Abstract
Ig class switch recombination (CSR) requires expression of activation-induced cytidine deaminase (AID) and transcription through target switch (S) regions. Here we show that knockdown of the histone chaperone facilitates chromatin transcription (FACT) completely inhibited S region cleavage and CSR in IgA-switch-inducible CH12F3-2A B cells. FACT knockdown did not reduce AID or S region transcripts but did decrease histone3 lysine4 trimethylation (H3K4me3) at both the Sμ and Sα regions. Because knockdown of FACT or H3K4 methyltransferase cofactors inhibited DNA cleavage in H3K4me3-depleted S regions, H3K4me3 may serve as a mark for recruiting CSR recombinase. These findings revealed an unexpected evolutionary conservation between CSR and meiotic recombination.
Antigen stimulation of B lymphocytes induces the expression of activation-induced cytidine deaminase (AID), which is responsible for generation of antibody memory (1, 2). Somatic hypermutation and class switch recombination (CSR) are two genetic events that engrave antibody memory into the Ig heavy-chain (H) locus of the B cell genome. CSR takes place between two switch (S) regions, located upstream of the individual H constant regions (CH) and converts the isotype from IgM to another class by bringing the specific CH region close to the H variable region (VH) exons and looping out the intervening DNA segment (3).
Gene-targeting experiments in the IgH locus have shown that active transcription through the S regions is an essential requirement of CSR (4, 5). This transcription initiates from the I promoter, located upstream of each S region, and proceeds through the I exon, the intronic S region, the CH exons, and the CH introns. The mature transcripts, designated as germline transcripts (GLTs), are generated by splicing out the S region and CH intronic sequences (3). However, it is not well understood whether the transcription itself, the transcription products, or both are important for CSR. The original chromatin-opening hypothesis suggested that transcription of the S region causes its chromatin structure to be relatively open, which increases its accessibility to a putative recombinase (6, 7). In fact, the migration of the transcription machinery accumulates positive and negative supercoil in its front and rear, respectively.
During this process, R-loop formation was detected in the DNA from switching B cells by the bisulfite sensitivity assay (8). The R-loop formation was considered to support the DNA deamination hypothesis proposed for the function of AID, as the single-strand DNA can serve as an efficient substrate of cytidine deamination by AID, as demonstrated in vitro (9). This hypothesis postulates that dU generated by AID deamination is recognized as a dU/dG mismatch and excised by uracil DNA glycosylase (10). The abasic sites thus formed is then cleaved by apyrimidinic/apurinic endonuclease. It has been also proposed that dU/dG mismatches are recognized and cleaved by mismatch repair proteins such as Msh2 and Msh6.
On the other hand, AID was recently shown to reduce the translation of Topoisomerase 1 (Top1) mRNA and thus decrease its protein level (11). The decrease in Top1 causes inefficient recovery of the excessive negative supercoil of the transcribed S region because Top1 removes the excessive supercoil by nicking and transient covalent binding to DNA, followed by rotation and religation. It was postulated that the resultant prolongation of the negative supercoil can induce the formation of non-B form DNA in the S region (12). Top1 can cleave non-B form DNA but not rotate efficiently because of its aberrant structure, resulting in irreversible single-stranded cleavage. According to this model, Top1 is the enzyme that cleaves the S region during CSR.
Despite these studies, however, the transcription requirement for CSR has not been fully elucidated, partly because in vivo transcription occurs on a chromatin template, in which the DNA is wrapped around core histone octamers (H2A, H2B, H3, and H4) (13). The transcriptional migration of RNA polymerase II (Pol II) along the chromatin template requires the reorganization of nucleosomes and numerous histone posttranslational modifications (PTMs) that include H2B ubiquitination and H3 methylation in transcribed regions (14). Such nucleosomal reorganization and modifications require the orchestrated contribution of numerous accessory factors. One of the most important of these components is the facilitates chromatin transcription (FACT) complex, which has been proposed to facilitate the passage of Pol II through the chromatin (15). An in vitro chromatin transcription assay demonstrated that human FACT acts as a histone chaperone that can displace H2A and H2B (16) from a nucleosome in front of Pol II, and can replace them again behind it. Therefore, one of the major proposed functions of FACT is the rapid reassembly of nucleosomes on highly transcribed regions.
