Abstract
The absence of tools for mapping the forces that drive morphogenetic movements in embryos has impeded our understanding of animal development. Here we describe a unique approach, video force microscopy (VFM), that allows detailed, dynamic force maps to be produced from time-lapse images. The forces at work in an embryo are considered to be decomposed into active and passive elements, where active forces originate from contributions (e.g., actomyosin contraction) that do mechanical work to the system and passive ones (e.g., viscous cytoplasm) that dissipate energy. In the present analysis, the effects of all passive components are considered to be subsumed by an effective cytoplasmic viscosity, and the driving forces are resolved into equivalent forces along the edges of the polygonal boundaries into which the region of interest is divided. Advanced mathematical inverse methods are used to determine these driving forces. When applied to multiphoton sections of wild-type and mutant Drosophila melanogaster embryos, VFM is able to calculate the equivalent driving forces acting along individual cell edges and to do so with subminute temporal resolution. In the wild type, forces along the apical surface of the presumptive mesoderm are found to be large and to vary parabolically with time and angular position, whereas forces along the basal surface of the ectoderm, for example, are found to be smaller and nearly uniform with position. VFM shows that in mutants with reduced junction integrity and myosin II activity, the driving forces are reduced, thus accounting for ventral furrow failure.
Keywords: embryo morphogenesis, tissue mechanics, biomechanics, cinemechanometry
Intense study has been directed at the forces that drive specific morphogenetic movements in embryos. Although atomic force microscopy can measure surface forces in single cells or cell doublets (1, 2) and laser microsurgery provides information about the forces carried by ablated structures (3), comprehensive maps of driving forces have remained elusive. Computational modeling, an important complement to experiments, can rigorously propose spatial and temporal maps of forces sufficient to account for morphogenetic movements, but uniqueness issues remain (4, 5). Here, we detail a previously undescribed, noninvasive method, video force microscopy or VFM, that combines dynamic imaging with computational mechanics to spatially and temporally map the forces that drive morphogenetic movements. The detail available in such maps is limited only by image resolution, capture frequency, and noise. The method has the potential to become a standard tool for biologists studying forces within tissues.
Conceptually, the mechanical forces at work in a tissue can be considered to be either active or passive (6, 7). Active forces are generated by cortical actomyosin, microtubules, and other systems that do mechanical work to the system, whereas passive forces arise primarily from deformation of cytoplasmic constituents that absorb this input energy and convert it to heat (Fig. 1A). Conveniently, the latter are typically characterized by an effective viscosity μ at the strain rates characteristic of morphogenetic movements (5, 7, 8). In our method, the tissue is discretized into polygonal regions, corresponding to cells or their parts or to groups of cells (Fig. 1B). When the corners of these regions are tracked during the process of interest, a large dataset results from which the deformations in each region can be quantified as a function of time. Finite element procedures or other techniques can be used to calculate the forces that must act at the corners of each of these regions, based on their viscosity (or other material properties if such are known), to make them deform as observed (see SI Text). The appropriate corner forces from all of the regions that contact any given node are then added vectorially to determine the net force that must act at that node to produce the observed deformations of the passive components of the tissue (Fig. 1C). These required nodal forces are assumed to be generated by active forces from actomyosin and other systems—which, in epithelia, typically act perpendicular and parallel to the apical-basal cell axis (Fig. 1A)—that can be resolved easily into equivalent loads (9) along specified line segments—typically the edges of the regions (Fig. 1D). Using this framework, one can ask whether there is a unique set of active forces able to generate these required nodal forces and thereby reshape the passive components of the embryo as observed. The mathematical inverse problem implied by this question is often poorly conditioned (10), but it can be solved using the special methods described in SI Text.
