fig. S2). In the micronucleus, all analyzed genes occur in approximately the same copy number. The same is true for the developing anlage, and, altogether, no copy number differences can be observed that mirror the situation in the mature macronucleus (Fig. 2 and Fig. S1). This excludes that gene copy number is already determined in the micronucleus, or that differential amplification takes place during the first phase of DNA synthesis, which leads to the formation of polytene chromosomes.
Gene amplification in the second DNA synthesis stage is observed in a morphologically highly organized structure, the replication band. It is difficult to imagine that differential activation of origins could occur in this structure (31). We therefore considered the possibility that copy number control, DNA reorganization, and processing could be closely linked mechanisms.
Is Differential DNA Amplification Template Dependent?
In stichotrichous ciliates, DNA reorganization and processing during macronuclear development are thought to be regulated both by scan RNAs and by a template, either RNA or DNA, derived from the old macronucleus, which directs MDS reordering and corrects imprecise excision of IESs according to the parental nanochromosomes (Fig. 1C) (12).
To examine whether differential DNA amplification is dependent on the concentration of a template, we tested whether selective degradation of such a template would affect the copy number of the respective nanochromosome in the new macronucleus. In a second set of experiments, we tested whether injection of a DNA or RNA template into conjugating cells or exconjugants would lead to a specific increase in gene copy number.
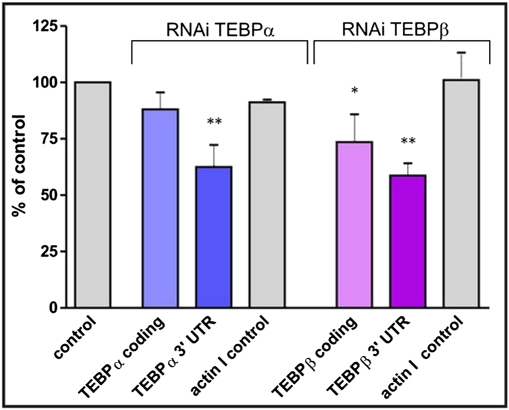
To investigate whether degradation of an RNA template leads to a decrease in gene copy number in the new macronucleus, we decided to analyze this effect on the two nanochromosomes encoding TEBPα and TEBPβ, which both are present at a high copy number in the macronucleus (Fig. 2). Vegetative cells were fed bacteria expressing dsRNA directed against these nanochromosomes for 5 d; cells were then allowed to conjugate, proceed through macronuclear differentiation, and grow vegetatively for several generations. By using five of these cells as a template, macronuclear gene copy number was determined by qRT-PCR. Because a template should not only comprise the coding region of the nanochromosome but also the noncoding regions, cells were fed bacteria expressing either dsRNA directed against the coding region of tebpα or tebpβ, or dsRNA directed against the noncoding untranslated region of these nanochromosomes (Fig. S3). The results are summarized in Fig. 3. dsRNA complementary to the coding region resulted in a ∼15–25% decrease in the copy number of the addressed nanochromosomes. The effect was significantly stronger when RNAi against the noncoding region was applied. The copy number of both nanochromosomes dropped to ∼60% of the copy number of control cells. Cells with this decreased copy number seemed to be fully viable over many vegetative generations; and, as determined by qRT-PCR analyses after about additional 100 vegetative generations, this decreased copy number was stably retained during vegetative growth. Controls were included to show that the observed effect was specific and not due to a nonspecific copy number reduction. When using RNAi directed against tebpα or tebpβ the effect on the copy number of the nanochromosome encoding the actin I gene was analyzed and shown not to be substantially affected (Fig. 3).
Fig. 3.
Reduction in copy numbers of tebpα and tebpβ nanochromosomes following RNAi treatment. Columns show the observed reduction in copy numbers. RNAi was directed either against the coding region of tebpα or tebpβ (“TEBPα/β coding”) or the 3′untranslated region (“TEBPα/β 3´UTR”) (Fig. S3 and Table S2). “Control” cells were fed with bacteria containing the vector L4440 (without insert) and expressing a 184 bp dsRNA from the polylinker region. This control was set to 100%. “Actin I controls” represent experiments in which actin I primers instead of tebp primers were used in “TEBPα/β 3´UTR”-cells. Actin I control after tebpα silencing, fourth column; after tebpβ silencing, seventh column. For each approach, three individual qRT-PCR experiments were performed and quadruplicates were analyzed. n = 12. Error bars represent SD. *P < 0.05, **P < 0.01.
