Abstract
Acetyl-CoA carboxylases (ACCs) are crucial metabolic enzymes and have been targeted for drug development against obesity, diabetes, and other diseases. The carboxyltransferase (CT) domain of this enzyme is the site of action for three different classes of herbicides, as represented by haloxyfop, tepraloxydim, and pinoxaden. Our earlier studies have demonstrated that haloxyfop and tepraloxydim bind in the CT active site at the interface of its dimer. However, the two compounds probe distinct regions of the dimer interface, sharing primarily only two common anchoring points of interaction with the enzyme. We report here the crystal structure of the CT domain of yeast ACC in complex with pinoxaden at 2.8-Å resolution. Despite their chemical diversity, pinoxaden has a similar binding mode as tepraloxydim and requires a small conformational change in the dimer interface for binding. Crystal structures of the CT domain in complex with all three classes of herbicides confirm the importance of the two anchoring points for herbicide binding. The structures also provide a foundation for understanding the molecular basis of the herbicide resistance mutations and cross resistance among the herbicides, as well as for the design and development of new inhibitors against plant and human ACCs.
Keywords: fatty acid metabolism, metabolic syndrome, structure-based drug design
Acetyl-CoA carboxylases (ACCs or ACCases) are biotin-dependent enzymes required for fatty acid metabolism. ACCs catalyze the carboxylation of acetyl-CoA to malonyl-CoA in two steps, through the action of three distinct protein components (1, 2). In the first step, a biotin moiety covalently linked to the biotin carboxyl carrier protein (BCCP) component enters the biotin carboxylase (BC) active site for carboxylation of the biotin. In the second step, the carboxylated biotin translocates to the carboxyltransferase (CT) active site to transfer the carboxyl group to the acetyl-CoA substrate, producing malonyl-CoA. In bacterial species, BC, BCCP, and CT reside in separate subunits, and ACC is a multisubunit enzyme. In contrast, ACCs from most eukaryotes are large, multidomain enzymes, with all three domains residing on a single polypeptide.
Two isoforms of ACC exist in mammals. ACC1 is a cytosolic enzyme that catalyzes the first and committed step in long-chain fatty acid biosynthesis. ACC2 is associated with the outer mitochondrial membrane, where its malonyl-CoA product is an inhibitor of carnitine palmitoyltransferase I (CPT-I), which controls the transport of long-chain acyl-CoAs into the mitochondria for oxidation (3, 4). Mice deficient in ACC2 have higher rates of fatty acid oxidation and reduced body fat (5), implicating ACC2 as a drug discovery target against obesity, type 2 diabetes, and other diseases associated with the metabolic syndrome (6, 7).
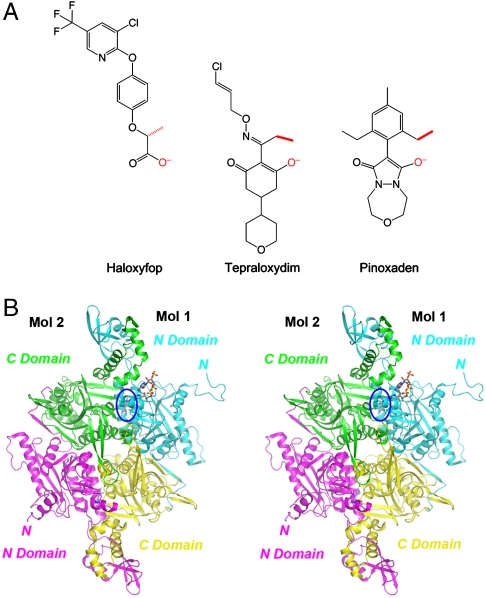
Plant ACCs are found in both the plastids, for fatty acid biosynthesis, and the cytosol, for synthesis of very long-chain fatty acids and flavonoids. The cytosolic ACCs exist in the multidomain form, while the plastid ACCs exist in either the multidomain (grasses) or the multisubunit (dicots) form. Three classes of commercially available herbicides target the CT activity of the multidomain form of plastid ACCs (Fig. 1A). The aryloxyphenoxy propionates (APPs or FOPs, represented by haloxyfop in Fig. 1A) and the cyclohexandiones (CHDs or DIMs, represented by tepraloxydim in Fig. 1A) have been widely used in the field since the late 1970s. The phenylpyrazolines (DENs) are relatively new, with pinoxaden being introduced in 2006 (8, 9).