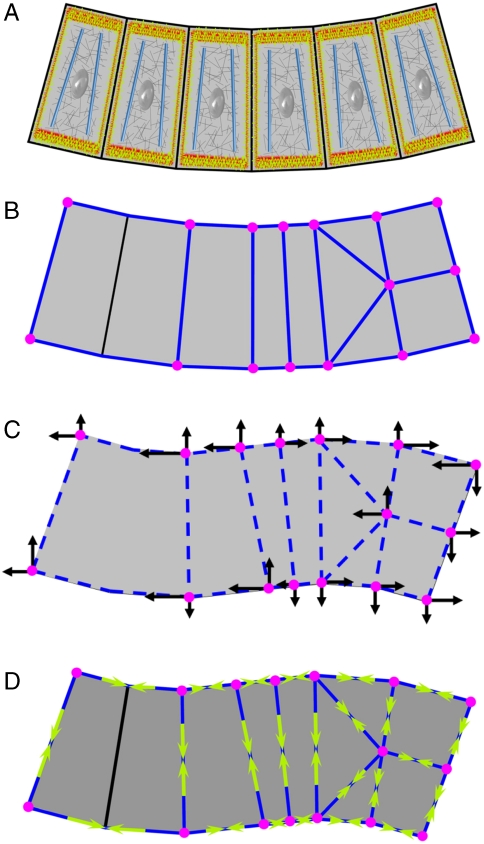
Fig. 1.
An illustration of the VFM method. A shows a cross-section through a generic epithelium. Active components in the cells are shown in color and passive ones in gray. (B) A polygonal partitioning of the epithelium at time t. (C) At time t + Δt, the example epithelium has adopted a new, deformed geometry. To deform the passive cellular components from geometry (B to C), the forces shown as black arrows must be applied at the registration points (magenta) for duration Δt. These forces are computed using a finite element procedure (24). (D) VFM calculates the edge forces (yellow arrows) and intracellular pressures (not shown) that must act in concert to produce the set of forces shown in C.
The method presented here, which might also be called cinemechanometry (CMM) (11), is fundamentally different from typical computational models. In those models, the researcher uses rational techniques to estimate the forces at work in the system and fine-tunes them so as to make the model output match the experimentally observed motions as closely as possible (4, 5), a process that can be quite tedious. Modeling of this type can be quite informative, but the uniqueness of the proposed set of driving forces usually cannot be proven. In contrast, the present method uses the large dataset of observed deformations as its primary input and from it directly calculates the forces that must act to drive those motions.
Results and Discussion
To demonstrate the potential of the method, we used it to investigate ventral furrow formation in Drosophila melanogaster, a relatively simple and well-understood system that can be imaged at high spatial and temporal resolution (Fig. 2, first column of Fig. 3, and Movies S1–S4). A multiphoton microscope was used to image WT embryos expressing Sqh (myosin II)-GFP. Embryos were mounted end on, and optical cross-sections were imaged every 45 s during the course of ventral furrow formation. Because cell neighbor changes do not occur (12) and tissue motions normal to the image plane are negligible, individual cells remain essentially confined to the imaging plane.
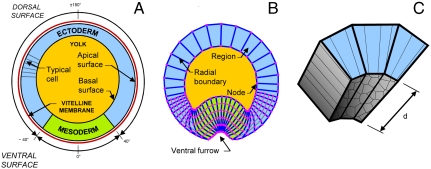
Fig. 2.
VFM as applied to Drosophila cross-sections. A shows a transverse cross-section through a Drosophila embryo. The tissue spanning an angle of approximately ± 40° in this figure will eventually form part of the ventral furrow (B), and it is denoted mesoderm, whereas the balance of the epithelium is called ectoderm. B shows the associated VFM mesh several minutes later, when the ventral furrow is becoming evident. The mesh is “Lagrangian,” following the tissue it represents. The thin gray edges that divide the cells on the ventral aspect of the embryo are assumed to carry zero load. C shows the cells in a volume of length d along the anterior–posterior axis of the embryo corresponding to three of the regions shown in cross-section in B. Note that even if the regions are made exactly one cell wide, they will always represent parts of cells and possibly multiple cells (depending on the value of d). The VFM edge forces reported in the cross-sections of Fig. 3 are those generated by the cells in the corresponding extruded volumes.
