Abstract
The most frequent infectious diseases in humans—and those with the highest potential for rapid pandemic spread—are usually transmitted via droplets during close proximity interactions (CPIs). Despite the importance of this transmission route, very little is known about the dynamic patterns of CPIs. Using wireless sensor network technology, we obtained high-resolution data of CPIs during a typical day at an American high school, permitting the reconstruction of the social network relevant for infectious disease transmission. At 94% coverage, we collected 762,868 CPIs at a maximal distance of 3 m among 788 individuals. The data revealed a high-density network with typical small-world properties and a relatively homogeneous distribution of both interaction time and interaction partners among subjects. Computer simulations of the spread of an influenza-like disease on the weighted contact graph are in good agreement with absentee data during the most recent influenza season. Analysis of targeted immunization strategies suggested that contact network data are required to design strategies that are significantly more effective than random immunization. Immunization strategies based on contact network data were most effective at high vaccination coverage.
Keywords: disease dynamics, network topology, public health, human interactions
Pandemic spread of an infectious disease is one of the biggest threats to society because of the potentially high mortality and high economic costs associated with such an event (1, 2). Understanding the dynamics of infectious disease spread through human communities will facilitate the development of much needed mitigation strategies (3). Schools are particularly vulnerable to infectious disease spread because of the high frequency of close proximity interactions (CPIs) that most infectious disease transmission depends on (3, 4). Infections that are transmitted predominantly via the droplet route, such as influenza, common colds, whooping cough, severe acute respiratory syndrome (SARS), and many others, are among the most frequent infectious diseases. Droplets from an infected person can reach a susceptible person in close proximity, typically a distance of less than 3 m (5, 6), making CPIs highly relevant for disease spread. Very little is known about the dynamic patterns of CPIs in human communities, however [but see Cattuto et al. (7)]. Here, we present data collected with a wireless sensor network deployment using TelosB motes (Crossbow Technologies Inc.) (8) to detect high-resolution proximity (up to 3 m) between subjects in a U.S. high school. The dataset represents a high-resolution temporal contact network relevant to the spread of infectious diseases via droplet transmission in a school.
Previous attempts to capture the contact networks relevant for infectious disease transmission have mostly been based on data collection using surveys, sociotechnological networks, and mobile devices like cell phones. Each of these approaches has advantages and disadvantages. Surveys manage to capture the interactions relevant for disease transmission but are often limited by small sample sizes (9) and are subject to human error (10). Sociotechnological networks can provide large long-term datasets (11) but fail to capture the CPIs relevant for disease transmission. The use of mobile devices aware of their location (or of other mobile devices in proximity) represents a promising third alternative. Using mobile phones to detect spatial proximity of subjects is possible with repeated Bluetooth scans (10), but the resolution is too coarse for diseases that are transmitted through the close contact route. Our approach is free of human error, captures the vast majority (94%) of the community of interest, and allows us to collect high-resolution contact network data relevant for infectious disease transmission.
Most efforts to understand and mitigate the spread of pandemic diseases (influenza in particular) have made use of large-scale spatially explicit models parameterized with data from various sources, such as census data, traffic/migration data, and demographic data (3, 4, 12–15). The population is generally divided into communities of schools, workplaces, and households, but detailed data on mixing patterns in such communities are scarce. In particular, very little is known about the contact networks in schools (16) even though schools are known to play a crucially important role in pandemic spread, mainly owing to the intensity of CPIs at schools. In what follows, we describe and analyze the contact network observed at a U.S. high school during a typical school day. Using an SEIR (susceptible, exposed, infectious, and recovered) simulation model, we investigate the spread of influenza on the observed contact network and find that the results are in very good agreement with absentee data from the influenza A (H1N1) spread in the fall of 2009. Finally, we implement and test various immunization strategies to evaluate their efficacy in reducing disease spread within the school.
