Abstract
Astrocyte-elevated gene-1 (AEG-1) expression increases in multiple cancers and plays a crucial role in oncogenic transformation and angiogenesis, which are essential components in tumor cell development, growth, and progression to metastasis. Moreover, AEG-1 directly contributes to resistance to chemotherapeutic drugs, another important hallmark of aggressive cancers. In the present study, we document that AEG-1 mediates protective autophagy, an important regulator of cancer survival under metabolic stress and resistance to apoptosis, which may underlie its significant cancer-promoting properties. AEG-1 induces noncanonical autophagy involving an increase in expression of ATG5. AEG-1 decreases the ATP/AMP ratio, resulting in diminished cellular metabolism and activation of AMP kinase, which induces AMPK/mammalian target of rapamycin-dependent autophagy. Inhibition of AMPK by siAMPK or compound C decreases expression of ATG5, ultimately attenuating AEG-1–induced autophagy. AEG-1 protects normal cells from serum starvation-induced death through protective autophagy, and inhibition of AEG-1–induced autophagy results in serum starvation-induced cell death. We also show that AEG-1–mediated chemoresistance is because of protective autophagy and inhibition of AEG-1 results in a decrease in protective autophagy and chemosensitization of cancer cells. In summary, the present study reveals a previously unknown aspect of AEG-1 function by identifying it as a potential regulator of protective autophagy, an important feature of AEG-1 that may contribute to its tumor-promoting properties.
Keywords: AMPK/mTOR, LC3, doxorubicin
Autophagy is an evolutionarily conserved process by which stressed cells catabolize their own contents, including long-lived proteins and cytoplasmic organelles, to provide cellular energy and building blocks for biosynthesis. This process is one of the cellular mechanisms regulating maintenance of homeostasis. Autophagy takes place when cells encounter environmental stressors, such as nutrient starvation, oxidative stress, accumulation of misfolded protein, xenobiotic treatment, and pathogen infection (1–3). During periods of intracellular metabolic stress, a reduced cellular energy (ATP) level is sensed by AMPK (5′-AMP-activated protein kinase). Active AMPK leads to phosphorylation and activation of the TSC1/2 complex, which inhibits mammalian target of rapamycin (mTOR) activity. The inhibition of mTORC1 activity leads to the activation of a set of evolutionarily conserved autophagy-regulating proteins (Atg proteins) and formation of autophagosomes, which fuse with lysosomes to form autolysosomes. The cargo in autolysosomes are then digested to metabolites and released back into the cytosol for recycling.
Although autophagy can function as a tumor-suppression mechanism (4), it has a more prominent role in sustaining cell viability in cancer cells with defects in apoptosis and undergoing long-term metabolic stress conditions. As tumors grow, cancer cells may require autophagy to survive nutrient-limited and low-oxygen conditions, especially in the central area of the tumor, which is often poorly vascularized. Survival through autophagy may be a key mechanism to enable long-term, tumor-cell viability and eventual regrowth and tumor recurrence (4–6). In addition, it has been reported that several cancer types, such as gastrointestinal, melanoma, and prostate, show a high frequency of autophagy markers, indicating a potential role of autophagy in tumor progression (7, 8).
Astrocyte elevated gene (AEG)-1 was cloned by rapid subtraction hybridization as a gene induced in primary human fetal astrocytes (PHFA) infected with HIV-1 or treated with TNF-α (9–12). Intriguingly, expression analysis revealed that AEG-1 was significantly elevated in subsets of breast carcinoma, hepatocellular carcinoma, melanoma, neuroblastoma, and malignant glioma cell lines compared with their normal cellular counterparts (9, 10, 13–19). These observations in cell lines have been confirmed and extended in patient-derived tumor samples mainly by immunohistochemistry in tissue microarrays. A prior study showed that strong AEG-1 staining was evident in 17 of 31 samples of breast carcinoma patients, whereas AEG-1 staining was absent in 18 of 20 samples of normal breast tissue (14). AEG-1 expression directly associates with a higher risk of metastasis, indicating that AEG-1 might be a prognostic factor that inversely correlates with patient survival in multiple types of cancer (14–19). Overexpression and inhibition studies in both in vitro and in vivo models emphasize the importance of AEG-1 in regulating multiple physiologically and pathologically relevant processes, including proliferation, invasion, metastasis, and gene expression. Overexpression of AEG-1 augmented the anchorage-independent growth of HeLa cells and human glioma cell lines and increased their migration and invasion properties (9, 20). AEG-1 also increased angiogenesis with expression of angiopoietin-1, matrix metalloprotease-2, and hypoxia-inducible factor 1-α (21). In PHFA, FM516-SV, and cloned rat embryo fibroblasts (CREF), AEG-1 protected cells from serum starvation-induced apoptosis by activating the PI3K/Akt signaling pathway (22). Conversely, inhibition of AEG-1 by siRNA significantly inhibited migration and invasion of malignant glioma and prostate cancer cells and suppressed in vivo lung metastasis of breast cancer cells (23, 24). Inhibition of AEG-1 in prostate cancer cells down-regulated Akt activation and lead to up-regulation of forkhead box 3a activity, resulting in apoptosis. AEG-1 promotes chemoresistance and inhibition of AEG-1 enhances response of tumor cells to chemotherapy (14, 25–27). In total, these observations indicate that AEG-1 may represent an essential gene regulating multiple signaling and biochemical pathways, leading to cell transformation and tumor progression in diverse target cells.