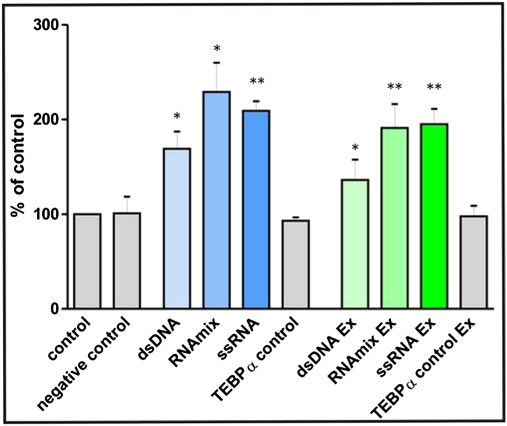
These results provide strong evidence that degradation of an RNA, complementary to a nanochromosome, during nuclear differentiation leads to a reduction of gene copy number in the new macronucleus, suggesting that the concentration of this RNA determines the copy number of the nanochromosome during macronuclear differentiation. Possibly this RNA is identical to the template required for correct processing of macronuclear precursor sequences during nuclear differentiation (12, 26). If this is true, delivery of large amounts of this template into cells of this stage should lead to an increase in copy number. Because the nanochromosome encoding the actin I gene is present in a relatively low copy number in the macronucleus, we decided to inject template molecules complementary to this gene. The following template molecules were injected either into both conjugating cells or into exconjugants immediately following conjugation at the very beginning of the first DNA amplification stage. In three distinct experiments, we injected one of the following: (i) dsDNA of the complete nanochromosome, including one telomeric repeat at each end; or (ii) an RNA mix containing the two ss RNAs complementary to both strands of the actin I nanochromosome, both terminated by one telomeric repeat at each end; or (iii) ssRNA complementary either to the 3′ or the 5′ strand of the nanochromosome, again including one telomeric repeat at each end. Approximately 12,000 molecules were injected into one cell. Cells were allowed to proceed through macronuclear differentiation and several rounds of vegetative cell division. Again, five of these cells were used as a template, and macronuclear copy number was determined by qRT-PCR. In all experiments, injection of template molecules (dsDNA, RNA mix, or ssRNA) into conjugating cells or early exconjugants led to a substantial copy number increase of the nanochromosome encoding actin I. Fig. 4 depicts representative examples for each template injected. Fig. S4 shows results of four separate experiments in which ssRNA was injected into exconjugant cells. In the case of dsDNA, the new macronucleus contained ∼60% more copies of the actin I nanochromosome compared with control cells. This effect was even more dramatic when injecting either the RNA mix or ssRNA. Here, an increase in gene copy number of up to 100% or more was observed. The effect was identical for both ssRNA and RNA mix and independent of the orientation of the ssRNA (Fig. S4). No effect on the copy number of the tebpα nanochromosome could be detected, showing the specificity of the effect observed (Fig. 4). As in the case of RNAi-treated cells, qRT-PCR analyses were performed after an additional 100 generations showing that these increased copy numbers were stably inherited during vegetative growth. Due to the lack of compatible mating types in experimentally manipulated cells, inheritance of copy changes could not be analyzed in this study. Interestingly, in these experiments, a number of cells with abnormal morphology and cell motility could be observed. These effects were not observed after injection of cells with Pringsheim medium, indicating that they are not due to experimental manipulation but, rather, are caused by the change in gene copy number. Finally, quantitative Western blot analyses showed an increase in actin I expression after template injection indicating that the concentration of the gene product correlates with the gene copy number (Fig. S5).
Fig. 4.