Results
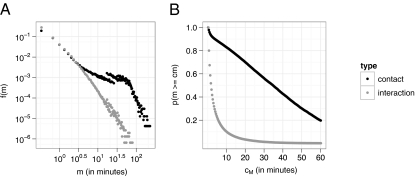
The dataset covers CPIs of 94% of the entire school population, representing 655 students, 73 teachers, 55 staff, and 5 other persons, and it contains 2,148,991 unique close proximity records (CPRs). A CPR represents one close (maximum of 3 m) proximity detection event between two motes. An interaction is defined as a continuous sequence (≥1) of CPRs between the same two motes, and a contact is the sum of all interactions between these two motes. Thus, a contact exists between two motes if there is at least one interaction between them during the day, and the duration of the contact is the total duration of all interactions between these two motes. Because the beaconing frequency of a mote is 0.05 s−1, an interaction of length 3 (in CPRs) corresponds to an interaction of about 1 min (SI Text and references therein). The entire dataset consists of 762,868 interactions with a mean duration of 2.8 CPRs (∼1 min), or 118,291 contacts with mean duration of 18.1 CPRs (∼6 min) (data available in SI Methods). Fig. 1A shows the frequency, f, of interactions and contacts of length m (in minutes) [f(m)]. The majority of interactions and contacts are very short (80th percentile of interactions at 3 CPRs, 80th percentile of contacts at 15 CPRs), and even though about 80% of the total time is spent in interactions that are shorter than 5 min, short contacts (<5 min) represent only about 10% of the total time (Fig. 1B).
Fig. 1.
(A) Normalized frequency, f, of interactions and contacts of duration m (in minutes) [f(m)] on a log-log scale. (B) Percentage, p, of total time of all CPIs by interactions and contacts with a minimum duration, cm (in minutes). Most CPI time is spent in medium-duration contacts consisting of repeated short interactions.
The temporal mixing patterns observed are in accordance with the schedule of the school day [i.e., the average degree (number of contacts) peaks between classes and during lunch breaks] (Fig. S1). The aggregate network for the entire day can be represented by a weighted undirected graph, wherein nodes represent individuals and edges represent contacts (edges are weighted by contact duration). The topology of the contact network is an important determinant of infectious disease spread (17, 18). Traditional infectious disease models assume that all subjects have the same number of contacts, or that the contact network of subjects is described by a random graph with a binomial degree distribution. Many networks from a wide range of applications, including contact networks relevant for infectious disease transmission (19, 20), have been found to have highly heterogeneous degree distributions, however. Such heterogeneity is important because it directly affects the basic reproductive number, R0, a crucially important indicator of how fast an infectious disease spreads and what fraction of the population will be infected. In particular, if ρ0 is the incorrect estimate for R0 in a heterogeneous network under the false assumption of a uniform degree distribution, the correct estimate is given by R0 = ρ0 (1 + CV2), where CV2 is the squared coefficient of variation of the degree distribution (17, 21). Thus, the CV quantifies the extent to which contact heterogeneity affects disease dynamics.
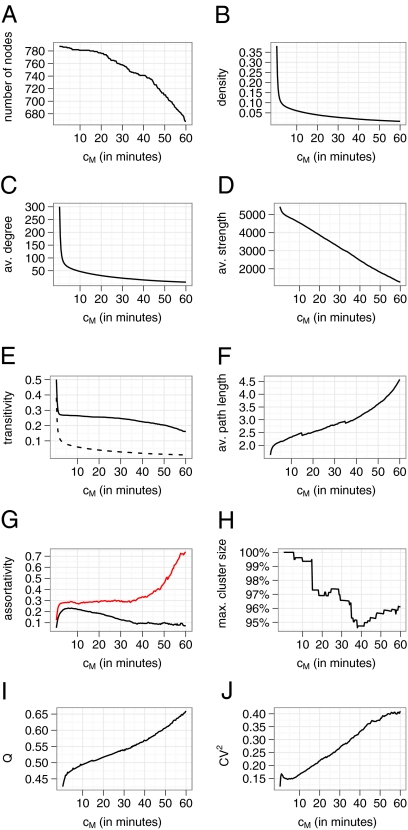
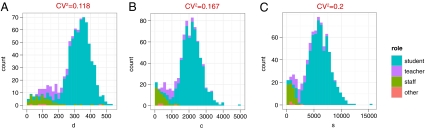
The descriptive statistics of the school network with different definitions of contact are shown in Fig. 2. To account for the fact that the majority of the contacts are relatively short (Fig. 1A), we recalculated all statistics of the network with a minimum requirement for contact duration, cm (i.e., all edges with weight <cm are removed from the graph). The network exhibits typical “small-world” properties (22), such as a relatively high transitivity (also known as clustering coefficient, which measures the ratio of triangles to connected triplets) and short average path length for all values of cm. Assortativity, the tendency of nodes to associate with similar nodes with respect to a given property (23), was measured with respect to degree and role of the person (e.g., student, teacher). Interestingly, although both measures are relatively high, degree assortativity decreases and role assortativity