Fig. 1.
Crystal structure of the CT domain in complex with pinoxaden. (A) Chemical structures of the herbicides haloxyfop, tepraloxydim, and pinoxaden. The two anchoring points for interactions with the CT domain (see text for details) are colored in red. (B) Schematic drawing of the structure of yeast CT domain dimer in complex with pinoxaden. The N domains of the two monomers are colored in cyan and magenta, while the C domains are colored in yellow and green. The inhibitor is shown in stick models, in slate blue for carbon atoms, and one inhibitor molecule is highlighted by the blue oval. The CoA molecule is shown for reference in gray (13). The structure figures were produced with PyMOL (www.pymol.org).
We have reported the crystal structures of the CT domain of yeast ACC and its complex with CoA (10), haloxyfop (11), and tepraloxydim (12). We have also reported the structure of the CT domain in complex with CP-640186 (13), a structurally distinct class of compounds that were identified as potent inhibitors of mammalian ACCs (14). The inhibitors occupy different regions of the CT active site, located at the interface of a dimer of this domain, and block catalysis by competing with the acetyl-CoA (haloxyfop and tepraloxydim) or the carboxybiotin (CP-640186) substrate. A large conformational change in the dimer interface and the active site is required for haloxyfop binding, while a much smaller change is observed for the tepraloxydim complex. Despite their chemical diversity (Fig. 1A), haloxyfop and tepraloxydim share two common points of contact (or anchoring points) with the CT domain (12), which are crucial for the inhibitory activity of these two classes of herbicides.
We report here the crystal structure at 2.8-Å resolution of the CT domain of yeast ACC in complex with pinoxaden, the third class of commercial herbicides. Even though pinoxaden is chemically rather distinct from the other two herbicides (Fig. 1A), it has a similar binding mode as tepraloxydim and requires a small conformational change in the dimer interface for binding. Crystal structures of the CT domain in complex with all three classes of herbicides are now available. They confirm the importance of the two anchoring points for herbicide binding. They also provide a foundation for understanding the molecular basis of the herbicide resistance mutations and cross resistance among the herbicides, as well as for the design and development of new ACC inhibitors.
Results and Discussion
The Overall Structure.
The crystal structure of the CT domain of yeast ACC in complex with pinoxaden (Fig. 1A) has been determined at 2.8-Å resolution (Table 1 and Fig. 1B). The current atomic model contains residues 1480–2051 and 2080–2195 in chain A, 1480–2047 and 2081–2192 in chain B, 1491–2051 and 2080–2193 in chain C, for the three CT domain molecules as well as three pinoxaden molecules in the crystallographic asymmetric unit. The atomic model has good agreement with the X-ray diffraction data and the expected values for geometric parameters (Table 1). The majority of the residues (88%) lie in the most favored region of the Ramachandran plot. Only residue Gln1744 of all three monomers is in the disallowed region, as is the case for all other structures of this domain.
Table 1.
Summary of crystallographic information
| Resolution range for refinement (Å) | 30–2.8 (2.9–2.8) * |
| No. of observations | 495,940 |
| Rmerge (%) | 9.9 (43.8) |
| Redundancy | 4.7 (4.4) |
| I/σI | 9.8 (2.1) |
| No. of reflections | 106,428 |
| Completeness (%) | 99 (99) |
| R factor (%) | 19.7 (31.3) |
| Free R factor (%) | 23.9 (37.3) |
| rms deviation in bond lengths (Å) | 0.012 |
| rms deviation in bond angles (°) | 1.3 |
*The numbers in parentheses are for the highest resolution shell.
The three CT domain monomers in the asymmetric unit have essentially the same conformation, with rms distance of about 0.5 Å for their equivalent Cα atoms. The conformations of the inhibitors in the three CT domain molecules are highly similar to each other as well.
Binding Mode of Pinoxaden.