A number of questions remain concerning the potential role of AEG-1 in promoting survival of cancer cells in the tumor microenvironment. In the present study, we investigated the potential role of AEG-1 in induction of protective autophagy, which might directly contribute to AEG-1–induced transformation and tumor development. We found that AEG-1 promotes noncanonical autophagy by increasing expression of ATG5 in normal cells. AEG-1 reduces the ATP/AMP ratio, resulting in a decrease in cellular metabolism, and activates AMP kinase, which induces AMPK/mTOR-dependent autophagy. We also observed that AEG-1 induces autophagy to protect normal cells from apoptosis. Decreasing AEG-1 expression, thereby attenuating autophagy might represent a viable and effective approach for treating cancers resulting from and addicted to high AEG-1 expression.
Results
Ad.AEG-1 Induces Autophagy in Normal Cells.
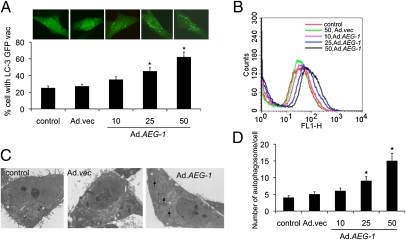
We first determined whether Ad.AEG-1 induces autophagy in immortalized primary human fetal astrocyte (IM-PHFA) cells. LC3, a mammalian homolog of yeast atg8, is essential for autophagosome formation. The intracellular localization of LC3 in autophagic vacuoles induced by Ad.AEG-1 was studied by transient transfection of IM-PHFA cells with a plasmid-expressing GFP fused with LC-3 (GFP-LC3) followed by Ad.AEG-1 infection. In control and Ad.vec-treated cells, GFP-LC3 was found predominantly as a diffuse green fluorescence in the cytoplasm. However, in Ad.AEG-1–treated cells, characteristic punctate fluorescent patterns were observed, indicating the recruitment of GFP-LC3 during autophagosome formation (Fig. 1A, Upper). The numbers of cells with punctate GFP-LC3 increased significantly in dose-dependent manner after 48 h of Ad.AEG-1 infection (Fig. 1A, Lower).
Fig. 1.
AEG-1 induces autophagy in IM-PHFA cells. (A) IM-PHFA cells were transfected with GFP-LC3 and infected with different pfu/cell of Ad.AEG-1 for 48 h, localization of LC3 in transfected cells was examined by confocal microscopy (magnification 100×), and autophagosome formation was quantified and data presented as percentage of GFP-LC3–transfected cells with punctate fluorescence to autophagosome formation. A minimum of 100 GFP-LC3–transfected cells were counted. *P < 0.05, compared with control. (B) The MDC fluorescent intensity of Ad.AEG-1–treated IM-PHFA cells was analyzed by flow cytometry. This result was representative of three different experiments. (C) IM-PHFA cells were infected with Ad.AEG-1 or Ad.vec for 48 h, fixed, and processed for electron microscopy (single arrow, autophagosome; control and others, 2 μm). (D) The numbers of autophagosomes in IM-PHFA cells 48 h after Ad.AEG-1 infection, the values are the means ± SD of three independent experiments. Asterisk indicates statistically significant change vs. corresponding control.
Because monodansylcadaverine (MDC) accumulates in mature autophagic vacuoles, such as autophagolysosomes, we confirmed Ad.AEG-1–induced autophagy by MDC-staining of IM-PHFA cells followed by flow cytometry. Similar to GFP-LC3 staining, we observed that MDC staining increased in a dose-dependent manner following 48 h of Ad.AEG-1 infection compared with control or Ad.vec-infected cells (Fig. 1B). We further verified that Ad.AEG-1 induced autophagy in IM-PHFA cells by electron microscopy. Electron micrographs of untreated and Ad.vec-infected cells showed normal morphology of all organelles, with mitochondria scattered homogenously throughout the cell (Fig. 1C). Images captured 48 h after Ad.AEG-1 infection indicated a marked accumulation of membrane-bound electron-dense structures sequestering cellular components, a distinctive feature of autophagosomes (Fig. 1C). In addition, there were many autophagic vacuoles or vesicles. Counting autophagic vesicles in the micrographs confirmed dose-dependent increases in Ad.AEG-1–infected cells (Fig. 1D).