increases with higher values of cm. Because of the very high density of the contact network, a giant component exists for all values of cm. Community structure (or modularity) is relatively high, increasingly so with higher values of cm, indicating that more intense contacts tend to occur more often in subgroups and less often between such groups (24). We find a very homogeneous degree distribution with a CV2 = 0.118 for the full network and slightly increased heterogeneity in the network with higher cutoff values cm (Fig. 2J). The distributions of number of interactions, c, and the strength, s (the weighted equivalent of the degree) (25) are equally homogeneous (Fig. 3). Overall, the data suggest that the network topology is best described by a low-variance small-world network.
Fig. 2.
Various statistics on the contact graph with minimum contact duration, cm (i.e., the left-most point in each panel represents the full contact graph, the right-most point represents the contact graph that contains only contacts that are at least 60 min long). With increasing cm, nodes drop out of the network if they have no contact that satisfies the minimum duration condition. (A) Hence, the reduction in the number, V, of nodes. (B) Density of the graph (2E/[V*(V − 1)]), where E is the number of edges. (C) Average (av.) degree. (D) av. strength, where the strength of a node is the total number of CPRs of the node. (E) Transitivity (i.e., cluster coefficient) as defined by Barrat et al. (25) and expected value (mean degree/V) in a random network (dashed line). (F) Average path length. (G) Assortativity (23) with respect to degree (black line) and role (red line). (H) Size of the largest component as a fraction of total network size. max., maximum. (I) Modularity, Q, as defined by Reichardt and Bornholdt (39). (J) CV2 of degree.
Fig. 3.
Distribution and CV2 of degree, d (A); number of interactions, c (B); and strength, s (C), based on the full contact network and colored by the role of individuals.
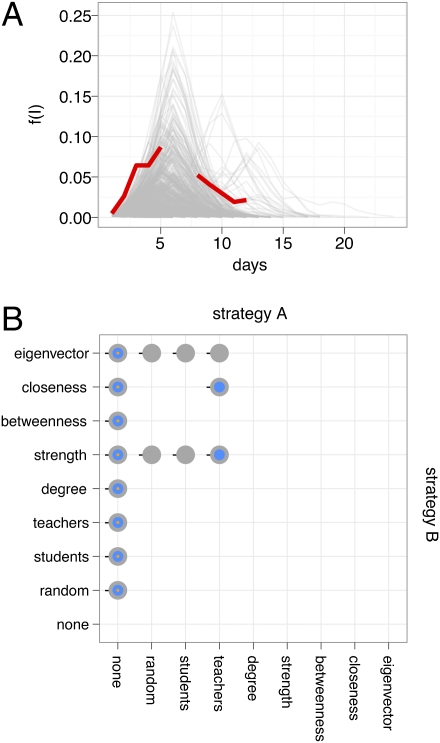
To understand infectious disease dynamics at the school, we used an SEIR simulation model (parameterized with data from influenza outbreaks; details presented in SI Methods), wherein an index case becomes infected outside of the school on a random day during the week and disease transmission at the school occurs during weekdays on the full contact network as described by the collected data. Each individual is chosen as an index case for 1,000 simulation runs, resulting in a total of 788,000 epidemic simulation runs. This simulation setting represents a base scenario, wherein a single infectious case introduces the disease into the school population. In reality, multiple introductions are to be expected if a disease spreads through a population, but the base scenario used here allows us to quantify the predictive power of graph-based properties of individuals on epidemic outcomes. We assume that symptomatic individuals remove themselves from the school population after a few hours. We find that in 67.7% of all simulations, no secondary infections occur and thus there is no outbreak, whereas in the remaining 32.3% of the simulations, outbreaks occur with an average attack rate of 3.87% (all simulations = 1.33%, maximum = 46.19%) and the average R0, measured as the number of secondary infections caused by the index case, is 3.85 (all simulations = 1.24, maximum = 18). Recent work on disease spread on networks has identified the relationship between R0, the network degree distribution, and the average probability that an infectious individual transmits the disease to a susceptible individual, T (18, 26). Based on this, R0 would be valued at 4.52 (SI Methods). This value is higher than what we measure in the simulations because it is based on the assumption of continuous transmission, whereas the simulations exhibit discontinuous transmission attributable to weekends; during that time, the school is closed and the chain of transmission is effectively cut for 2 d. Finally, absentee data from the school during the fall of 2009 (i.e., during the second wave of H1N1 influenza in the northern hemisphere) are in good agreement with simulation data generated by the SEIR model running on the contact network (Fig. 4A).