In the present study, we demonstrated that the histone3 lysine4 trimethylation (H3K4me3) mark of the S-region chromatin, which is regulated by the FACT core components (SSRP1 and SPT16), was required for DNA cleavage of the target S region during CSR. This finding revealed a surprising conservation of the H3K4me3 mark requirement for V(D)J, meiotic, and class switch recombination (17–19).
Results
Requirement of the SSRP1 and SPT16 Heterodimer for CSR.
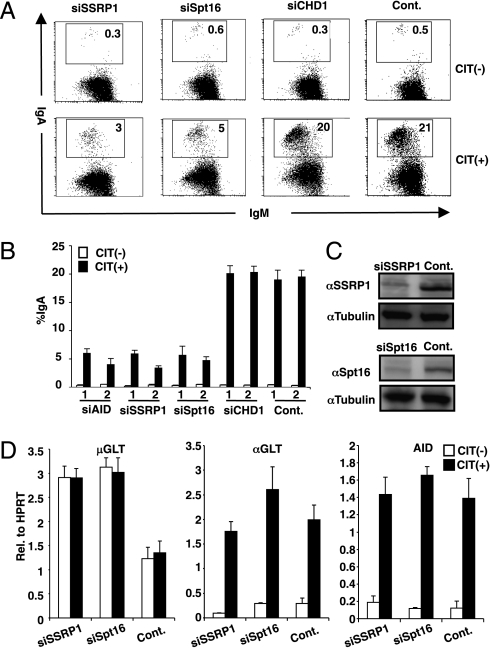
To determine the role of the chromatin structure during transcription of the S region in CSR, we knocked down the FACT components (SSRP1 and SPT16) in the CH12F3-2A cell line, which undergoes IgM to IgA switching with high frequency upon stimulation with CD40L, IL-4, and TGFβ (CIT) (20). The FACS profiles showed that the knockdown of SSRP1 or SPT16 dramatically inhibited the IgA switching in CH12F3-2A cells stimulated with CIT (3% vs. 21%) (Fig. 1 A and B). This inhibitory effect persisted for at least 2 d without significant cell death or proliferation defect. The introduced siRNAs for SSRP1 and SPT16 specifically reduced their target transcripts and proteins (Fig. 1C and Fig. S1A). However, neither GLTs of Sμ and Sα nor AID transcripts were significantly reduced by FACT knockdown (Fig. 1D). Furthermore, Sμ GLTs were rather augmented, probably due to histone loss from the sites, which may allow uncontrolled transcription (discussed below). Consistently, ChIP analysis showed that Pol II occupancy was not reduced at the Sμ region (Fig. S2). As expected, the SSRP1 knockdown also strongly inhibited IgG1 switching in splenic B cells stimulated with lipopolysaccharide and IL-4 (Fig. S1B).
Fig. 1.
SSRP1 and Spt16 play a critical role in CSR. (A) Flow cytometry (FACS) profile of the IgA switching population following the introduction of the indicated RNAi oligonucleotide introduction under CIT(−) and (+) conditions. (B) Summarized data of IgA switching population of various gene knockdowns with respect to AID in CIT(−) and (+) conditions. Numbers 1 and 2 represent two independent RNAi oligonucleotides. SD values were determined from three independent experiments. (C) Knockdown efficiency of the indicated gene was quantified by immunobloting. (D) μGLT, αGLT, and AID transcripts were quantified by real-time PCR normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) following the introduction of the indicated RNAi oligonucleotide. SD values were derived from three independent experiments.