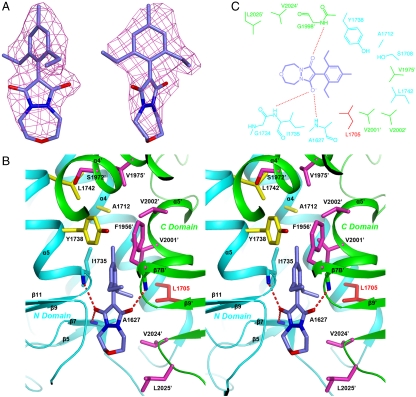
The crystal structure of the pinoxaden complex was obtained by soaking free enzyme crystals of yeast CT domain with the inhibitor at 1-mM concentration for 80 min. Longer soaking times and/or higher inhibitor concentrations (up to 2.5 mM, due to solubility limits) invariably led to significant reduction in the diffraction quality of the crystals. Clear electron density for the inhibitor molecules was observed based on the crystallographic data (Fig. 2A). The average temperature factor values of the three inhibitor molecules are 70, 74, and 94 Å2, while that for all three protein molecules is 33 Å2. This suggests that the occupancy of the inhibitor may not be 100% in the crystal, especially for chain C. The lower occupancy is also consistent with our kinetic studies, which showed only minor inhibition of the CT domain of yeast ACC by pinoxaden at 1-mM concentration. In comparison, the Ki of haloxyfop for this CT domain is about 0.5 mM (10).
Fig. 2.
The binding mode of pinoxaden. (A) Final omit Fo-Fc electron density at 2.8-Å resolution for pinoxaden, contoured at 3σ, in two views. (B) Stereographic drawing showing the binding site for pinoxaden. The N domain of one monomer is colored in cyan, and the C domain of the other monomer in green. The side chains of residues in the binding site are shown in yellow and magenta, respectively. Leu1705 is shown and labeled in red, because it is equivalent to a site of resistance mutations in plants. Hydrogen bonds from the inhibitor to the protein are indicated with dashed red lines. (C) Schematic drawing of the interactions between pinoxaden and the CT active site.
Pinoxaden is bound in the active site of the CT domain at its dimer interface (Fig. 2B). The central pyrazoline ring is situated between residues Gly1734 and Val2024′ (primed residue numbers indicate the other monomer). One oxygen atom on this ring is hydrogen-bonded to the main-chain amides of Ala1627 (2.9 Å) and Ile1735 (3.0 Å) (Fig. 2C). The other oxygen is hydrogen-bonded to the main-chain amide of Gly1998′ (2.8 Å). The electron density shows that this ring is planar (Fig. 2A), indicating that one of the carbonyl groups is in the enol form (Fig. 1A). It is likely that this enol, in its ionized state (enolate), is the one hydrogen-bonded to the main-chain amides of Ala1627 and Ile1735, because an oxyanion in haloxyfop (11) and tepraloxydim (12) is also positioned here (see below).
The 4-methyl-(2,6)-diethyl phenyl ring lies between the amide bonds of Gly1734-Ile1735 and Gly1997′-Gly1998′, showing π-π interactions (Fig. 2B). One of the ethyl substituents points in the direction of a hydrophobic pocket that is composed of the side chains from Ala1627, Leu1705, Ser1708, Val2001′, and Val2024′, with its closest neighbor being the side-chain CD2 atom of Leu1705. The other ethyl group is positioned near the side chains of Tyr1738, Leu1756, and Phe1956′. The methyl group is located deepest in the dimer interface, surrounded by residues Ile1735, Tyr1738, Val2001′, and Val2002′.
The oxadiazepane ring is mostly solvent exposed and has few strong interactions with the enzyme. This ring, especially its oxygen atom, is likely to be flexible, because it has weaker electron density compared to the rest of the inhibitor (Fig. 2A). This region of the binding site is unlikely to be covered by the BC domain in the full-length ACC enzyme, based on observations on the structure of the propionyl-CoA carboxylase holoenzyme (15).
The observed binding mode of pinoxaden is consistent with the structure-activity relationships of this series of compounds (9). For example, replacing the 2,6-diethyl substituents with dimethyl groups or removing the 4-methyl group on the phenyl ring both led to large losses in activity. The presence of an enol in this compound is also supported by the fact that pinoxaden is actually marketed as a pivaloyl ester prodrug with one of the oxygen atoms on the pyrazoline ring (8, 9).
Pinoxaden Has a Similar Binding Mode as Tepraloxydim.