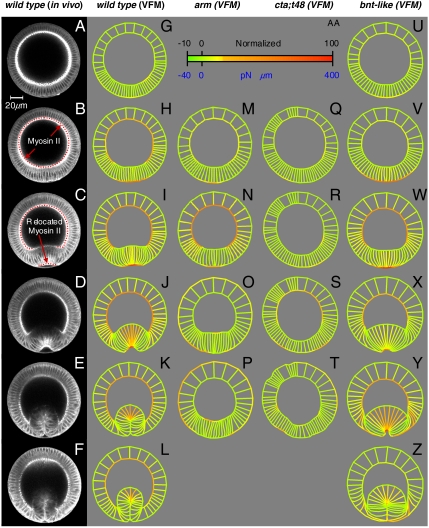
Fig. 3.
Driving forces as determined by VFM. (A–F) Selected multiphoton images of transverse cross-sections of a WT embryo during ventral furrow formation labeled with Sqh-GFP. Embryos are oriented with their dorsal surface upward, and the time interval between successive frames is 4.5 min. G–L show the driving forces in a WT embryo as determined by VFM. The meshes in G–L were formed by placing a grid on image A and tracking its corners throughout the image sequence (see also Movie S1). The region boundaries were then colored according to the spectrum shown in AA to represent the driving forces (Fig. 1D) calculated by VFM. The spectrum is calibrated so as to give forces in terms of arbitrary units and so as to give dimensioned forces per microns of embryo length on the basis of the μ value shown in the text. Radial forces have been normalized to the length of the circumferential region they represent in the initial geometry. (M–P) Forces in an arm mutant, which lacks strong apical junctions. (Q–T) Forces in a cta;t48 mutant, in which apical constriction is completely abolished. Forces in a bnt mutant (U–Z), where germ-band extension and posterior midgut invagination are suppressed. Embryos were synchronized using apical–basal cell height profiles (see Figs. S1 and S2).
To apply VFM to these images, we partitioned most of the embryo into regions nominally one cell wide and the full thickness of the epithelium (Figs. 2 and 3). Cells at the lateral boundaries of the presumptive ventral furrow area, however, were broken into regions one-fifth the thickness of the epithelium so that they could conform to the strongly curved cell geometries that develop there (Figs. 2B and 3 C–F), and edges interior to the cells (shown in gray in Fig. 2B) were assumed to carry zero tension. In addition, the dorsal ectoderm was broken into contiguous quadrilateral regions several cells wide (Fig. 3G) because the mechanical behavior of cells within such groups was indistinguishable from one another and in practice one does not want to digitize any more points than necessary. The registration points used to track in vivo movements were chosen to correspond to the corners of these regions. The passive components of the cells and yolk were assumed to have the same effective viscosity μ (8) because a short time earlier they constituted a common, uncellularized mass. Though relatively small, forces associated with intracellular pressures were crucial to a proper analysis (see SI Text).
The discretized regions were then tracked over time and VFM was used to calculate the edge forces that must act during the intervals between each successive pair of images to produce the observed motions. The forces associated with selected frames are illustrated in Fig. 3 according to the color spectrum in AA. The chosen discretization facilitates correlation of these edge forces with the relatively simple organization of the gastrula cytoskeleton (13): Tensions along the apical and basal edges of the cells represent actomyosin contractility and membrane tension along those surfaces (14, 15); tensions along the lateral cell edges represent the net contribution of lateral cell edge contractions that are likely to be the result of actomyosin and microtubule-based forces. To confirm the validity of using multicell regions, all of the embryos shown in Fig. 3 were also digitized with one cell-wide regions throughout the ectoderm. In the refined meshes, the edge tensions, including the radial ones when reported per unit of circumferential width, were visually indistinguishable from those shown, and the rms coarse-fine difference (typical differences in key driving forces were 1 to 2%) was less than half of that of the left–right differences in the WT and bicoid nanos torso-like (bnt) embryos.
Although these figures bear similarities to those reporting computational model results, the forces reported here arise instead from VFM analysis of the in vivo dynamics. The distinctive shapes and fine spacing of the tension profiles (Fig. 4A–D) speak to the accuracy of the method. If the solutions contained significant errors, the curves would be expected to cross like tangled spaghetti. Instead, closely spaced curves are produced with almost no crossover.