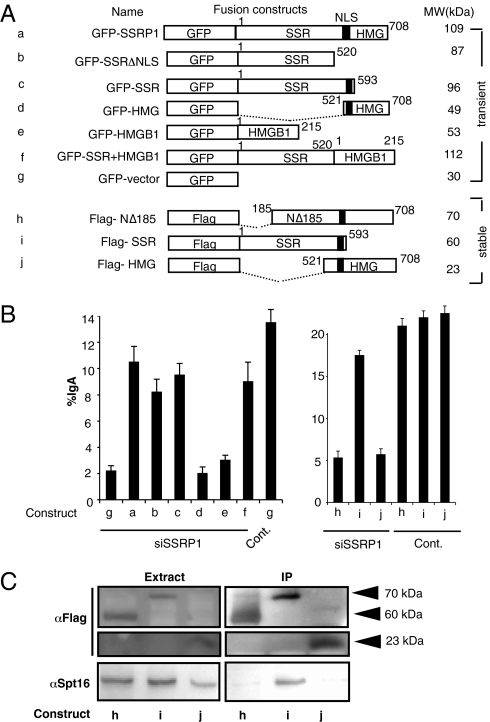
To confirm the role of SSRP1 as the FACT complex in CSR, we constructed siRNA-resistant version of SSRP1 and its mutants. Introduction of GFP-SSRP1 successfully complemented the effect of SSRP1 knockdown to almost the control level, whereas introducing GFP alone was not effective (Fig. 2 A and B). SSRP1 (708 residues) possesses two distinct domains: an N-terminal SSR (structure-specific recognition) domain of 520 residues, and a C-terminal HMG (high- mobility group)-like domain of 188 residues, which interact with SPT16 and CHD1, respectively (21, 22). Rescue experiments with various deletion constructs showed that the SSR but not HMG domain (or its paralogue HMGB1) is necessary for CSR, in agreement with the requirement of SPT16 but not CHD1 (Fig. 1 A and B, knockdown experiments).
Fig. 2.
Rescue of CSR in SSRP1 knockdown cells expressing various SSRP1 deletion mutants and coimmunoprecipitation of SSRP1 and Spt16. (A) Representation of the various SSRP1 mutants and related constructs used in the CSR complementation experiment. Amino acid positions are indicated above each construct. All constructs used were well expressed (Fig. S3), and the expected molecular weights are shown on the right. NLS, nuclear localization signal. (B) Percentages of the IgA population as determined by FACS following the introduction of SSRP1 RNAi oligonucleotide along with various GFP- and Flag-tagged constructs. CSR efficiency was measured as the percentage of surface IgA expression in GFP-positive cells. SD values were determined from three independent experiments. (C) Immunoprecipitation (IP) by anti-Flag and the immunoblot analysis of Flag constructs and Spt16. Arrowheads indicate the expected molecular weights.
Flag-SSR (residues 1–593) efficiently pulled down SPT16 and almost fully complemented the reduction of IgA switching by the knockdown of endogenous SSRP1 (Fig. 2 B and C). On the other hand, Flag-NΔ185 (N-terminal 184-residue deletion) and Flag-HMG (residues 521–708) failed to pull down SPT16 or to rescue IgA switching. We therefore conclude that the N-terminal but not the C-terminal region of SSRP1 is critical for the SPT16 interaction. Interestingly, the NLS motif located between the SSR and HMG domains is not essential to CSR, probably because SSRP1 migrates with Spt16 to the nucleus. Taken together, these results indicate that the FACT complex consisting of the SSRP1 and SPT16 heterodimer is required for efficient CSR.
FACT Depletion Alters the Chromatin Landscape of the Sμ and Sα Regions Differentially.
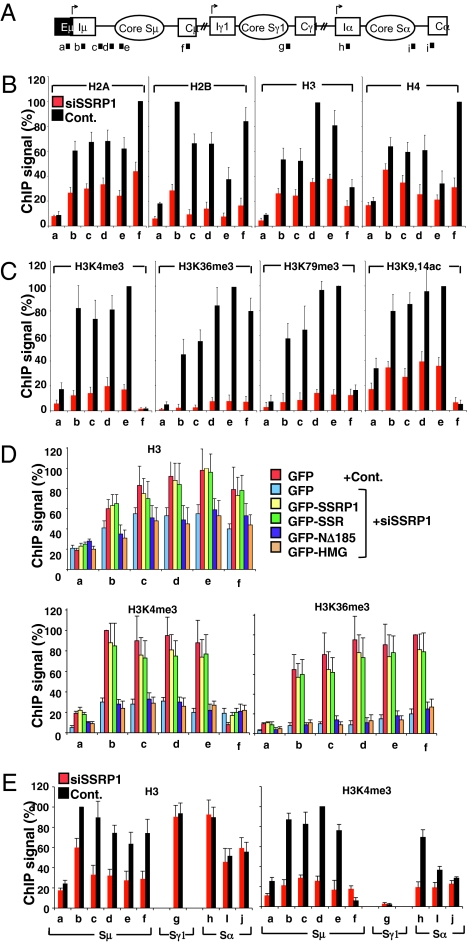
We investigated the chromatin state in the FACT-deficient condition by ChIP analysis and observed a reduction of the core histone levels in the Sμ region and its surroundings upon SSRP1 knockdown (Fig. 3 A and B). This observation was supported by MNase-based nucleosome mapping experiments in which recovered DNA from SSRP1 knockdown samples gave a lower signal intensity than control samples (Fig. S4). Although the FACT complex is known to target H2A and H2B in vitro (16), nearly 50% of all of the core histones were displaced in the absence of SSRP1. This suggests that, in vivo, FACT maintains the balance of all four core histones in the Sμ region; this agrees with a recent report using FACT-deficient yeast (23).