Fig. 4.
(A) Absentee data (red) and data generated by the SEIR model (gray; 1,000 runs with R0 >1 shown). Gray lines show frequency of infectious individuals, f(I); red lines show the combined frequency of students who reported, or were diagnosed with, a fever and teachers who were absent (gap in the line attributable to weekend). (B) Differences in effect of vaccination strategies. Colors represent vaccination coverage of 5% (orange), 10% (blue), and 20% (gray). A point at the intersection of strategy A and strategy B indicated that between those strategies, there was a significant difference (P < 0.05, two-sided Wilcoxon test) in the outbreak size at all transmission probability values at the given vaccination coverage. A black horizontal or vertical line points in the direction of the strategy that resulted in smaller outbreak sizes. Because of the symmetry of the grid, data points below the left bottom and top right diagonal line are not shown.
A strong correlation exists between the size of an outbreak caused by index case individual i and the strength of the node representing individual i (r2 = 0.929). The correlation between outbreak size and degree is substantially weaker (r2 = 0.525) because at the high temporal resolution of the dataset, the degree contains many short-duration contacts whose impact on epidemic spread is minimal. To estimate the sampling rate at which degree has maximal predictive power, we systematically subsampled our original dataset to yield lower resolution datasets. Fig. S2 shows that sampling as infrequently as every 100 min would have resulted in the same predictive power for degree as sampling every 20 s, whereas the maximum predictive power for degree would have been attained at ∼20 min. At this sampling rate, the 95% confidence intervals for the correlation between degree and outbreak size and the correlation between strength and outbreak size start to overlap (because of the high correlation between degree and strength; Fig. S2, blue line). These results suggest that high-resolution sampling of network properties such as the degree of nodes might be highly misleading for prediction purposes if used in isolation (i.e., without the temporal information that allows for weighting).
To mitigate epidemic spread, targeted immunization interventions or social distancing interventions aim to prevent disease transmission from person to person. Finding the best immunization strategy is of great interest if only incomplete immunization is possible, as is often the case at the beginning of the spread of a novel virus. In recent years, the idea of protecting individuals based on their position in the contact network has received considerable attention (11, 27, 28). Graph-based properties, such as node degree and node betweenness centrality (29), have been proposed to help identify target nodes for control strategies, such as vaccination; however, because of the lack of empirical contact data on closed networks relevant for the spread of influenza-like diseases, such strategies could only be tested on purely theoretical networks [or on approximations from other empirical social networks that did not measure CPIs directly (11)]. To understand the effect of partial vaccination, we measured outbreak size for three different levels of vaccination coverage (5%, 10%, and 20%) and a number of different control strategies based on node degree, node strength, betweenness centrality, closeness centrality, and eigenvector centrality (so-called “graph-based strategies”). In addition, we tested vaccination strategies that do not require contact network data (random vaccination, preferential vaccination for teachers, and preferential vaccination for students; SI Methods). To ensure robustness of the results to variation in transmission probabilities, all simulations were tested with three different transmission probabilities (Methods). Ten thousand simulations for each combination of vaccination strategy, vaccination coverage, and transmission probability with a random index case per simulation were recorded (i.e., total of 810,000 simulations) to assess the effect of vaccination. Fig. 4B shows which strategies led to significantly (P < 0.05, two-sided Wilcoxon test) different outcomes at all transmission probability values (results separated by transmission probability are presented in Fig. S3). As expected, all strategies managed to reduce the final size of the epidemic significantly. Compared with the random strategy, graph-based strategies had an effect only at higher vaccination coverage. Graph-based strategies did not differ much in their efficacy; in general, strength-based strategies were the most effective. Overall, two main results emerge: (i) in the absence of information on the contact network, all available strategies, including random immunization, performed equally well and (ii) in the presence of information on the contact network, high-resolution data support a strength-based strategy, but there was no significant difference among the graph-based strategies.