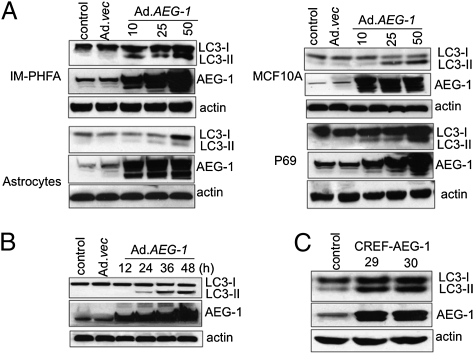
To rule out nonspecific aggregations of ectopically expressed GFP-LC3, we monitored changes in the expression of endogenous LC3. Infection of IM-PHFA cells with Ad.AEG-1 led to a rapid accumulation of the LC3-II form (corresponding to the characteristic lipidation of this protein during autophagosome formation) in a dose- and time-dependent manner compared with control and Ad.vec-infected cells (Fig. 2 A and B). Similar accumulation of LC3-II upon Ad.AEG-1 infection was also observed in primary human fetal astrocytes and other normal human cells, including MCF10A (immortal mammary epithelial cells) and P69 (immortal prostate epithelial cells), indicating that Ad.AEG-1 induces autophagy in normal cells of diverse genetic background (Fig. 2A). These results were extended using CREF cells stably overexpressing AEG-1, clones 29 and 30, indicating that AEG-1 induces autophagy both in normal human and rodent cells (Fig. 2C).
Fig. 2.
AEG-1 induces accumulation of LC3-II in normal cells. (A) IM-PHFA, Primary human fetal astrocytes, P69, and MCF10A cells were infected with Ad.AEG-1 or Ad.vec for 48 h and LC3 expression was analyzed by Western blotting. (B) IM-PHFA cells were infected with Ad.AEG-1 or Ad.vec for different times and LC3 expression was analyzed by Western blotting. (C) LC3 expression was examined in stable AEG-1–overexpressing CREF clones 29 and 30 (13).
Autophagy Induction by Ad.AEG-1 in IM-PHFA Cells Proceeds Through the Noncanonical Pathway.
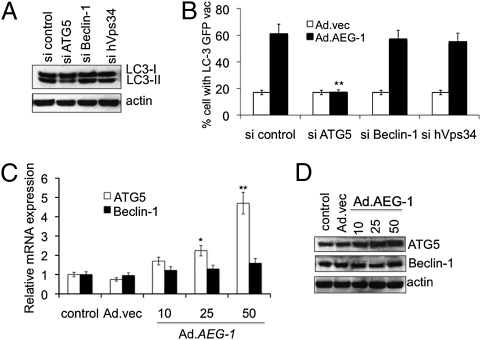
Autophagy can be induced by the canonical pathway, in which Beclin-1 initiates the generation of the autophagosome by forming a multiprotein complex with class III phosphatidylinositol-3-kinase or hVps34 (28). Alternatively, autophagy can occur by the noncanonical pathway independent of Beclin-1 and hVps34. To determine which pathway mediates Ad.AEG-1–induced autophagy, we used an siRNA approach to selectively knockdown essential autophagy (atg) genes, such as Beclin-1, atg5, and hVps34, and quantified GFP-LC3 punctate formation and LC3-II accumulation. The targeted siRNAs resulted in a significant down-regulation of their corresponding encoded proteins (Fig. S1A). Among these genes, inhibition of Beclin-1 and hVps34 did not decrease the percentage of GFP-LC3–positive cells or LC3-II levels. However, knockdown of atg5 significantly inhibited Ad.AEG-1–induced LC3-II accumulation (Fig. 3A) and punctate staining of GFP-LC3 vacuoles (Fig. 3B), indicating that Ad.AEG-1 triggers autophagy via the noncanonical pathway. The role of ATG5 was further confirmed by examining expression profiles of ATG5 at the mRNA and protein levels, supporting the potential involvement of ATG5 in Ad.AEG-1–induced autophagy (Fig. 3 C and D).
Fig. 3.