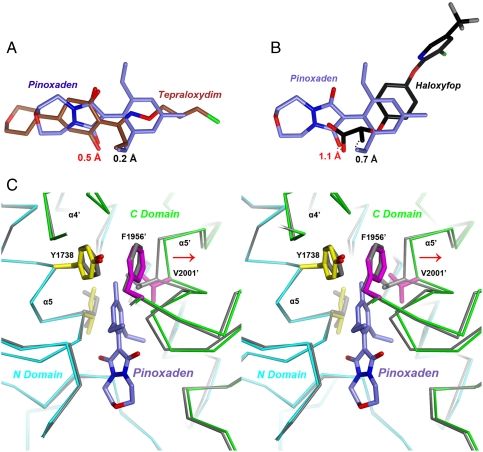
The binding mode of pinoxaden is similar to that of tepraloxydim (12) (Fig. 3A), even though the two compounds are rather distinct from each other chemically (Fig. 1A). The pyrazoline ring of pinoxaden is comparable to the central cyclohexanedione ring of tepraloxydim. The two oxygen atoms on the cyclic structures in both compounds are tethered to the enzyme in exactly the same way, with the most important interactions being those between the main-chain amides of Ala1627 and Ile1735 with one of the oxygen atoms, located within 0.5 Å of each other in pinoxaden and tepraloxydim (Fig. 3A). The position of this oxygen atom is also occupied by one of the carboxylate oxygen atoms of haloxyfop (Fig. 3B) (12), suggesting that it is an oxyanion in all three inhibitors (enolate in tepraloxydim and pinoxaden, Fig. 1A). In fact, a hydroxyl group at this position is unlikely to be able to maintain hydrogen bonds to both main-chain amides.
Fig. 3.
Comparison of the binding modes of pinoxaden with haloxyfop and tepraloxydim. (A) Overlay of the binding modes of pinoxaden (in slate blue for carbon atoms) and tepraloxydim (brown). The distances between the oxyanions and methyl groups of the two inhibitors are indicated. (B) Overlay of the binding modes of pinoxaden and haloxyfop (black). (C) Structural overlay of the CT domain free enzyme (in gray) and the pinoxaden complex (in cyan and green for the N and C domains) near the inhibitor binding site. Side chains in the binding site with large conformational changes are also shown. Phe1956′ is shown for reference. The shift in the position of the α5′ helix is indicated with the red arrow.
One of the ethyl groups on the phenyl ring of pinoxaden has the same conformation as the ethyl group of tepraloxydim (Fig. 3A), and both are located in the hydrophobic pocket that contains residue Leu1705 (Fig. 2C). The phenyl ring of pinoxaden overlaps with the planar oxime moiety of tepraloxydim (Fig. 3A), while the other ethyl group of pinoxaden replaces a water molecule that is observed in the tepraloxydim complex. The methyl group on the phenyl ring of pinoxaden does not reach as deeply into the dimer interface as tepraloxydim. Interestingly, replacing the 4-methyl with an ethyl group led to a loss in activity (9), possibly because the ethyl group does not have the correct geometry to maintain interactions with the enzyme. The structure suggests that a linear substituent at the 4-position may best mimic the binding mode of tepraloxydim in this region.
Consistent with their similar binding modes, the overall conformation of the CT domain in the pinoxaden complex is also similar to that of the tepraloxydim complex, as evidenced by an rms distance of 0.36 Å between their equivalent Cα atoms. Compared to the structure of the free CT domain, there are small but important conformational changes upon pinoxaden binding (Fig. 3C). Especially, helix α5′ (residues 1996′–2003′) moves by about 1 Å, and the side chains of Ile1735, Tyr1738 and Val2001′ adopt a different conformation to accommodate the inhibitor. These conformational changes are similar to those observed for the tepraloxydim complex (12).
On the other hand, the binding mode of pinoxaden has sharp differences to that of haloxyfop (Fig. 3B), which requires a large conformational change in the active site and the dimer interface for binding (11). Nonetheless, a comparison of the binding modes of the three compounds highlights the two anchoring points that are important for their interactions with the CT active site (12). One anchoring point involves the double hydrogen bonds between an oxyanion of the inhibitor and the main-chain amides of Ala1627 and Ile1735. The other involves hydrophobic interactions between a methyl (or ethyl) group of the inhibitor and the pocket that contains residue Leu1705. The functional importance of the latter anchoring point is illustrated by the fact that residue 1705 is a major site of mutations in plants that confer resistance to these herbicides (see next).