Discussion
In summary, we present high-resolution data from the CPI network at a U.S. high school during a typical school day. Notably, the month of the experiment (January) is associated with the second highest percentage of influenza cases in the United States for the 1976–1977 through 2008–2009 influenza seasons (second only to February). The data suggest that the network relevant for disease transmission is best described as a small-world network with a very homogeneous contact structure in which short repeated interactions dominate. The low values of the coefficients of variation in degree, strength, and number of interactions (Fig. 3) suggest that the assumption of homogeneity in traditional disease models (21) might be sufficiently realistic for simulating the spread of influenza-like diseases in communities like high schools. Furthermore, we do not find any “fat tails” in the contact distribution of our dataset, corroborating the notion (9) that the current focus on networks with such distributions is not warranted for infectious disease spread within local communities.
It is important to recognize the limitations of the data presented here, particularly in light of the fact that transmission of influenza-like diseases also occurs via other routes, for example, via contact with contaminated surfaces (30). Moreover, different pathogens as well as different strains of a particular pathogen might have different minimum requirements (both spatial and temporal) that need to be met for person-to-person transmission. At present, the data capture the contact network during a single day only. This is not an inherent shortcoming of the approach presented here, however, and long-term studies in the future could address how the large-scale structure of the contact network in a high school changes over time. Data collection at different schools with different demographic compositions would be helpful in clarifying if and how demographic compositions affect the properties of the network relevant for disease transmission. Wireless sensor network technology certainly allows further elucidation of the contact networks not only at different schools but in households, hospitals, workplaces, and other community settings.
With regard to immunization strategies, our simulation results suggest that contact network data are necessary to design strategies that are significantly more effective than random immunization to minimize the number of cases at the school caused by a single index case. Great care needs to be taken in interpreting these results for various reasons. First, the limitations of the data as discussed above mean that these results may not hold in other settings, underlining the need for further empirical network studies. Second, the simulations assume neither multiple introductions nor ongoing interactions of participants outside of the school. To what extent these assumptions, particularly the latter, are violated when a disease spreads through a community is unknown and remains to be measured. Third, future work needs to establish the robustness of the effect of vaccination strategies against errors in the measurement of graph-based properties. Fourth, and perhaps most importantly, a particular immunization strategy may be optimal for reducing the number of cases in one particular school but, at the same time, may not be optimal from the perspective of an entire community. Immunization strategies must also take into account medical, social, and ethical aspects (31). Thus, although we believe that data of the kind reported here can help to inform public health decisions, particularly as more data become available in the future, it is clear that one cannot derive public health recommendations at this stage directly from this study alone. We note, however, that our findings support the notion that graph-based immunization strategies could, in principle, help to mitigate disease outbreaks (11, 28).
Methods
The mote deployment is described in detail in SI Methods.
Epidemic Simulations.
To simulate the spread of an influenza-like disease, we used an SEIR simulation model parameterized with data from influenza outbreaks (12, 32, 33). In the following, we describe the model in detail.