AEG-1 induces noncanonical autophagy in IM-PHFA cells. (A) IM-PHFA cells were transfected with the indicated siRNAs and LC3-GFP followed by infection with 50 pfu/cell Ad.AEG-1 or Ad.vec and cytoplasmic aggregation of LC3-GFP was determined. A minimum of 100 GFP-LC3–transfected cells were counted. *P < 0.05; **P < 0.001, compared with sicontrol. (B) LC3-II expression was determined 48 h after administration of the indicated siRNAs and Ad.AEG-1 infection (50 pfu/cell) by immunoblotting. IM-PHFA Cells were infected with Ad.AEG-1 or Ad.vec for 48 h and mRNA (C) and protein (D) expression of Beclin-1 and ATG5 were analyzed using Taqman real-time PCR and Western blotting, respectively. Values are the mean ± SD of three independent experiments and *P < 0.05; **P < 0.001 vs. control cells.
Ad.AEG-1–Induced Cellular Energy Deficiency Leads to AMPK/mTOR Pathway-Dependent Autophagy.
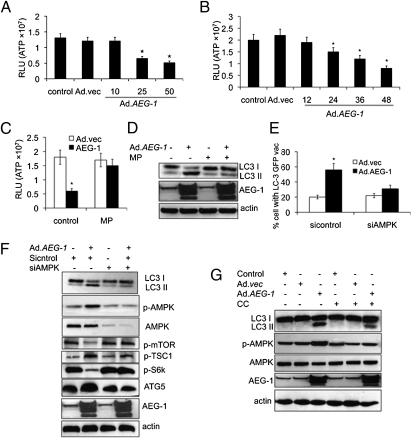
Akt is believed to contribute to the activation of mTOR, which is a crucial autophagy regulator, either directly or indirectly through phosphorylation and suppression of TSC2 (29). AEG-1 was previously shown to activate Akt (22), a pivotal prosurvival and pro-oncogenic protein that is activated by serine and threonine phosphorylation and activation of Akt by AEG-1 nullified its role in AEG-1–induced autophagy (Fig. S1B). Autophagy is also activated in response to cellular oxidative stress from reactive oxygen species (ROS) (29, 30) and we hypothesized that AEG-1–induced ROS might activate autophagy. However, we observed that AEG-1 did not increase ROS, suggesting that this process does not contribute to Ad.AEG1-induced autophagy (Fig. S1C). Alternatively, we investigated whether a decrease in ATP/AMP ratio might induce autophagy by affecting cellular bioenergetics (2, 3, 29). To test this possibility, we monitored changes in cellular ATP levels in response to AEG-1. IM-PHFA cells infected with Ad.AEG-1 displayed a dose- and time-dependent decrease in the levels of ATP (Fig. 4 A and B), suggesting that cellular energy depletion might be responsible for AEG-1–induced autophagy. For that reason, we next determined whether the depletion of ATP production in AEG-1–overexpressing cells could be reversed by pretreating cells with a cell-permeable form of pyruvate, methylpyruvate (MP), which can be oxidized in the tricarboxylic acid cycle to produce NADH, which fuels the electron transport system and ATP production. MP restored ATP production in AEG-1–infected cells to levels that paralleled those observed in untreated cells (Fig. 4C). Moreover, LC3 II accumulation was reduced after addition of MP in AEG-1–overexpressing IM-PHFA cells (Fig. 4D). Collectively, our data demonstrate that AEG-1–induced disruption of cellular bioenergetics contributes to autophagy formation.
Fig. 4.
AEG-1 induces cell energy deficiency and activates the AMPK/mTOR-dependent pathway by autophagy. IM-PHFA Cells were infected with Ad.AEG-1 or Ad.vec at different doses (pfu/cell) (A) and times (h) (B) followed by measurement of ATP levels using Bioluminescence Assay Kit. Asterisks indicate statistically significant change vs. corresponding control. IM-PHFA cells were infected with 50 pfu/cell of Ad.AEG-1 or Ad.vec for 48 h and then pretreated with 1 mM MP for 2 h followed by measurement of ATP levels in cells (C) and analysis of LC3 expression by immunoblotting (D). (E) IM-PHFA Cells were transfected with siAMPK and LC3-GFP followed by infection with 50 pfu/cell of Ad.AEG-1 or Ad.vec and cytoplasmic aggregation of LC3-GFP was determined. A minimum of 100 GFP-LC3–transfected cells were counted. *P < 0.05; **P < 0.001, compared with sicontrol. (F) LC3-II expression and other protein expressions were determined 48 h after administration of the siAMPK and Ad.AEG-1 infection (50 pfu/cell) by immunoblotting. (G) IM-PHFA cells were infected with 50 pfu/cell of Ad.AEG-1 or Ad.vec in the presence of compound C (10 μmol/L for 1 h) for 48 h, followed by analysis of LC3 expression and other proteins by immunoblotting.