Fig. 3.
SSRP1 regulates the histone status of the S regions. (A) Schematic diagram of the position of the PCR products for ChIP assay. (B, C, and E) The ChIP assay was performed in SSRP1 knockdown and control samples using the indicated antibodies. No difference was observed between CIT(−) and (+) conditions. Data shown are under CIT(−) condition. (D) Rescue experiments of histone H3, H3K4me3 and H3K36me3 using various SSRP1 mutants in SSRP1 knockdown and control samples. Background values from controls with no antibody were subtracted (generally <5% of antibody signal). Values were normalized to the DNA input signals, followed by the maximum value in each data set. SD values were derived from three independent experiments.
Because FACT knockdown altered the chromatin landscape, we examined known histone modifications in chromatin of the actively transcribed Sμ region: three trimethylation marks (H3K4Me3, H3K79Me3, H3K36Me3) and the total histone H3 acetylation mark (H3K9,14Ac). Remarkably, all of the histone H3 methylation marks were dramatically decreased in the Sμ region by SSRP1 knockdown, whereas the acetylation status remained higher than the methylation status and consistent with the level of histone H3 at the sites (Fig. 3C).
Expression of the intact SSR domain, which is capable of interacting with SPT16, fully restored the histone occupancy, as judged by histone H3 levels in the absence of endogenous SSRP1 (Fig. 3D). Similarly, the defective histone PTM status of H3K4Me3 and H3K36Me3 in the Sμ region was also fully restored. On the other hand, an SPT16 interaction-defective mutant (NΔ185) of SSRP1 or its HMG domain alone failed to rescue the histone modification-status defect. These findings demonstrate that FACT-regulated chromatin status plays a critical role in CSR.
Next, we examined the chromatin status of the Sα region. In contrast to the Sμ region, the Sα region did not show any obvious loss of core histone H3, but the H3K4me3 mark was severely reduced upon SSRP1 knockdown (Fig. 3E). These results indicate that FACT differentially regulates chromatin reassembly and the histone PTM at these two loci. We suspect the histone PTM reduction in the Sμ region by FACT knockdown may be due to the loss of core histones, whereas the reduction of H3K4me3 in Sα is probably caused by the role of FACT in H2B-ubiquitination–mediated transhistone modification cascade (Discussion). This idea is supported by the fact that knockdown of Bre1, a specific H2B ubiquitin ligase, affected H3K4me3 of the Sα specifically but not the Sμ region, resulting in abrogation of CSR (Fig. S5 A and B).
H3K4me3 Is Essential for S Region Cleavage in CSR.