Increase in copy numbers of the actin I nanochromosome following microinjection. Columns show the observed increases in copy numbers. dsDNA, ssRNA, or a mixture of both RNA strands (“RNAmix”) was injected, either into the cytoplasm of both conjugating cells (blue columns) or into early exconjugants (green columns), as described in Material and Methods. “Control” cells were not injected but did conjugate. This control was set to 100%. Only Pringsheim medium was injected into “negative control” cells. ”TEBPα controls” represent experiments in which tebpα primers instead of actin I primers were used in ssRNA-injected cells. For each approach, three individual qRT-PCR experiments were performed and quadruplicates were analyzed. n = 12. Error bars represent SD. *P < 0.05, **P < 0.01.
Discussion
In this report, we exploit the unique genome organization of a ciliate model system to analyze the control of amplification of genes to specific copy numbers during a nuclear differentiation process. Earlier work indicated that, in Stylonychia, the average nanochromosomal copy number is ∼15,000 per macronucleus; however, copy numbers of different anochromosomes can vary significantly (11). Copy numbers have been roughly estimated for a few nanochromosomes by quantitative Southern blot analyses (20–22), but no exact numbers have been obtained. In this study, we determined the copy numbers of various nanochromosomes by qRT-PCR. Our results show that copy numbers of macronuclear nanochromosomes can vary from a few thousands to several hundred thousands (Fig. 2). The nanochromosomal copy number seems to correlate with the number of gene product present in the cell. For example, in our analyses, the nanochromosome encoding rRNA has the highest copy number, or the nanochromosomes encoding the telomere end-binding proteins TEBPα and TEBPβ are highly abundant and occur at a similar copy number. The TEBPs are prominent nuclear proteins and are required in equimolar amounts in the cell (29). Existence of a correlation between copy number and expression level is further indicated by the observation that increasing copy number by template injection leads to an increased expression level of the protein (Fig. S5). Therefore, it seems that in addition to promoter strength, gene amplification is used to ensure high levels of expression, very similar to what has been described in other organisms (22, 32).
Copy numbers determined in micronuclear and anlagen DNA do not mirror the situation in the mature macronucleus, showing that macronuclear precursor sequences are not present in different copy numbers in the micronucleus, and that differential amplification does not take place during the formation of polytene chromosomes but, rather, during or after DNA elimination. This is in agreement with studies in Euplotes, in which the timing of differential amplification during macronuclear development was examined by Southern blot analyses (21). The authors concluded that the degree of amplification correlates with the timing of gene excision during macronuclear differentiation, and speculated that early processed macronuclear nanochromosomes can enter the second DNA amplification stage earlier than nanochromosomes that are processed late during differentiation.
It is now generally believed that correction of imprecise excision events and reordering of macronucleus-destined sequences is guided by a template derived from the old macronucleus (15, 25). Injection of artificial templates, either DNA or RNA, with disordered MDSs led to incorrectly processed nanochromosomes (26). Hence, it seemed reasonable to investigate whether DNA processing and differential DNA amplification are closely linked processes. To test this hypothesis, we either specifically degraded a possible RNA template by RNAi treatment, or increased concentration of such a template by injection of a DNA or RNA template into differentiating cells and subsequent determination of nanochromosomal copy numbers.
We decided to study the effect of template degradation by RNAi treatment on highly amplified genes, as it could be assumed that a reduction of the copy number of these genes still could lead to viable offspring. The two nanochromosomes encoding the telomere-end binding proteins TEBPα or TEBPβ are present in ∼40,000 copies per macronucleus in Stylonychia (Fig. 2.). RNAi was directed against either the coding region or the 3′ untranslated region. In all cases, a reduction in copy numbers was observed (Fig. 3); interestingly, however, the observed effect was stronger when RNAi directed against the noncoding regions of the nanochromosomes was performed. In this case, a reduction in tebp copy number of ∼40% could be detected, whereas feeding of dsRNA directed against the coding region of these genes led to a reduction in copy numbers of only 15–25%. A possible explanation for this result could be that RNAi directed against the ORF leads not only to a degradation of an RNA template but also to silencing of expression of these genes, which are required for progression through nuclear differentiation and for vegetative growth (28). In this case, only viable cells in which the effect of RNAi is less pronounced would be analyzed. In contrast, RNAi directed against the noncoding untranslated region would not induce silencing of gene expression. Altogether, however, a cell probably tolerates only a limited reduction in gene copy number, as observed by the high (∼50%) mortality of exconjugant cells after RNAi treatment.