AMPK is activated as intracellular AMP/ATP ratios rise (20). AEG-1 caused a significant increase of AMPK phosphorylation at Thr-172 (Fig. S1D). Phospho-AMPKThr-172 phosphorylates and activates TSC2, further inhibiting the activation of downstream targets, such as mTOR and S6. Phosphorylation of mTOR and S6 proteins was substantially decreased in AEG-1–treated cells. To address whether AMPK activation plays an essential role in AEG-1–induced autophagy formation, we determined the ability of AEG-1 to induce autophagy in AMPK suppressed cells by (i) using siAMPK and (ii) treating the cells with a pharmacological inhibitor of AMPK (compound C, CC), a cell-permeable pyrrazolopyrimidine compound that acts as a potent, selective, reversible, and ATP-competitive inhibitor of AMPK. AEG-1–induced autophagy as measured by LC3 II accumulation was significantly decreased in AMPK knockdown IM-PHFA cells compared with scrambled sicontrol-treated cells. This finding was demonstrated by both confocal microscopy and Western blotting (Fig. 4 E and F). Furthermore, all predicted downstream molecules were altered in siAMPK cells, resulting in a decrease in expression of ATG5 and ultimately a decrease in AEG-1–induced autophagy. Similarly, Compound C also inhibited AEG-1–induced phosphorylation of AMPK and autophagy (Fig. 4G). These data indicate that AMPK or its downstream targets are important mediators of AEG-1–induced autophagy in normal cells.
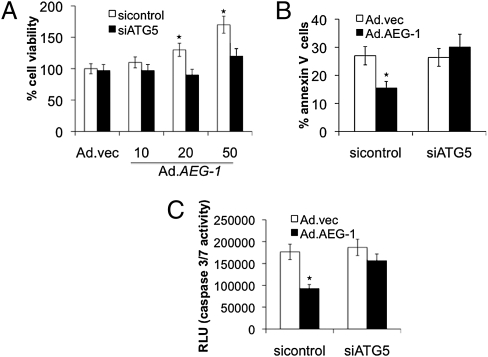
Blocking ATG5 Inhibits AEG-1–Mediated Survival and Enhances Apoptosis of IM-PHFA Cells Grown Under Serum-Starvation Conditions.
Previous experiments indicate that AEG-1 enhances cell survival of normal cells under serum-starvation conditions by inhibiting apoptosis (22). To investigate the role of AEG-1–induced autophagy in survival of normal cells under serum-starvation conditions, we analyzed cell survival by standard MTT assay of AEG-1–infected ATG5-deficient IM-PHFA cells grown in 0.1% FBS (Fig. 5A). Infection of ATG5-deficient (siATG5-treated) IM-PHFA cells with Ad.AEG-1 prevented increased cell survival under low-serum conditions compared with Ad.AEG-1–infected parental sicontrol-treated IM-PHFA cells. We also analyzed cell death using Annexin V binding and propidium iodide staining by flow cytometry (Fig. 5B) in siATG5- and sicontrol-treated IM-PHFA cells infected with Ad.AEG-1. The results showed that ATG5-deficient IM-PHFA cells infected with Ad.AEG-1 were sensitive to serum starvation-induced apoptosis, whereas AEG-1 overexpression in sicontrol IM-PHFA cells protected these cells from apoptosis. The caspase3/7 assay corroborated the data obtained using Annexin V staining (Fig. 5C). Additionally, caspase-3/7 activity was increased in ATG5-deficient Ad.AEG-1–infected IM-PHFA cells, indicating that AEG-1 promotes cell survival by altering both autophagy and apoptotic mechanisms in normal cells grown under serum-starvation conditions.
Fig. 5.
Inhibition of autophagy resulted in serum starvation-induced cell death. IM-PHFA cells were transfected with siATG5 and MTT assay for cell viability (A), Annexin V assay for apoptosis (B), and Caspase-Glo(R) 3/7 assay for caspase-3 expression (C) was performed 48 h after infection with Ad.AEG-1 (10, 20, or 50 pfu/cell) or Ad.vec (50 pfu/cell), followed by 4 d after serum starvation (0.1%). *P < 0.05, compared with sicontrol.
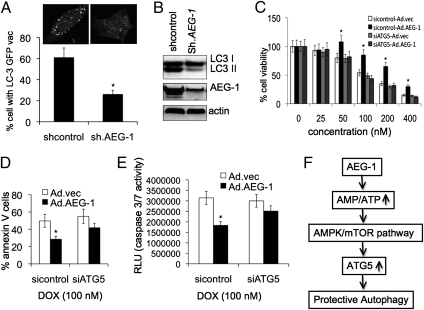
Inhibition of AEG-1–Induced Autophagy in Cancer Cells Enhances Sensitivity to Chemotherapeutic Agents.