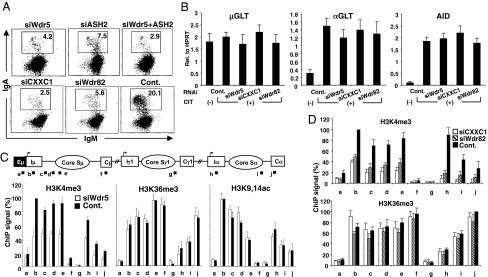
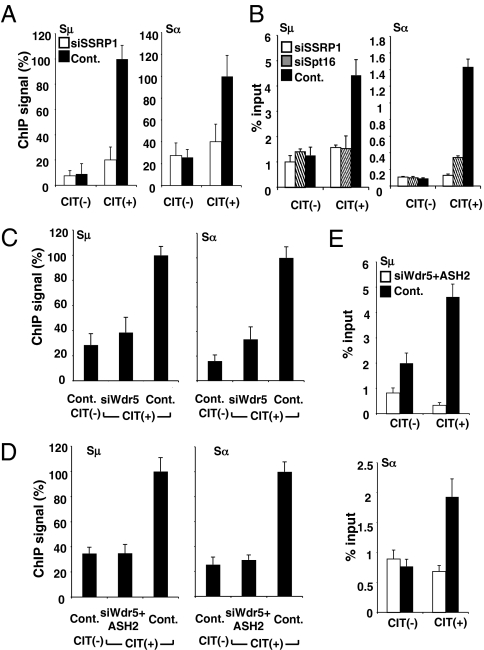
To directly demonstrate the association between H3K4me3 and CSR, we measured CSR after knocking down the common components of H3K4 methyltransferases (Wdr5 and ASH2) as well as the Set1-methyltransferase–specific factors (CXXC1 and Wdr82) (24). CSR was dramatically reduced by the knockdown of any of the Set1 complex components (Wdr5, ASH2, CXXC1, and Wdr82), although the GLTs and AID transcript were all unaffected by these knockdowns (Fig. 4 A and B). By contrast, knockdown of MLL1, which belongs to another group of H3K4 methyltransferase (24), did not significantly affect CSR (Fig. S5C). We also confirmed by ChIP analysis that H3K4me3 was decreased in the Sμ and Sα regions by the knockdown of Wdr5, CXXC1 or Wdr82, whereas other PTMs, such as H3K36me3 and H3K9,14Ac, were unaffected (Fig. 4 C and D). These results indicated that H3K4me3 itself is critically important for CSR.
Fig. 4.
H3K4me3 is critical for CSR. (A) Profile of the IgA switching population as determined by FACS in CH12F3-2A cells following the introduction of the indicated RNAi oligonucleotide. (B) Quantification of GLTs and AID transcripts by real-time PCR derived from the RNA of the indicated RNAi oligonucleotide samples normalized to HPRT. SD values were determined from three independent experiments. (C and D) (Top) Schematic diagram of the position of PCR products for the ChIP assay. The ChIP assay was performed in various knockdown and control samples using the indicated antibodies. Background values from controls with no antibody were subtracted (generally <5% of the antibody signal). Values were normalized to the DNA input signals followed by the maximum value in each data set. SD values were derived from three independent experiments.
We examined whether the CSR inhibition by the knockdown of FACT or H3K4 methyltransferase components was due to the blockade of DNA cleavage in the S region. We carried out two types of DNA cleavage assays: (i) detection by ChIP of γH2AX foci that accumulate at the flanking regions of double-strand breaks (DSBs) and spread over a long distance, and (ii) quantification of the broken DNA ends by direct labeling with biotin-dUTP and T4 polymerase, followed by pull-down of the labeled DNA fragments. The knockdown of SSRP1 reduced cleavage of the Sμ and Sα regions to the control level by both assays (Fig. 5 A and B). Similarly, SPT16 knockdown dramatically reduced the Sμ and Sα cleavage by the end labeling assay.
Fig. 5.
H3K4me3 is essential for S region DNA cleavage. (A) DNA break assay by γH2AX ChIP. The ChIP assay was performed in SSRP1 knockdown and control samples using an anti-γH2AX antibody. The pulled-down DNA was subjected to S- and Sα-specific detection by real-time PCR normalized to the fraction of γH2AX in H2AX, followed by the maximal value in each dataset. (B) The biotin-labeling break assay was performed in SSRP1 knockdown, Spt16 knockdown, and control samples. The pulled-down DNA was subjected to Sμ- and Sα specific detection by real-time PCR normalized to the input DNA. SD values were derived from three independent experiments. (C and D) The ChIP assay was performed in Wdr5 or Wdr5 and ASH2 knockdown and control samples using the anti-γH2AX antibody. (E) Biotin-labeling break assay was performed in Wdr5 and ASH2 knockdown and control samples.
We then asked directly whether the H3K4me3 mark itself is critical for DNA cleavage. Indeed, a defect in H3K4 trimethylation, achieved by the knockdown of methyltransferease components (Wdr5 and Ash2) inhibited the γH2AX focus formation in both the Sμ and Sα regions (Fig. 5 C and D). Similar results were obtained with the biotin-dUTP incorporation assay (Fig. 5E). Taken together, the presence of trimethylated H3K4 in the S region chromatin is indeed critical for both S region cleavage and CSR. Thus, the major CSR defect accompanying FACT deficiency may be attributable to a reduction in the H3K4 trimethylation status in the S region.