A more dramatic effect on the copy number was observed when additional template molecules (either RNA or DNA) were injected into differentiating Stylonychia cells. For this experiment, a lower copy number nanochromosomal sequence, actin I, was chosen (Fig. 2). The highest increase in copy number was observed after injection of an RNA template, either as a mixture of both strands or ssRNA complementary to either strand of the nanochromosomal DNA (Fig. 4). The fact that copy number was similarly affected after injection of ssRNA complementary to each of the nanochromosomal DNA strands (Fig. S4) is compatible with the template-guided model proposed by Prescott et al. (25). The effect on copy numbers after RNAi treatment together with the fact that injection of RNA templates leads to the highest increase in copy number strongly suggest that the template consists of RNA. The observed effect after injection of DNA might be due to its transient transcription, as already suggested by Nowacki et al. (26) when injecting artificial templates in Oxytricha. Interestingly, injection of these template molecules did not lead only to an increased copy number of the actin I nanochromosome but also to cells with abnormal morphology, demonstrating that a stringent copy number control is essential for correct functioning of the vegetative cell.
In summary, our results strongly argue that DNA processing and specific gene amplification are closely related mechanisms and depend on an RNA-template derived from the old macronucleus. Our results can now be incorporated into a mechanistic model for copy number control during macronuclear differentiation. The rate of DNA processing most likely depends on the concentration of the RNA-template, and, as it is derived from the old macronucleus, it should be proportional to the copy number of a nanochromosome present in the macronucleus. Therefore, highly amplified genes could be processed much faster than low copy number genes and could enter a second DNA amplification stage immediately after complete processing. Alternatively, the amount of available template could affect the number of macronuclear precursor sequences present in the DNA of the polytene chromosome stage that are correctly processed. Endoreplication of correctly processed nanochromosomes would then lead to specific gene copy numbers. Nanochromosomes not completely or incorrectly processed would not enter the second DNA amplification stage. In both cases, any change in template concentration leads to a change in nanochromosomal copy number, as demonstrated in our analyses. According to this model, no selective activation of replication origins is required at any stage, but gene copy number directly correlates with the rate of DNA processing during nuclear differentiation. This rate is dependent on the number of template molecules, which in turn corresponds to the number of nanochromosomes in the parental macronucleus.
Finally, our results show that RNA is involved not only in the translation of the genetic information and the regulation of gene expression, but also in the specific amplification of DNA sequences. As such, we describe a previously uncharacterized biological function of RNA.
Materials and Methods
Cells and DNA.
Growth of Stylonychia lemnae an isolation of macronuclear, micronuclear, and anlagen DNA was performed as described earlier (13). To achieve conjugation, cells of two different mating types were mixed and kept at RT; mating efficiency was greater than 80%. Anlagen DNA was isolated during the first amplification phase but before reaching full polyteny, which is ∼100–200 (Fig. 1A).
RNAi Treatment.
RNAi treatment was performed by feeding Stylonychia with Escherichia coli-expressing, double-stranded (ds) RNA directed against the gene of interest, as described elsewhere (28). For both TEBPs, we cloned one fragment amplified from the coding region and one fragment from the 3′noncoding region (Fig. S3) into the vector L4440. The vectors were transfected into the RNaseIII-deficient E. coli strain HT115, and dsRNA expression was induced by adding IPTG to a final concentration of 0.4 mM at an OD600 of 0.4. A mixture of Chlorogonium elongatum and heat-inactivated (10 min, 65 °C) E. coli was fed to opposite mating types of Stylonychia for 5 d. As a control, cells were fed induced E. coli containing the vector L4440 without an insert (producing a 184 bp dsRNA from the L4440 polylinker). On day 5, Stylonychia cells were mixed for conjugation. To ensure that copy numbers were exclusively determined in cells that did indeed conjugate, ∼200 pairs of conjugating cells were isolated and grown for 10–15 generations.