We recently observed that AEG-1 is frequently overexpressed in a number of histologically distinct human tumors and AEG-1 regulates multiple physiologically and pathologically relevant processes, including cell proliferation, invasion, metastasis, angiogenesis, and defined programs of gene expression (9). Because AEG-1 induces ATG5-mediated autophagy in normal cells, the higher expression of AEG-1 might be associated with elevated autophagy in many cancer cells. To investigate this hypothesis, we developed shAEG-1-T98G cells, a stable AEG-1 knockdown clone in T98G, a human malignant glioma cell line, and observed that shAEG-1-T98G cells had decreased accumulation of LC3 II compared with shAEG-1-T98G control cells, confirmed by confocal microscopy and Western blotting (Fig. 6 A and B). A similar decrease in accumulation of LC3-II upon Ad.shAEG-1 infection was also observed in human HeLa (cervical carcinoma), MDA-MB-231 (breast carcinoma), HO-1 (melanoma), and MIA PaCa 2 (pancreatic carcinoma) cells, indicating that higher expression of AEG-1 may be a general phenomenon associated with autophagy in cancer cells (Figs. S2 A and B, and S3 A and B). Previous observations showed that knockdown of AEG-1 significantly enhanced chemosensitivity. To determine whether AEG-1–induced chemoresistance is mediated through autophagy, we evaluated growth inhibition potential of chemotherapeutic reagents including doxorubicin in atg5-deficient T98G cells in the presence of AEG-1. The results demonstrated that Ad.AEG-1 infection increased the resistance of T98G cells toward doxorubicin as measured by standard MTT assays, whereas atg5-deficient cells exhibited chemosensitivity toward this drug (Fig. 6C). This finding was confirmed by measuring apoptotic cells using Annexin V binding and caspase 3/7 assays (Fig. 6 D and E). Moreover, the chemosensitizing effect of AEG-1–mediated autophagy inhibition was also demonstrated in HeLa, MDA-MB-231, HO-1, and MIA PaCa 2 cells (Figs. S2 C and D, and S3 C and D), indicating that AEG-1 chemoresistance of diverse human cancer cells might also be mediated in part through protective autophagy induced by AEG-1.
Fig. 6.
Inhibition of AEG-1–induced autophagy in cancer cells enhances sensitivity to chemotherapeutic agents. (A) Stable shAEG-1-T98G cells were transfected with GFP-LC3, localization of LC3 in transfected cells was examined by confocal microscopy (magnification 100×) and autophagosome formation was quantified and data presented as percentage of GFP-LC3–transfected cells with punctate fluorescence to autophagosome formation. A minimum of 100 GFP-LC3–transfected cells were counted. *P < 0.05, compared with control. (B) LC3 expression was analyzed in shAEG-1-T98G cells by Western blotting. T98G cells were transfected with siATG5, and MTT assays for cell viability (C), Annexin V assays for apoptosis (D), and Caspase Glo(R) 3/7 assays for caspase 3 expression (E) were performed 24 h after infection with Ad.AEG-1 (50 pfu/cell) or Ad.vec (50 pfu/cell), followed by 4-d treatment with doxorubicin. *P < 0.05, compared with sicontrol.Ad.vec. (F) Model illustrating the possible molecular mechanism of AEG-1–mediated protective autophagy, which leads to escape from apoptosis and chemotherapy toxicity.
Discussion
Cancer is a progressive process culminating in transformed cells that develop both qualitatively and quantitatively distinct properties as a tumor expands and evolves. An initial step in conversion of a cancer cell from normal to transformed frequently involves the activation of a positive-acting oncogene that significantly alters cellular phenotype (31, 32). Recent studies provide compelling evidence that during cancer progression, autophagy can serve as a protective mechanism in sustaining cell viability in apoptosis-defective and long-term metabolic-stressed cancer cells (5, 6). AEG-1 plays a central role in oncogenesis, tumor progression, and metastasis (9, 10). In the present study, we define a previously unrecognized role of AEG-1 in promoting protective autophagy and we unravel AEG-1’s physiological significance in cell survival under restrictive/apoptosis-inducing conditions. These studies provide important insights and a unique perspective on this multifunctional oncogene, AEG-1.