Discussion
It is well established that transcription of the S region is required for CSR. The Pol II transcription machinery migrates along the chromatin template by very intricate biochemical steps that include DNA unwinding, superhelix control, and nucleosomal reorganization. During the elongation phase, an extensive histone PTM cascade takes place. The FACT complex has been proposed to be a histone chaperone that facilitates transcription through the chromatin template by promoting histone disassembly and reassembly (16). Several recent reports also suggest that there is an interdependency between FACT and histone PTM (23, 25). In the present study, we showed that transcription per se of the S region is not sufficient for CSR, but histone modification especially the H3K4me3 mark generated by the assistance of the FACT complex and specific histone methyltransferase components in B cells is required. Needless to say, other marks on chromatin or DNA structure may be also required to determine the fine specificity for the S-region cleavage.
The depletion of SSRP1 altered the nucleosomal structure surrounding the Sμ region by decreasing the level of the core histones and consequently histone PTM. SSRP1 knockdown also depleted the H3K4me3 level in the Sα region. Unlike the Sμ region, however, this was not accompanied by nucleosomal loss. The reduction of histone PTM in this case could be caused by a defective transhistone modification pathway—a cascade in which FACT involvement has been proposed by regulating the level of the cascade initiator—H2B ubiquitination (23, 25, 26). Corroborating our speculation, knockdown of Bre1/RNF20 H2B ubiquitin ligase also affected H3K4me3 in the Sα region and simultaneously blocked CSR to IgA. It is intriguing that the FACT knockdown exhibited the different chromatin architecture in the two S regions. This could be related with the inherent nature between the constitutive (Sμ) and inducible (Sα) locus in CH12F3-2A cells. Indeed, FACT appears to regulate the chromatin status in a locus-specific manner, because FACT deficiency affects only a limited number of genes (<0.5%), the majority of which were up-regulated rather than down-regulated (Table S1).
Because H3K4me3 depletion had no adverse effects on transcription and cell viability (25, 27), the direct requirement of H3K4me3 for CSR was demonstrated by knockdown of shared components of H3K4 methyltransferase (ASH2 and Wdr5) as well as specific components of Set1 methyltransferase (CXXC1 and Wdr82). Subsequently, we showed that depletion of H3K4me3 strongly inhibited DNA cleavage in both Sμ and Sα regions. These results, taken together, suggest that H3K4me3 may serve as a marker of DNA cleavage in CSR. It is likely that H3K4me3 may be involved in recruitment of DNA cleaving enzyme, as discussed below.
H3K4me3 is frequently found not only in active genes but also in lymphoma-associated breakpoints, and recombination-prone sites (28, 29). RAG2 is known to interact directly with H3K4me3 through its noncanonical plant homeo domain (PHD) (19). This association is required for efficient V(D)J recombination in vivo, most likely by stabilizing the association of the RAG complex with the recombination signal sequence. This observation provides an explanation for Ommen immune deficiency syndrome, in which a mutation in the PHD domain disrupts the interaction between RAG2 and H3K4me3 (30). Thus, RAG-mediated V(D)J recombination in pro-B cells requires not only the conserved recombination signal sequence but also the specific H3K4me3 modification of chromatin.