Microinjection of DNA and RNA.
Injection of DNA, single-stranded (ss) RNA, or a mixture or of both RNA strands (all homologous to the actin I nanochromosome) in proximity of nuclei of either both of the two conjugating cells or of early exconjugants (Fig. 1A) was performed as described before (33). Injection time of 1.0 s and three injection pulses per cells were applied. DNA or the RNA mix (consisting of ssRNA homologous to both strands of the actin I nanochromosome) was diluted in Pringsheim solution (13) to 50 μg/mL; ssRNA to a concentration of 25 μg/mL DNA consisted of purified PCR products using primer pair P5′tel/ P3′tel and a cloned actin I nanochromosome as a template (Table S1). RNA was prepared by transcribing the cloned actin I nanochromosome, containing one telomeric repeat at each end by T7 or SP6 RNA polymerase (Fermentas), respectively. Before injection, the RNA mix containing both ssRNA strands was incubated at 65 °C for 10 min and cooled to room temperature. It can be estimated that ∼12,000 copies of DNA, RNA mix, or ssRNA were injected. Injection of each template was repeated at least 15 times in independent experiments. Greater than 70% of cells survived the injection procedure. Separated injected cells were allowed to complete macronuclear development and several rounds of vegetative cell division. The copy number of the actin I nanochromosome was then determined in three independent experiments for each template.
Determination of Gene Copy Numbers by qRT-PCR.
qRT-PCR analyses were performed using a Light Cycler 1.5 instrument (Roche Diagnostics) as described before (24). To generate standard curves for amplicons, plasmid DNA with cloned insert (specific sequences of macronuclear or micronuclear DNA) and of known concentration was serially diluted and analyzed in triplicate. Using individual standard curves for each amplicon, the amounts of specific macronuclear, micronuclear, or anlagen DNA could be calculated. Analyzed genes were as follows: actin I, tebpα, tebpβ, hsp70, mdp2, piwi (mdp1), DNA pol α, the 1.1kb gene, and rDNA (Table S3).
For the determination of copy numbers following RNAi treatment or microinjection, treated Stylonychia cells were starved for 2 d to ensure an arrest in G1 phase. Subsequently, cells were transferred to deionized water, and 5 cells were used in a single qRT-PCR. Here, quadruplicates were analyzed. Each qRT-PCR experiment was repeated three times, resulting in a total of 12 independent data points. Various controls were included; copy number determination following RNAi treatment was also performed in control cells which were fed bacteria containing the vector without an insert. Furthermore, copy numbers were determined using actin I primers in cells where RNAi was directed against the noncoding regions of tebpα or -β to exclude unspecific copy number variations.
Following microinjections, we used tebpα primers in cells with injected actin I templates (ssRNA). Moreover, copy numbers were determined in noninjected cells as well as in cells where only Pringsheim solution was injected.
Statistical Analyses.
Data were analyzed using the unpaired two-tailed t test with the GraphPad Prism Program. Asterisks indicate the degree of significant differences compared with controls (*P < 0.05, **P < 0.01).
Supplementary Material
Acknowledgments
We thank Katharina Heyse for technical and intellectual support. We also thank Mariusz Nowacki, Joanna E. Haye, Wenwen Fang, Vikram Vijayan, and Laura F. Landweber for sharing their data on DNA amplification in Oxytricha trifallax. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to H.J.L.).
Footnotes
See Commentary on page 21951.
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009284107/-/DCSupplemental.