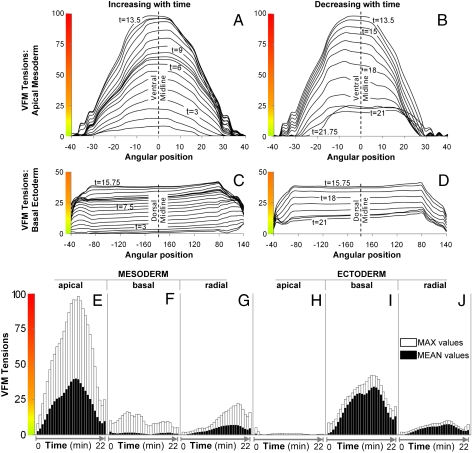
Fig. 4.
Tension profiles. Tension along the apical surface of the ectoderm (A, B, and E) is approximately parabolic with respect to space and time and the apical–basal lengths of the dorsal cells (Fig. S2) peak at the same time as the driving forces. In contrast, forces along the basal surface of the ectoderm (C and D) are highly uniform along the most dorsal 70% of the tissue. They are substantially smaller than those in the mesoderm and they peak later (E and I). Maximum cell height and peak force again coincide. The histograms (E–J) show the maximum forces (white bars) and mean forces (black bars) present along the apical, basal, and radial edges of the mesodermal and ectodermal cells over time. Substantial differences in magnitude and profile are apparent.
VFM shows that the highest tensile forces arise on the apical surface of the presumptive mesodermal cells on the ventral side of the embryo (Fig. 3G–L). In contrast to patterns commonly assumed in computational models (4, 5), VFM shows that the forces vary smoothly with time and position. These apical contractions arise over a period of 13.5 min and are initially concentrated over the six most medial cells on the ventral aspect of the embryo, tapering down over the neighboring five cells on either side to encompass approximately 16 cells in all. With time, the driving forces become stronger and more medially concentrated—partly because the apices of the cells in which they act constrict and partly because the ventral furrow cells themselves become more densely packed toward the ventral midline. The reported forces are generated by the cells and portions thereof contained in volumes (Fig. 2C) formed by extruding the regions visible in the cross-section in Fig. 3 a distance d along the anterior–posterior axis of the embryo. The forces are proportional to dimension d (Fig. 2C) and the viscosity used, and μ values reported for Drosophila range from 4.3 × 10-3 (8) to 105 Pa·s (16). Fig. 3 shows forces in arbitrary units and, for reference purposes only, in dimensional units based on d = 1 μm and the higher viscosity because it gives forces that seem more realistic.
Surprisingly, VFM also reveals moderate and uniform contraction of the basal surface of the dorsal and lateral ectoderm during the process—forces that could assist inward bending of the furrow (17). In these same dorsal–lateral ectodermal cells, forces along apical–basal edges are significantly smaller (Fig. 4E–J), suggesting that combined contractile and elongation forces along the apical–basal axis remain modest. In contrast, significant tension is seen along the lateral edges of presumptive mesodermal cells near the ventral midline—especially during the latter part of furrow formation. These active forces are concentrated in the ventral-most cells and drive shortening of the apically constricted cells, again assisting with cell internalization. The existence of such forces had been hypothesized (18, 19), but direct evidence was not available until now. The VFM analysis module receives no information regarding embryo symmetry, topology (e.g., whether an edge is apical, basal, or lateral), or cytoskeletal morphology. Therefore, one way of validating VFM is to compare the forces it calculates with the locations of force generators. The extent of contraction of apical cell surfaces calculated by VFM correlates (Fig. S3) with local myosin II levels revealed using Sqh-GFP (Fig. 3A–F and Movie S1). Significantly, the exception to this correlation between contractile force and myosin II localization is apical–basal shortening in cells of the furrow, which occurs without a corresponding accumulation of lateral myosin II, suggesting that other molecular processes underlie this process, as previously suggested (18, 19).