Transmission occurs exclusively along the contacts of the graph as collected at the school. Each individual (i.e., node of the network) can be in one of four classes: susceptible, exposed, infectious, and recovered. Barring vaccination, all individuals are initially susceptible (more information on vaccination is presented below). At a random time step during the first week of the simulation, an individual is chosen as the index case and his or her status is set to exposed. A simulation is stopped after the number of both exposed and infectious individuals has gone back to 0 (i.e., all infected individuals have recovered). Each time step represents 12 h and is divided into day and night. Transmission can occur only during the day and only on weekdays (i.e., apart from the initial infection of the index case, we do not consider any transmission outside of the school; although this assumption will not hold in reality, it allows us to focus exclusively on within-school transmission and to analyze the spread of a disease starting from a single infected case).
Transmission of disease from an infectious to a susceptible individual occurs with a probability of 0.003 per 20 s of contact (the interval between two beacons). This value has been chosen because it approximates the time-dependent attack rate observed in an outbreak of influenza aboard a commercial airliner (32). In particular, the probability of transmission per time step (12 h) from an infectious individual to a susceptible individual is 1 − (1 − 0.003)w, where w is the weight of the contact edge (in CPRs). On infection, an individual will move into the exposed class (infected but not infectious). After the incubation period, an exposed individual will become symptomatic and move into the infectious class. The incubation period distribution is modeled by a right-shifted Weibull distribution with a fixed offset of half a day [power parameter = 2.21, scale parameter = 1.10 (12)]. On the half day that the individual becomes infectious, the duration of all contacts of the infectious individual is reduced by 75%. This reduction ensures that if an individual becomes symptomatic and starts to feel ill during a school day, social contacts are reduced and the individual leaves the school or is dismissed from school after a few hours. In the following days, all contacts are reduced by 100% until recovery (i.e., the individual stays at home). Once an individual is infectious, recovery occurs with a probability of 1 − 0.95t per time step, where t represents the number of time steps spent in the infectious state [in line with data from an outbreak of H1N1 at a New York City school (33)]. After 12 d in the infectious class, an individual will recover if recovery has not occurred before that time.
Based on these simulation settings and the finding that the average contact duration is 18.1 CPRs (Results), the transmissibility, T, as defined by Newman (18) and Meyers et al. (26), is 1 − (1 − 0.003)18.1*0.25 = 0.0135. Furthermore, based on the framework established by Newman (18) and Meyers et al. (26), R0 can be calculated as R0 = T × <ke>, where the average excess degree, <ke>, is <k2>/ <k>− 1 = 334.76.
We assume that all exposed individuals developed symptoms. A high incidence of asymptomatic spread may affect infectious disease dynamics (34), but reports of asymptomatic individuals excreting high levels of influenza virus are rare (35). In addition, a recent community-based study investigating naturally acquired influenza virus infections found that only 14% of infections with detectable shedding at RT-PCR were asymptomatic and viral shedding was low in these cases (36), suggesting that the asymptomatic transmission plays a minor role. Similar patterns were observed for SARS-CoV, another virus with the potential for rapid pandemic spread: Asymptomatic cases were infrequent, and lack of transmission from asymptomatic cases was observed in several countries with SARS outbreaks (37).
Vaccination.
The efficacy of vaccination strategies was tested by simulation. Vaccination occurs (if it occurs at all) before introduction of the disease by the index case. Vaccinated individuals are moved directly into the recovering class. We assume that the vaccine provides full protection during an epidemic.
Three vaccination strategies are implemented that do not require measuring graph-based properties; these strategies are called “random,” “students,” and “teachers.”
Random.
Individuals are chosen randomly until vaccination coverage is reached.
Students.
Students only are chosen randomly until vaccination coverage is reached.
Teachers.
Teachers only are chosen randomly until vaccination coverage is reached. If vaccination coverage is so high that all teachers get vaccinated before the coverage is reached, the strategy continues as the student strategy (see above) for the remaining vaccinations.
Five vaccination strategies are implemented that require measuring graph properties: These strategies are called “degree,” “strength,” “betweenness,” ”closeness,” and “eigenvector.” In all cases, individuals are ranked according to the specific graph property and chosen according to that ranking (in descending order) until vaccination coverage is reached.
Degree.
Degree is calculated as the number of contacts during the day of measurement.
Strength.