The unexpected requirement of H3K4me3 for CSR also reveals its interesting similarity with meiotic recombination, in which H3K4me3 mark is also associated with DNA cleavage sites (28, 31). The hotspot motif of DNA cleavage in meiotic recombination consists of a weakly conserved 13-mer flanked by a transcription initiation site (32). In mammals, PRDM9 is a histone methyl-transferase that generates H3K4me3 and binds to the 13-mer hotspot motif (17, 18). Because PRDM9 is essential for generation of DSBs in meiotic recombination, H3K4me3 is assumed to indirectly recruit Spo11 (Top2). In yeast meiotic recombination, H3K4me3 mark and consequent DSB are dependent on Set1 histone methyltransferase, which is regulated by Bre1, as we showed for CSR (28, 33). Thus, both CSR and meiotic recombination appear to require transcription of the weakly conserved target DNA and the H3K4me3 mark for recruiting the cleavaging enzyme (Table S2). Second, both appear to use the topoisomerase as a cleaving enzyme: DSB by Spo11 (Top2) in meiotic recombination and nicking by Top1 in CSR. Third, after Spo11 cleaves DNA, it forms a covalent bond with the 5′ end of the cleaved DNA, and this Spo11-DNA fragment is resected by CtIP and the MRN complex (Mre11, Rad50, and Nbs1) to expose the 5′ DNA end of the DSB. Interestingly, the MRN complex is also known to be required for CSR (34). Although the precise function of the MRN complex is unknown in CSR, we speculate that it may resect Top1 bound to the 3′ end of cleaved DNA, generating the single-stranded gap in the S region. Forth, both meiotic recombination and CSR require members of the mismatch repair protein family which are known to serve as recombinant structure stabilizers in the meiotic recombination. It may be interesting to test similar possibility in CSR because their roles in CSR are not well demonstrated.
Although single-stranded DNA is an efficient substrate for AID deamination in vitro, the current in vivo findings suggest that DNA targeted by AID may be recognized in the context of chromatin. Therefore, the in vitro assay for DNA deamination by AID should be reexamined using mammalian Pol II and its cofactors on chromatin templates; previous studies have used naked DNA and T7 polymerase (9, 35). It would also be important to examine how FACT and nucleosomes with H3K4me3 are influencing the DNA deamination reaction, and whether AID or uracil DNA glycosylase is preferentially recruited to chromatin with H3K4me3.
Materials and Methods
RNAi Oligonucleotide Transfection and CSR Rescue Experiment.
CH12F3-2A cells were transfected with various RNAi oligonucleotides (Invitrogen) by electroporation (Amaxa). In the rescue experiment, GFP-SSRP1 or other mutant plasmids derived from pEGFP-C2 were also introduced, with a final concentration of 1.5 μg. The mixture was subjected to electroporation, and the cells were resusupended in the CH12F3-2A medium. The cells were cultured for 24 h, stimulated by CIT, and further cultured for another 24 h. The codons of mouse SSRP1 constructs were modified so as to prevent their RNAi-mediated degradation by the SSRP1#1 oligonucleotide. The primers, antibodies, and RNAi oligonucleotides used are summarized in Table S3.
ChIP.
The ChIP assay was performed using ActiveMotif ChIP-IT Express Kit according to the manufacturer's instructions. In brief, 4 × 106 cells were fixed in the presence of 1% formaldehyde for 5 min at room temperature. The reaction was stopped by the addition of glycine to a final concentration of 0.125 M. A soluble chromatin fraction containing fragmented DNA of 500–2,000 bp was obtained after cell lysis and sonication. Immunoprecipitation was performed by incubating the lysate with 2–3 μg antibody. The pulled-down DNA was subjected to detection by real-time PCR normalized to the amount of input followed by the maximum value in each data set, as described elsewhere (36).
Other materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors are grateful to I. Okazaki for RNAi system development in CH12 cells, and to Ms. Y. Shiraki for the preparation of the manuscript. This research was supported by a Grant-in-Aid for Specially Promoted Research 17002015 of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016923108/-/DCSupplemental.
References
- 1.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: Activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 4.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yancopoulos GD, et al. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 10.Rada C, et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi M, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci USA. 2009;106:22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 14.Fingerman IM, Du HN, Briggs SD. Controlling histone methylation via trans-histone pathways. Epigenetics. 2008;3:237–242. doi: 10.4161/epi.3.5.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 16.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 17.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 21.VanDemark AP, et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 22.Kelley DE, Stokes DG, Perry RP. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999;108:10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- 23.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 24.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavri R, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Gomez CA, et al. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–5664. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Székvölgyi L, Nicolas A. From meiosis to postmeiotic events: Homologous recombination is obligatory but flexible. FEBS J. 2010;277:571–589. doi: 10.1111/j.1742-4658.2009.07502.x. [DOI] [PubMed] [Google Scholar]
- 33.Kniewel R, Keeney S. Histone methylation sets the stage for meiotic DNA breaks. EMBO J. 2009;28:81–83. doi: 10.1038/emboj.2008.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen HM, et al. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J Exp Med. 2009;206:1057–1071. doi: 10.1084/jem.20082678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.