The structural information on the three herbicides suggests that the main-chain amides of Ala1627 and Ile1735 may be optimally positioned to recognize an oxyanion. They may also play a crucial role in CT catalysis, stabilizing the enolate of the acetyl group in the transition state of the reaction (16). Therefore, these two amides may be the oxyanion hole for the CT reaction, and the herbicides may function in part as transition-state mimics to inhibit this enzyme. Especially, an enolate moiety is present in the structures of DIMs and pinoxaden (Fig. 1A). Resistance mutations have not been observed for either of these two residues, probably because such mutations will also lead to a significant loss in catalytic activity of the enzyme.
Molecular Basis for Cross Resistance Among the Three Classes of Herbicides.
The application of FOPs and DIMs in the field over the years has led to the emergence of resistant plants. Resistance to herbicides can be mediated by mutations in the target enzyme or by changes in the uptake and/or degradation of the herbicides. Seven mutations in the CT domain of plastid ACCs have been identified from resistant plants: I1781L (equivalent to Leu1705 in yeast ACC), W1999C (Trp1924), W2027C (Trp1953), I2041N or I2041V (Val1967), D2078G (Asp2004), C2088R (Met2014), and G2096A (Ala2022) (17–20). Of these, the D2078G and C2088R mutations confer resistance to both FOPs and DIMs, and the I1781L mutation confers resistance to FOPs and some DIMs. The remaining mutations confer resistance to the FOPs but have smaller to moderate effects on the DIMs.
Although pinoxaden was only introduced in 2006, resistance plants have already been identified due to cross resistance with the other two classes of herbicides (21–23). In fact, in most cases the resistance is preexistent in the plants, before their exposure to pinoxaden. The I1781L, D2078G, and possibly the W2027C mutations confer resistance to pinoxaden (21). Overall, the pattern of resistance to pinoxaden appears to parallel that of the DIMs (22), although there are also distinct differences. The effects of the C2088R mutation on pinoxaden have not been studied yet.
Crystal structures of the CT domain in complex with all three classes of herbicides provide a foundation for understanding the molecular basis of these resistance mutations (11, 12). The similar binding modes of pinoxaden and the DIMs explain the mechanism of cross resistance between these two classes of herbicides. The I1781L mutation (Leu1705 in yeast ACC), which produces resistance to all three classes of herbicides, is located in a binding pocket that is occupied by a methyl or ethyl group in all these compounds (Fig. 3 A and B), which in fact represents one of the anchoring points for their binding. This mutation is the most frequently identified in resistant plants, underscoring the functional importance of this residue for herbicide binding.
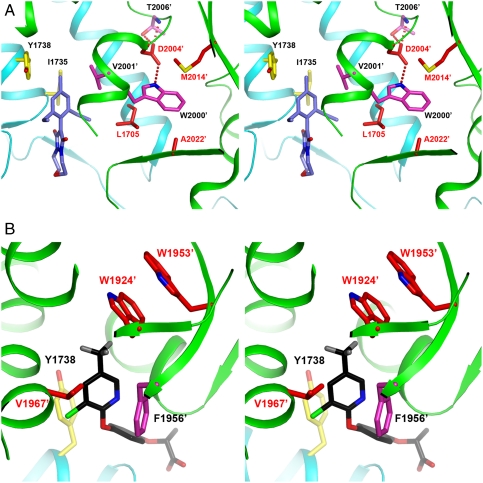
The D2078G (Asp2004 in yeast ACC) mutation also confers resistance to all three classes of herbicides. This residue is not in direct contact with the herbicides. It is located just after the α5′ helix (Fig. 4A), which must undergo a conformational change for pinoxaden (Fig. 3C) and tepraloxydim binding (12). The side chain of Asp2004′ makes hydrogen bonds with the side chain of Trp2000′ (in helix α5′) and the main-chain amide and side-chain hydroxyl of Thr2006′ (Fig. 4A). The interactions may hold these residues (especially the α5′ helix) in a conformation necessary for inhibitor binding. In fact, another mutation that is known to confer resistance to both FOPs and DIMs, C2088R, is equivalent to Met2014′ in yeast ACC, whose side chain has van der Waals interactions with the Trp2000′ side chain (Fig. 4A). Therefore, it is likely that the C2088R mutation will also confer resistance to pinoxaden. In addition, another resistance mutation, G2096A (A2022′ in yeast CT), is also located near the Trp2000′ side chain (Fig. 4A).
Fig. 4.