We then applied VFM analysis to three sets of mutant embryos in which we expect sustained junctional forces, apical constriction, and external forces in the embryo to be compromised. For this we chose, respectively, germ-line clones lacking both maternal and zygotic copies of armadillo (arm), double mutants for concertina and t48 (cta;t48), and bnt embryos. In arm embryos (Fig. 3 M–P and Movie S2), ventral furrow cells correctly localize myosin apically and begin constricting to yield a small ventral indentation; however, this architecture then collapses due to failure of the mutant’s weak apical junctions. VFM analysis indicates only a slight apical contraction in the ventral-most cells—as one would expect—but it also indicates that basal contraction of the dorsal and lateral ectoderm remains strong. In cta;t48 embryos (Fig. 3 Q–T and Movie S3), the ventral furrow cells neither constrict apically nor accumulate myosin (20). VFM analysis of these embryos shows no apical contraction in mesodermal cells, but moderate levels of basal contraction are again seen in ectodermal cells (Fig. 3 R and S). This suggests that actomyosin contraction in the basal ectoderm and apical mesoderm are independent of one another. More surprisingly, a failure to induce sustained apical constriction in both cases leads to a loss of all active apical–basal shortening. This correlation is also borne out in WT embryos, where shortening is confined to cells that have an altered form as the result of apical constriction. Thus, in contrast to most previous suggestions, shortening appears to be dependent on previous morphological events. Finally, we analyzed bnt embryos (Fig. 3 U–Z and Movie S4), where interference is minimized between the process of ventral furrow formation and normally concomitant germ-band extension and posterior midgut invagination (21). As expected for active forces, the general patterns of forces calculated using VFM in bnt embryos were similar to those in WT embryos, even though other coincident morphogenetic movements fail in these mutants and despite the somewhat longer and deeper furrow in the absence of anterior–posterior patterning (21). Thus, the driving forces calculated using VFM are borne out both by the localization of myosin II-GFP and by the mutant analysis.
Preliminary tests show that VFM can be applied to 3D cellular systems and that the higher dimensionality of these systems improves the conditioning of the VFM equations. VFM could also be applied to systems containing substantial elastic or viscoelastic components, although in such cases, the stress-free configuration and deformation history, respectively, must be known and appropriate materials models used in the finite element analysis. Subject to image quality, the approach could be applied to systems of arbitrary granularity, potentially allowing the action of single cytoskeletal components, the forces relieved by laser ablation experiments, or the bulk forces in whole tissues to be determined.
As this study illustrates, video force microscopy is able to resolve forces that drive morphogenetic movements with subcellular spatial detail and subminute temporal resolution. VFM makes it possible to identify force irregularities in single cells, quantify cell-to-cell variability, and correlate force generation with cell-level gene expression. This information has the potential to change the way morphogenetic movements associated with embryogenesis, wound healing, and tissue engineering are studied and the way that computational models are applied. As the present analysis of ventral furrow formation suggests, VFM has the potential to become an important biomechanical tool for modelers and biologists alike.
Methods and Materials
At midcellularization stage, embryos were dechorionated for 2 min at room temperature in 50% bleach, followed by several washes with PBS. They were kept in PBS for several hours without apparent developmental delays or problems. To genetically abolish anterior–posterior polarity, germ-band extension, and posterior midgut invagination, we used the stock w; Sqh-GFP42; bicoidE1 nanosL7 torso-like146/TM3 Sb and analyzed progeny from mothers homozygous for bicoidE1 nanosL7 torso-like146 (in short: Sqh-GFP; bnt embryos). To specifically block apical constriction, we used w, Sqh-GFP42; ctaR10/CyO; Df(3R)CC1.2/TM3 Sb stocks. The Df(3R)CC1.2 deletion uncovers the t48 locus. Twenty-five percent of progeny from homozygous ctaR10 parents showed a complete absence of apical constriction as described by ref. 20 and were thus assumed to be Sqh-GFP; cta;t48 embryos. We generated armadillo germ-line clones using the Flipase-Recombination-Target Dominant-Female Sterile (FLP-DFS) system (22). By crossing arm043A01 FRT101/FM7; Sqh-GFP42 females to w ovoD FRT101/Y; flp-138 males, we obtained arm043A01FRT101/w ovoD FRT101; flp-138/+ females. These females were heat shocked as larvae for 2 h at 37 °C to induce mitotic recombination in the germ line and crossed to FM7/+; flp-138/+ males.