We now demonstrate that AEG-1 induces autophagy in a Beclin-1- and class III PI3 kinase-independent manner, and this effect is mediated through up-regulation of ATG5. Moreover, AEG-1 promoted autophagy in different normal human cell types derived from prostate, breast, and brain tissue, and in normal immortal CREF cells, indicating that this phenomenon is not restricted to a specific normal cell type or species, but rather represents a generalized property associated with elevated AEG-1 expression. Evidence is provided that decreased ATP levels in cells play a central role in AEG-1–induced autophagy. AMPK is sensitive to the cytosolic AMP-to-ATP ratio and metabolic stress activates autophagy by suppression of mTOR signaling. AMPK plays a major role in energy homeostasis by coordinating a number of adaptive responses under ATP-depleting metabolic stresses (29). Upon activation, AMPK down-regulates several anabolic enzymes by phosphorylation and shuts down the ATP-consuming metabolic pathways, whereas its activation facilitates energy-producing pathways. Our study confirms that the decrease in ATP causes a type of metabolic stress, resulting in activation of AMPK followed by alterations in downstream molecules and induction of autophagy. We confirmed that inhibition of AMPK activity, either by compound C or by siAMPK, significantly reduced AEG-1–induced autophagy. Furthermore, inhibition of AMPK induced activation of mTOR and S6K and inactivation of TSC1, followed by a decrease in ATG5 expression and promotion of autophagy. Taken together, these data provide relevant insights into the complex nature of AMPK and downstream responses that may be involved in AEG-1–induced protective autophagy and cancer progression.
Autophagy plays an important protective mechanism in cell survival during nutrient and growth factor deprivation (29). It maintains the viability of starved cells by temporally digesting portions of cytoplasm to supply vital substrates and energy. Additionally, oncogenic autophagy contributes to maintenance of cancer cell viability during these stressful conditions. A recent study indicated that overexpression of the MUC1 oncoprotein, found in many human cancers, induced autophagy and provided a survival advantage in microenvironments with low glucose levels (33). Similarly, the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process in response to growth factor deprivation (34). In another study, simian virus 40 small T-antigen enabled cancer cell survival under glucose deprivation via inhibition of protein synthesis and activation of autophagy as an alternate energy source (35). Our previous studies demonstrated that AEG-1 could induce serum-independent cell growth, another property of oncogenes, by inhibiting apoptosis. Here we provide evidence that the serum-starvation protective mechanism of AEG-1 is also controlled by AEG-1–mediated autophagy and inhibition of AEG-1–induced autophagy resulted in serum starvation-induced cell death. This study explains the multiple oncogenic properties of AEG-1 for protection of cancer cells in unfavorable conditions, such as nutrition deprivation.
Autophagy has been shown to provide resistance to therapy-mediated tumor cell death (36, 37). When tumor cells induce protective autophagy, inhibition of autophagy could provide a way of sensitizing tumor cells to therapy by activating apoptosis. Here we demonstrate that chemoresistance promoted by AEG-1 may be mediated through the ability of this gene to induce protective autophagy. Our study indicated that inhibition of AEG-1 results in a decrease in protective autophagy and causes chemosensitization of cancer cells. These results are in agreement with previous observations that inhibiting AEG-1 expression suppresses both in vitro and in vivo tumor phenotypes, indicating that the ability of AEG-1 to confer chemoresistance is mediated by protective autophagy induced by this cancer-promoting gene. In summary, the present study reveals distinctive aspects of AEG-1 function and identifies this gene as a unique regulator of protective autophagy, which may directly contribute to the tumor-promoting potential of AEG-1 under metabolic stress and apoptosis-deficient conditions.
Materials and Methods
Cell Lines, Culture Conditions, and Viability Assays.
PHFA, IM-PHFA, P69 (an SV40-immortalized human prostate epithelial cell line), and CREF cells stably overexpressing AEG-1 were cultured as described (14). MCF10A cells were obtained from the American Type Culture Collection and cultured as described. Cells were infected with 50 pfu/cell of Ad.AEG-1 or Ad.vec and analyzed, as described. Cell viability by MTT was performed as described (28).
Measurement of Autophagy.
After infection of Ad.AEG-1 for 48 h, cells were cultured with 0.05 mmol/L MDC and analyzed by FACScan flow cytometry. IM-PHFA cells were transfected with GFP-labeled LC3 fusion protein followed by infection with Ad.AEG-1 for different times and analyzed by confocal laser-scanning microscopy (Zeiss 510 Meta confocal imaging system) (28).
Transmission Electron Microscopy.
For transmission electron microscopy, IM-PHFA cells were processed and analyzed by transmission electron microscopy (Joel JEM-1230 equipped with a Gatan UltraScan 4000SP 4K × 4K charge-coupled device camera), as described (28).
Annexin V Binding and Caspase Activity Assays.
Annexin V-binding assays were performed as previously described (19). Caspase 3/7 activities in IM-PHFA were measured using Caspases-Glo 3/7 Assay kits (Promega) according to the manufacturer's instructions.
Western Blot Analysis.
Preparation of whole-cell lysates and Western blotting for AEG-1, LC3, ATG5, hVps34, Beclin-1, total AMPK, phospho-AMPK, phospho-mTOR, phospho-TSC-1, and phospho-S6K protein levels were performed as described (28).