Molecular basis for herbicide resistance mutations. (A) Stereographic drawing showing the structure of yeast CT domain near the mutation sites I1781L (L1705 in yeast ACC), D2078G (D2004′), C2088R (M2014′), and G2096A (A2022′). The bound position of pinoxaden is also shown. (B) Stereographic drawing showing the structure of yeast CT domain near the mutation sites W1999C (W1924′ in yeast ACC), W2027C (W1953′), and I2041N (V1967′). The bound position of haloxyfop is also shown.
Therefore, four of the resistance mutations, I1781L (Leu1705 in yeast ACC), D2078G (Asp2004′), C2088R (Met2014′), and G2096A (A2022′), are clustered close to each other in the structure, distributed around the side chain of the strictly conserved Trp2000′ residue (Fig. 4A). Three of the mutations in this cluster, I1781L, D2078G, and C2088R, confer resistance to all three classes of herbicides, and Ile1781 (Leu1705) has direct contacts with these compounds. The other three resistance mutations, W1999C (W1924′), W2027C (W1953′), and I2041N (V1967′), are clustered near the first aryl ring of haloxyfop (11) (Fig. 4B). This is a unique feature of that class of inhibitors (Fig. 1A), which may explain why these mutations primarily confer resistance to the FOPs while having only small effects on the DIMs. A recent computational study on the W2027C, I2041N, D2078G, and G2096A mutations suggests that they may introduce large conformational changes in the binding pocket, which may also contribute to the reduced sensitivity of these mutants to the herbicides (24).
Among the seven mutation sites that are known to confer resistance to herbicides, yeast ACC has a different residue at four of them compared to plant ACC—Leu1705, Val1967, Met2014, and Ala2022—and therefore behaves mostly like a resistant ACC. We have used the CT domain of yeast ACC as a model system to define the binding modes of the three classes of herbicides by crystallographic analysis because we have so far not been able to obtain crystals of the CT domain of a sensitive ACC. It may be expected, however, that the overall binding modes are likely similar between the two types of ACCs.
Our attempts at introducing mutations into yeast CT to make it more sensitive to herbicides have so far not been successful. Our earlier studies showed that the L1705I/V1967I double mutant, which converts two of the four residues to their equivalents in plant ACCs, was not more sensitive to haloxyfop (11). We produced the L1705I/V1967I/M2014C/A2022G quadruple mutant of the yeast CT domain but found that it was not more sensitive either. Like the L1705I/V1967I double mutant (11), the quadruple mutant also had much lower catalytic activity compared to the wild-type enzyme. Compensatory mutations in other regions of the CT domain may be important for restoring the catalytic activity of the mutants. It is likely that additional differences between the CT domains of plant and yeast ACCs are also determinants of herbicide sensitivity, and some of these positions may be identified as sites of resistance mutations in the future.
Materials and Methods
Protein production and crystallization.
The CT domain (residues 1476–2233) of Saccharomyces cerevisiae ACC was expressed and purified according to protocols described previously (11). Free enzyme at 10 mg/ml concentration was crystallized using the hanging-drop method at 4 °C. The reservoir solution contained 0.1 M sodium citrate (pH 5.5), 9% (w/v) PEG8000, 0.2 M NaCl, and 10% (v/v) glycerol. The pinoxaden complex was obtained by soaking the free enzyme crystal with 1 mM of the compound for 80 min. Cryoprotection was achieved by the addition of 25% (v/v) glycerol and the crystal was then flash frozen in liquid nitrogen for data collection at 100 K.
Data Collection and Structure Determination.
X-ray diffraction data were collected at the X29A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the HKL package (25). The crystal belongs to space group C2, with unit cell parameters of a = 247.2 Å, b = 123.4 Å, c = 145.7 Å, and β = 94.3°. There are three CT molecules in the asymmetric unit, forming a noncrystallographic dimer and a crystallographic dimer. The structure refinement was carried out with the programs CNS (26) and Refmac (27). The atomic model was built with the programs O (28) and Coot (29). The crystallographic information is summarized in Table 1.
Acknowledgments.
We thank Neil Whalen for setting up the X29A beamline, and Shi-Xian Deng for help with removing the pivalate group from pinoxaden prodrug. This research is supported in part by National Institutes of Health (NIH) Grant DK067238 (L.T.), and the Organic Chemistry Collaborative Center and the Clinical and Translational Science Award of Columbia University, supported by NIH Grant UL1 RR024156 and the NIH Roadmap for Medical Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3PGQ).