Transgenic Sqh-GFP embryos were mounted vertically in 1% agarose in PBS on Mattek culture dishes, with the posterior end facing the objective. They were imaged on a custom-built two-photon microscope, using a Nikon 40× NA 0.8 objective and simultaneous epi- and transdetection as described in ref. 23. We used laser lines between 890 and 910 nm. A 488-nm laser with an output power of 15 W generated 150-mW laser pulses between 890 and 910 nm at the level of the objective. Embryos imaged under these conditions were able to develop normally afterward. Imaging depth was 110 μm from the posterior end of the embryo, and images were acquired every 45 s with MATLAB. Note that the furrow is extended in bnt embryos.
Custom software was used to generate a reference grid that was adjusted manually to fit the first image of each set. A combination of automated and manual methods was used to track the registration points from image to image. Custom software was used to play multiple frames forward and back so that the location of the registration points could be placed with precision. Many of the tracked points fell at the corners of cells. The spacing of multiple points along radial edges was kept even with the aid of software. Constant-volume algorithms facilitated placement of basal registration points when the basal edge of the cell was difficult to discern.
To calculate the forces needed to deform the passive components of the cells, a finite element procedure based on the approach of Brodland et al. (24) was used. Because the contents of the epithelial and mesodermal cells were still contiguous with the yolk, the cells and the yolk were assumed to have the same viscosity μ. These forces fi at time step i were assumed to be equal to the matrix product 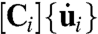 , where Ci is a finite element-based damping matrix based on μ and the current geometry of each region, and
, where Ci is a finite element-based damping matrix based on μ and the current geometry of each region, and 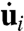 is the vector of nodal velocities between successive images. The strain rates are assumed to be sufficiently low that inertial and elastic forces can be ignored (7, 17). Further details of the calculations are given in SI Text and in ref. 11. To determine the active forces acting along each region boundary, matrix equations relating edge forces and intracellular pressures to nodal forces were written. These took the general form
is the vector of nodal velocities between successive images. The strain rates are assumed to be sufficiently low that inertial and elastic forces can be ignored (7, 17). Further details of the calculations are given in SI Text and in ref. 11. To determine the active forces acting along each region boundary, matrix equations relating edge forces and intracellular pressures to nodal forces were written. These took the general form
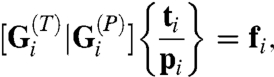 |
[1] |
where 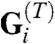 is a matrix of direction cosines that converts ti a vector of edge tensions into equivalent nodal loads, whereas
is a matrix of direction cosines that converts ti a vector of edge tensions into equivalent nodal loads, whereas 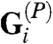 is a matrix that calculates the equivalent joint (nodal) loads produced by the intracellular pressures pi. In essence, VFM determines the tensions ti and pressures pi, given the current geometry—from which the G matrices are calculated—and the viscous nodal forces fi. To improve the quality of the solutions to these equations, they were solved over multiple steps using a variable forgetting factor approach (10, 25, 26).
is a matrix that calculates the equivalent joint (nodal) loads produced by the intracellular pressures pi. In essence, VFM determines the tensions ti and pressures pi, given the current geometry—from which the G matrices are calculated—and the viscous nodal forces fi. To improve the quality of the solutions to these equations, they were solved over multiple steps using a variable forgetting factor approach (10, 25, 26).
Supplementary Material
Acknowledgments.
We thank Adam Martin (Massachusetts Institute of Technology, Boston, MA) for providing Sqh-GFP arm GLC embryos and Maria Leptin (Cologne University, Cologne, Germany) for providing w; ctaR10/CyO; Df(3R)CC1.2 stocks. We also thank Eric Wieschaus (Princeton University, Princeton, NJ) in whose lab the biological data were collected and the Howard Hughes Medical Institution and National Institutes of Health that funded that work. Stephan Thiberge (Princeton University, Princeton, NJ) assisted with obtaining the time-lapse images and some of the images were digitized by Jordan Lui and Marianella Cascante (University of Waterloo, Waterloo, ON, Canada). Funding was provided by the Human Frontiers Science Program (to G.W.B., W.S.H., and A.J.), by the Natural Sciences and Engineering Research Council of Canada (to G.W.B. and S.N.), by the Engineering and Physical Science Research Council of the United Kingdom (to M.M. and V.C.), by the New Jersey Commission of Cancer Research (to F.U.), and by the Royal Society (to B.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006591107/-/DCSupplemental.