Extraction of Total RNA and Real-Time PCR.
Total RNA was extracted using a RNeasy Mini kit (Qiagen). Real-time PCR was performed using ABI 7900 Fast Real-Time PCR System and Taqman gene-expression assays (Applied Biosystems).
Statistical Analysis.
Data are represented as the mean ± SE and analyzed for statistical significance using one-way ANOVA followed by Newman-Keul's test as a post hoc test. A P value of <0.05 was considered significant.
Supplementary Material
Acknowledgments
The present study was supported in part by National Institutes of Health Grants R01 CA127641 and CA134721, the Samuel Waxman Cancer Research Foundation and the National Foundation for Cancer Research (to P.B.F.), National Cancer Institute Grant R01 CA138540 (to D.S.), and the National Research Foundation Medical Research Center Program of the Korean Ministry of Education, Science and Technology (2009-0063466) (to S.-G.L). D.S. is the Harrison Endowed Scholar in Cancer Research at the Virginia Commonwealth University Massey Cancer Center. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research in the Virginia Commonwealth University Massey Cancer Center and is a Samuel Waxman Cancer Research Foundation Investigator.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009479107/-/DCSupplemental.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 4.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 5.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshioka A, et al. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461–468. [PubMed] [Google Scholar]
- 8.Miracco C, et al. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol. 2010;41:503–512. doi: 10.1016/j.humpath.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar D, et al. Astrocyte elevated gene-1: Far more than just a gene regulated in astrocytes. Cancer Res. 2009;69:8529–8535. doi: 10.1158/0008-5472.CAN-09-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emdad L, et al. Astrocyte elevated gene-1: Recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther. 2007;114:155–170. doi: 10.1016/j.pharmthera.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su ZZ, et al. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- 12.Kang DC, et al. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 14.Hu G, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 16.Yoo BK, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SG, et al. Astrocyte elevated gene-1 contributes to the pathogenesis of neuroblastoma. Oncogene. 2009;28:2476–2484. doi: 10.1038/onc.2009.93. [DOI] [PubMed] [Google Scholar]
- 18.Song L, et al. Over-expression of AEG-1 significantly associates with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. J Pathol. 2009;219:317–326. doi: 10.1002/path.2595. [DOI] [PubMed] [Google Scholar]
- 19.Yu C, et al. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. doi: 10.1093/carcin/bgp064. [DOI] [PubMed] [Google Scholar]
- 20.Emdad L, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 21.Emdad L, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci USA. 2009;106:21300–21305. doi: 10.1073/pnas.0910936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-G, et al. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene. 2008;27:1114–1121. doi: 10.1038/sj.onc.1210713. [DOI] [PubMed] [Google Scholar]
- 23.Kikuno N, et al. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26:7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- 24.Emdad L, et al. Astrocyte elevated gene-1: A novel target for human glioma therapy. Mol Cancer Ther. 2010;9:79–88. doi: 10.1158/1535-7163.MCT-09-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo BK, et al. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 2010;70:3249–3258. doi: 10.1158/0008-5472.CAN-09-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo BK, et al. Identification of genes conferring resistance to 5-fluorouracil. Proc Natl Acad Sci USA. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, et al. Knockdown of astrocyte elevated gene-1 inhibits proliferation and enhancing chemo-sensitivity to cisplatin or doxorubicin in neuroblastoma cells. J Exp Clin Cancer Res. 2009;28:19–28. doi: 10.1186/1756-9966-28-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhutia SK, et al. Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res. 2010;70:3667–3676. doi: 10.1158/0008-5472.CAN-09-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 31.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 32.Fisher PB. Enhancement of viral transformation and expression of the transformed phenotype by tumor promoters. In: Slaga TJ, editor. Tumor Promotion and Cocarcinogenesis In Vitro, Mechanisms of Tumor Promotion, Enhancement of Viral Transformation and Expression of the Transformed Phenotype by Tumor Promoters. Boca Raton, FL: CRC Press; 1984. pp. 57–123. [Google Scholar]
- 33.Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein promotes autophagy in a survival response to glucose deprivation. Int J Oncol. 2009;34:1691–1699. doi: 10.3892/ijo_00000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Münger K. Expression of the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology. 2009;385:192–197. doi: 10.1016/j.virol.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar SH, Rangarajan A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol. 2009;83:8565–8574. doi: 10.1128/JVI.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann Surg Oncol. 2009;16:761–771. doi: 10.1245/s10434-008-0260-0. [DOI] [PubMed] [Google Scholar]
- 37.Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumour cells. J Cell Mol Med. 2009;13(9B):3644–3654. doi: 10.1111/j.1582-4934.2009.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.