References
- 1.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 2.Tong L. Acetyl-coenzyme A carboxylase: Crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay RR, Gandour RD, van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta. 2001;1546:21–43. doi: 10.1016/s0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- 5.Abu-Elheiga L, Matzuk MM, Abo-Hashema KAH, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 6.Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: Target for metabolic syndrome. J Lipid Res. 2009;50:S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong L, Harwood HJ., Jr Acetyl-coenzyme A carboxylases: Versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofer U, Muehlebach M, Hole S, Zoschke A. Pinoxaden—for broad spectrum grass weed management in cereal crops. J Plant Dis Protect. 2006;20:989–995. [Google Scholar]
- 9.Muehlebach M, et al. Aryldiones incorporating a [1,4,5]oxadiazepane ring. Part I: Discovery of the novel cereal herbicide pinoxaden. Bioorg Med Chem. 2009;17:4241–4256. doi: 10.1016/j.bmc.2008.12.062. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Yang Z, Shen Y, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science. 2003;299:2064–2067. doi: 10.1126/science.1081366. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Tweel B, Tong L. Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA. 2004;101:5910–5915. doi: 10.1073/pnas.0400891101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang S, Callaghan MM, Watson KG, Tong L. A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim. Proc Natl Acad Sci USA. 2009;106:20723–20727. doi: 10.1073/pnas.0908431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Tweel B, Li J, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with CP-640186. Structure. 2004;12:1683–1691. doi: 10.1016/j.str.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Harwood HJ, Jr, et al. Isozyme-nonselective N-substituted bipiperidylcarboxamide acetyl-CoA carboxylase inhibitors reduce tissue malonyl-CoA concentrations, inhibit fatty acid synthesis, and increase fatty acid oxidation in cultured cells and in experimental animals. J Biol Chem. 2003;278:37099–37111. doi: 10.1074/jbc.M304481200. [DOI] [PubMed] [Google Scholar]
- 15.Huang CS, et al. Crystal structure of the a6b6 holoenzyme of propionyl-coenzyme A carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles JR. The mechanism of biotin-dependent enzymes. Annu Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 17.Yu Q, et al. Diversity of acetyl-coenzyme A carboxylase mutations in resistant Lolium populations: Evaluation using clethodim. Plant Physiol. 2007;145:547–558. doi: 10.1104/pp.107.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, et al. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA. 2007;104:3627–3632. doi: 10.1073/pnas.0611572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delye C, Zhang X-Q, Michel S, Matejicek A, Powles SB. Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol. 2005;137:794–806. doi: 10.1104/pp.104.046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delye C, Matejicek A, Michel S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds.(black-grass), re-examined at the recommended herbicide field rate. Pest Manag Sci. 2008;64:1179–1186. doi: 10.1002/ps.1614. [DOI] [PubMed] [Google Scholar]
- 21.Petit C, Bay G, Pernin F, Delye C. Prevalence of cross- or multiple resistance to the acetyl-coenzyme A carboxylase inhibitors fenoxaprop, clodinafop and pinoxaden in black-grass (Alopecurus myosuroides Huds.) in France. Pest Manag Sci. 2010;66:168–177. doi: 10.1002/ps.1851. [DOI] [PubMed] [Google Scholar]
- 22.Kaundun SS. An aspartate to glycine change in the carboxyl transferase domain of acetyl CoA carboxylase and non-target-site mechanism(s) confer resistance to ACCase inhibitor herbicides in a Lolium multiflorum population. Pest Manag Sci. 2010;66:1249–1256. doi: 10.1002/ps.2003. [DOI] [PubMed] [Google Scholar]
- 23.Kuk YI, Burgos NR, Scott RC. Resistance profile of diclofop-resistant Italian ryegrass (Lolium multiflorum) to ACCase- and ALS-inhibiting herbicides in Arkansas, USA. Weed Sci. 2008;56:614–623. [Google Scholar]
- 24.Zhu X-L, Ge-Fei H, Zhan C-G, Yang G-F. Computational simulations of the interactions between acetyl-coenzyme A carboxylase and clodinafop: resistance mechanism due to active and nonactive site mutations. J Chem Inf Model. 2009;49:1936–1943. doi: 10.1021/ci900174d. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan KD. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]