References
- 1.Tower J. Developmental gene amplification and origin regulation. Annu Rev Genet. 2004;38:273–304. doi: 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- 2.Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- 3.Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oögenesis. Proc Natl Acad Sci USA. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka H, Yao MC. Palindromic gene amplification—an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- 5.Spradling AC. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27:193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- 6.Schimke RT. Gene amplification in cultured cells. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- 7.Hourcade D, Dressler D, Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proc Natl Acad Sci USA. 1973;70:2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trask BJ, Hamlin JL. Early dihydrofolate reductase gene amplification events in CHO cells usually occur on the same chromosome arm as the original locus. Genes Dev. 1989;3:1913–1925. doi: 10.1101/gad.3.12a.1913. [DOI] [PubMed] [Google Scholar]
- 9.Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammad MM, Donti TR, Sebastian Yakisich J, Smith AG, Kapler GM. Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J. 2007;26:5048–5060. doi: 10.1038/sj.emboj.7601919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juranek SA, Lipps HJ. New insights into the macronuclear development in ciliates. Int Rev Cytol. 2007;262:219–251. doi: 10.1016/S0074-7696(07)62005-1. [DOI] [PubMed] [Google Scholar]
- 13.Ammermann D, Steinbrück G, von Berger L, Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974;45:401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- 14.Landweber LF, Kuo TC, Curtis EA. Evolution and assembly of an extremely scrambled gene. Proc Natl Acad Sci USA. 2000;97:3298–3303. doi: 10.1073/pnas.040574697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Möllenbeck M, et al. The pathway to detangle a scrambled gene. PLoS ONE. 2008;3:e2330. doi: 10.1371/journal.pone.0002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prescott DM. Genome gymnastics: Unique modes of DNA evolution and processing in ciliates. Nat Rev Genet. 2000;1:191–198. doi: 10.1038/35042057. [DOI] [PubMed] [Google Scholar]
- 17.Chang WJ, Stover NA, Addis VM, Landweber LF. A micronuclear locus containing three protein-coding genes remains linked during macronuclear development in the spirotrichous ciliate Holosticha. Protist. 2004;155:245–255. doi: 10.1078/143446104774199628. [DOI] [PubMed] [Google Scholar]
- 18.Roth M, Prescott DM. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell. 1985;41:411–417. doi: 10.1016/s0092-8674(85)80014-3. [DOI] [PubMed] [Google Scholar]
- 19.Kraut H, Lipps HJ, Prescott DM. The genome of hypotrichous ciliates. Int Rev Cytol. 1986;99:1–28. doi: 10.1016/s0074-7696(08)61422-9. [DOI] [PubMed] [Google Scholar]
- 20.Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- 21.Dönhoff T, Klein A. Timing of differential amplification of macronucleus-destined sequences during macronuclear development in the hypotrichous ciliate Euplotes crassus. Chromosoma. 1996;105:172–179. doi: 10.1007/BF02509498. [DOI] [PubMed] [Google Scholar]
- 22.La Terza A, Miceli C, Luporini P. Differential amplification of pheromone genes of the ciliate Euplotes raikovi. Dev Genet. 1995;17:272–279. doi: 10.1002/dvg.1020170312. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 24.Postberg J, Heyse K, Cremer M, Cremer T, Lipps HJ. Spatial and temporal plasticity of chromatin during programmed DNA-reorganization in Stylonychia macronuclear development. Epigenetics Chromatin. 2008;1:3. doi: 10.1186/1756-8935-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prescott DM, Ehrenfeucht A, Rozenberg G. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J Theor Biol. 2003;222:323–330. doi: 10.1016/s0022-5193(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 26.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottschling DE, Zakian VA. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 28.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 29.Price CM, Cech TR. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- 30.Bender J, Kämpfer M, Klein A. Faithful expression of a heterologous gene carried on an artificial macronuclear chromosome in Euplotes crassus. Nucleic Acids Res. 1999;27:3168–3172. doi: 10.1093/nar/27.15.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postberg J, Alexandrova O, Cremer T, Lipps HJ. Exploiting nuclear duality of ciliates to analyse topological requirements for DNA replication and transcription. J Cell Sci. 2005;118:3973–3983. doi: 10.1242/jcs.02497. [DOI] [PubMed] [Google Scholar]
- 32.Claycomb JM, Orr-Weaver TL. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 2005;21:149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Jönsson F, Wen JP, Fetzer CP, Lipps HJ. A subtelomeric DNA sequence is required for correct processing of the macronuclear DNA sequences during macronuclear development in the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 1999;27:2832–2841. doi: 10.1093/nar/27.14.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.