Strength is calculated as the total time exposed to others during the day of measurement.
Betweenness.
Betweenness centrality, CB(i), of individual i is calculated as
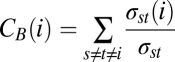 |
where s, t, and i are distinct individuals in the contact graph; σst is the total number of shortest paths between nodes s and t; and σst(i) is the number of those shortest paths that go through node i (29). The shortest path is calculated using inverse weights.
Closeness.
Closeness centrality, CC(i), of individual i is calculated as
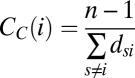 |
where s and i are distinct individuals in the contact graph, dsi is the shortest path between nodes s and i, and n is the number of individuals in the graph (29). The shortest path is calculated using inverse weights.
Eigenvector.
Calculation of eigenvector centrality is described by White and Smyth (38) through application of the page-rank algorithm with jumping probability 0. The measure captures the fraction of time that a random walk would spend at a given vertex during an infinite amount of time.
We tested three different levels of vaccination coverage: 5%, 10%, and 20%. These percentages apply to the entire population [i.e., a 10% vaccination coverage means that 10% of the entire school population is vaccinated, independent of the particular vaccination strategy (except for the strategy “none,” which means no vaccinations occur]. In addition to the default transmission probability per CPR interval described above (i.e., 0.003), we tested lower (0.002) and higher (0.0045) transmission probability values.
Supplementary Material
Acknowledgments
We thank Ignacio Cancino, Elena V. Jordan, Alison Brown, and Rahel Salathé as well as members of the M.W.F., P.L., and J.H.J. groups for help with the mote deployments; Marc Horowitz for providing a crucial link; and two anonymous referees for their valuable comments. We are particularly grateful to the staff members of the school who made this project possible. Special thanks to the creators and maintainers of the Java Universal Network/Graph Framework and the R package iGraph. This research was supported by a National Science Foundation award (BCS-0947132), a Branco Weiss fellowship (to M.S.), National Institute of Child Health and Human Development Award 1K01HD051494 (to J.H.J.), and National Institutes of Health Grant GM28016 (to M.W.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009094108/-/DCSupplemental.
References
- 1.Murray CJL, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918-20 pandemic: A quantitative analysis. Lancet. 2006;368:2211–2218. doi: 10.1016/S0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- 2.Meltzer MI, Cox NJ, Fukuda K. The economic impact of pandemic influenza in the United States: Priorities for intervention. Emerg Infect Dis. 1999;5:659–671. doi: 10.3201/eid0505.990507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halloran ME, et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci USA. 2008;105:4639–4644. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326:729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiore A, et al. Prevention and control of influenza: Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2008;59(RR-8):1–62. and erratum (2010) 59:1147. [PubMed] [Google Scholar]
- 6.Xie X, Li Y, Chwang AT, Ho PL, Seto WH. How far droplets can move in indoor environments—Revisiting the Wells evaporation-falling curve. Indoor Air. 2007;17:211–225. doi: 10.1111/j.1600-0668.2007.00469.x. [DOI] [PubMed] [Google Scholar]
- 7.Cattuto C, et al. Dynamics of person-to-person interactions from distributed RFID sensor networks. PLoS ONE. 2010;5:e11596. doi: 10.1371/journal.pone.0011596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polastre J, Szewczyk R, Culler D. IPSN ’05: Proceedings of the Fourth International Symposium on Information Processing in Sensor Networks. IEEE Press: Los Angeles; 2005. Telos: Enabling ultra-low power wireless research. [Google Scholar]
- 9.Read JM, Eames KT, Edmunds WJ. Dynamic social networks and the implications for the spread of infectious disease. J R Soc Interface. 2008;5:1001–1007. doi: 10.1098/rsif.2008.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eagle N, Pentland A, Lazer D. Inferring friendship network structure by using mobile phone data. Proc Natl Acad Sci USA. 2009;106:15274–15278. doi: 10.1073/pnas.0900282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salathé M, Jones JH. Dynamics and control of diseases in networks with community structure. PLoS Comput Biol. 2010;6:e1000736. doi: 10.1371/journal.pcbi.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson NM, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson NM, et al. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longini IM, Jr., et al. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 15.Brockmann D, Hufnagel L, Geisel T. The scaling laws of human travel. Nature. 2006;439:462–465. doi: 10.1038/nature04292. [DOI] [PubMed] [Google Scholar]