References
- 1.Puech P, et al. Measuring cell adhesion forces of primary gastrulating cells from zebrafish using atomic force microscopy. J Cell Sci. 2005;118:4199–4206. doi: 10.1242/jcs.02547. [DOI] [PubMed] [Google Scholar]
- 2.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 3.Hutson MS, et al. Combining laser microsurgery and finite element modeling to assess cell-level epithelial mechanics. Biophys J. 2009;97:3075–3085. doi: 10.1016/j.bpj.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte V, Muñoz JJ, Baum B, Miodownik M. Robust mechanisms of ventral furrow invagination require the combination of cellular shape changes. Phys Biol. 2009;6:016010,. doi: 10.1088/1478-3975/6/1/016010. 10.1088/1478-3975/6/1/016010. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Brodland GW. Multi-scale finite element modeling allows the mechanics of amphibian neurulation to be elucidated. Phys Biol. 2008;5:015003. doi: 10.1088/1478-3975/5/1/015003. 10.1088/1478-3975/5/1/015003. [DOI] [PubMed] [Google Scholar]
- 6.Brodland GW, Chen DI, Veldhuis JH. A cell-based constitutive model for embryonic epithelia and other planar aggregates of biological cells. Int J Plasticity. 2006;22:965–995. [Google Scholar]
- 7.Lecuit T, Lenne P. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Bio. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 8.Gregor T, Bialek W, de Ruyter van Steveninck RR, Tank DW, Wieschaus EF. Diffusion and scaling during early embryonic pattern formation. Proc Natl Acad Sci USA. 2005;102:18403–18407. doi: 10.1073/pnas.0509483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodland GW, Clausi DA. Embryonic tissue morphogenesis modeled by FEM. J Biomech Eng. 1994;116:146–155. doi: 10.1115/1.2895713. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Betti R. On-line identification and damage detection in non-linear structural systems using a variable forgetting factor approach. Earthq Eng Struct D. 2003;33:419–444. [Google Scholar]
- 11.Cranston PG, Veldhuis JH, Narasimhan S, Brodland GW. Cinemechanometry (CMM): A method to determine the forces that drive morphogenetic movements from time-lapse images. Ann Biomed Eng. 2010;38:2937–2947. doi: 10.1007/s10439-010-9998-1. [DOI] [PubMed] [Google Scholar]
- 12.Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Young PE, Pesacreta TC, Kiehart DP. Dynamic changes in the distribution of cytoplasmic myosin during drosophila embryogenesis. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early drosophila embryos. Mol Biol Cell. 2004;15:838–850. doi: 10.1091/mbc.E03-06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawes-Hoang RE, et al. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 16.Bittig T, Wartlick O, Kicheva A, Gonzalez-Gaitan M, Julicher F. Dynamics of anisotropic tissue growth. New J Phys. 2008;10:063001. [Google Scholar]
- 17.Clausi DA, Brodland GW. Mechanical evaluation of theories of neurulation using computer simulations. Development. 1993;118:1013–1023. [Google Scholar]
- 18.Costa M, Sweeton D, Wieschaus E. In: The Development of Drosophila melanogaster. Michael Bate, Martinez Arias Alfonso., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab; 1993. p. 425. [Google Scholar]
- 19.Leptin M. Gastrulation in drosophila: The logic and the cellular mechanisms. EMBO J. 1999;18:3187–3192. doi: 10.1093/emboj/18.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 21.Desprat N, Supatto W, Pouille PA, Beaurepaire E, Farge E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in drosophila embryos. Dev Cell. 2008;15:470–477. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 24.Brodland GW, Viens D, Veldhuis JH. A new cell-based FE model for the mechanics of embryonic epithelia. Comp Met Biomech Biomed Eng. 2007;10:121–128. doi: 10.1080/10255840601124704. [DOI] [PubMed] [Google Scholar]
- 25.Fortescue TR, Kershenbaum LS, Ydstie BE. Implementation of self-tuning regulators with variable forgetting factors. Automatica. 1981;17:831–835. [Google Scholar]
- 26.Paleologu C, Benesty J, Ciochina S. A robust variable forgetting factor recursive least-squares algorithm for system identification. IEEE Signal Proc Let. 2008;15:597–600. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.