- 16.Glass L, Glass R. Social contact networks for the spread of pandemic influenza in children and teenagers. BMC Public Health. 2008;8:61. doi: 10.1186/1471-2458-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May RM. Network structure and the biology of populations. Trends Ecol Evol. 2006;21:394–399. doi: 10.1016/j.tree.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Newman ME. Spread of epidemic disease on networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;66:016128. doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- 19.Liljeros F, Edling CR, Amaral LA, Stanley HE, Aberg Y. The web of human sexual contacts. Nature. 2001;411:907–908. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- 20.Jones JH, Handcock MS. Social networks: Sexual contacts and epidemic thresholds. Nature. 2003;423:605–606. doi: 10.1038/423605a. discussion 606. [DOI] [PubMed] [Google Scholar]
- 21.Anderson RM, May RM. Infectious Diseases of Humans, Dynamics and Control. Oxford, UK: Oxford Science Publications; 1991. [Google Scholar]
- 22.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 23.Newman ME. Mixing patterns in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67(Pt 2):026126. doi: 10.1103/PhysRevE.67.026126. [DOI] [PubMed] [Google Scholar]
- 24.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrat A, Barthelemy M, Pastor-Satorras R, Vespignani A. The architecture of complex weighted networks. Proc Natl Acad Sci USA. 2004;101:3747–3752. doi: 10.1073/pnas.0400087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers LA, Pourbohloul B, Newman MEJ, Skowronski DM, Brunham RC. Network theory and SARS: Predicting outbreak diversity. J Theor Biol. 2005;232:71–81. doi: 10.1016/j.jtbi.2004.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YP, Paul G, Havlin S, Liljeros F, Stanley HE. Finding a better immunization strategy. Phys Rev Lett. 2008;101:058701. doi: 10.1103/PhysRevLett.101.058701. [DOI] [PubMed] [Google Scholar]
- 29.Freeman LC. Centrality in social networks—Conceptual clarification. Soc Networks. 1978;1:215–239. [Google Scholar]
- 30.Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. Lancet Infect Dis. 2007;7:257–265. doi: 10.1016/S1473-3099(07)70029-4. [DOI] [PubMed] [Google Scholar]
- 31.Medlock J, Galvani AP. Optimizing influenza vaccine distribution. Science. 2009;325:1705–1708. doi: 10.1126/science.1175570. [DOI] [PubMed] [Google Scholar]
- 32.Moser MR, et al. An outbreak of influenza aboard a commercial airliner. Am J Epidemiol. 1979;110:1–6. doi: 10.1093/oxfordjournals.aje.a112781. [DOI] [PubMed] [Google Scholar]
- 33.Lessler J, et al. New York City Department of Health and Mental Hygiene Swine Influenza Investigation Team Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med. 2009;361:2628–2636. doi: 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- 34.King AA, Ionides EL, Pascual M, Bouma MJ. Inapparent infections and cholera dynamics. Nature. 2008;454:877–880. doi: 10.1038/nature07084. [DOI] [PubMed] [Google Scholar]
- 35.Influenza Team, European Centre for Disease Prevention and Control Influenza transmission: Research needs for informing infection control policies and practice. Euro Surveill. 2007;12:E070510. [PubMed] [Google Scholar]
- 36.Lau LL, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis. 2010;201:1509–1516. doi: 10.1086/652241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilder-Smith A, et al. Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerg Infect Dis. 2005;11:1142–1145. doi: 10.3201/eid1107.041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White S, Smyth P. Algorithms for estimating relative importance in networks. International Conference on Knowledge Discovery and Data Mining. 2003 KDD '03 Proceedings of the ninth ACM SIGKDD international conference on knowledge discovery and data mining (Association for Computing Machinery, New York), pp 266–275. [Google Scholar]
- 39.Reichardt J, Bornholdt S. Statistical mechanics of community detection. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:016110. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.