Abstract
An important step during plant development is the transition from juvenile to adult growth. It is only after this transition that plants are reproductively competent. Given the great danger that transposon activity represents to the germ line, this may also be an important period during development with respect to transposon regulation and silencing. We demonstrate that a change in expression of a key component of the RNA silencing pathway is associated with both vegetative phase change and shifts in epigenetic regulation of a maize transposon.
Keywords: epigenetics, Mutator, methylation, MuDR
A primary problem facing all organisms is the proliferation of selfish DNA in the form of transposable elements. This proliferation is predominantly held in check via epigenetic silencing (1, 2). Recent work in both plants and animals suggests that there are key points in development during which silencing of transposons is reinforced (3, 4). In animals, mechanisms to reinforce silencing appear to be largely confined to the germ line. Indeed, recent work in Drosophila suggests that transposon regulation and germ-line differentiation are intimately connected through the activity of PIWI proteins, which are involved in processing of specific classes of small RNAs, most of which are derived from transposons (5). However, plants face a distinct challenge because, unlike animals, they do not have a sequestered germ line. Thus, changes in transposon activity in somatic tissues can be transmitted to subsequent generations (6). From this perspective, an important stage in plant development is the transition from the juvenile phase to the adult phase of growth, for it is only after this transition that plants become reproductively competent (7). Here we provide evidence that this transition in maize is associated with a transient loss of expression of a key regulator of the trans-acting siRNA (tasiRNA) pathway and with dramatic changes in epigenetic regulation of a transposon as it is being silenced.
Our model for plant transposon silencing is the MuDR element of maize. Heritable silencing of MuDR transposons can be triggered by a naturally occurring derivative of MuDR called Mu killer (Muk) (8). MuDR elements are flanked by two long terminal inverted repeats (TIRs) that carry the promoters for two convergently transcribed genes, mudrA and mudrB (Fig. 1A). The mudrA gene encodes the putative transposase; mudrB is a helper gene required for insertion of Mu elements in maize (9). Muk produces a long, double-stranded RNA (dsRNA) hairpin transcript with homology to mudrA (8, 10). Silencing of MuDR by Muk is triggered by a simple genetic cross between a plant carrying MuDR and one carrying Muk. Silencing of MuDR by Muk is associated with the amplification of substantial quantities of small RNAs homologous to mudrA in the second leaf of developing seedlings (10). By the immature ear stage of these F1 (MuDR;Muk) plants, mudrA becomes transcriptionally silenced, and some cytosines within the TIR immediately adjacent to mudrA (TIRA) become methylated. The natural MuDR;Muk system allows us to track the initiation and maintenance of silencing of a single transposable element in plants.
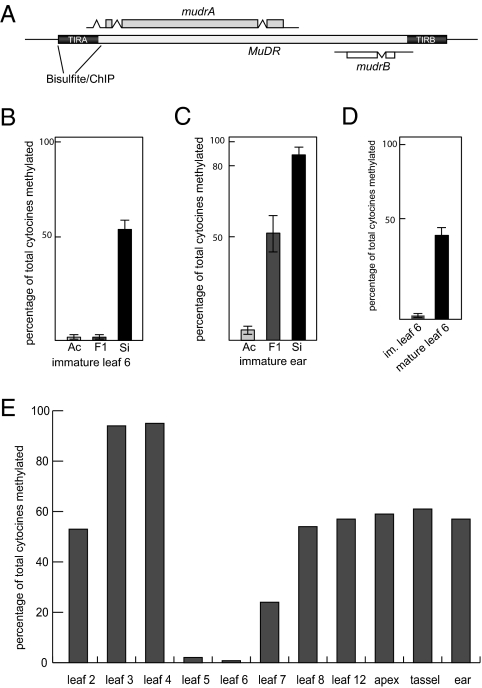
Fig. 1.
DNA methylation at TIRA. (A) Diagram of the structure of MuDR. The region of TIRA where DNA methylation and histone modifications were assayed is indicated. (B) Percentage of total TIRA cytosine methylation in active, F1, and stably silenced MuDR elements in immature leaf 6. (C) Percentage of total TIRA cytosine methylation in active, F1, and stably silenced MuDR elements in the immature ear. (D) Comparison of percentage of TIRA cytosine methylation in F1 plants in immature versus mature leaf 6. (E) Percentage of TIRA cytosine methylation in various tissues of F1 plants. Ac, active MuDR; F1, MuDR exposed to Muk in the first generation; Si, stably silenced MuDR.
Results
To more thoroughly document the effects of Muk on MuDR, we examined patterns of cytosine methylation and histone modifications in TIRA. These marks were assayed in active MuDR elements that had not been exposed to Muk, stably silenced MuDR elements in the absence of Muk, and F1 plants that carried both MuDR and Muk (see SI Materials and Methods for derivation of these plants). Cytosine methylation is a hallmark of epigenetic silencing in both plants and many animals (11, 12). In plants, cytosines can be methylated in all three sequence contexts, each one of which is maintained by distinct enzymes (12). To establish a baseline, we used bisulfite sequencing to determine methylation in the TIRA of active and stably silenced MuDR elements in two tissues, one somatic (developing leaves) and one germinal (immature ears). As expected, TIRA was hypomethylated in active MuDR elements and extensively methylated in stably silenced MuDR elements (Fig. 1 B and C). Cytosines in the silenced element were methylated in all three sequence contexts (Fig. S1), which is consistent with the involvement of both maintenance (CG and CHG) and de novo (CHH) DNA methylation. The degree of methylation in a stably silenced element was lower in a young leaf than it was in an immature ear (Fig. 1 B versus C), suggesting differences in the stability of the silent state in different tissues.
Analysis of methylation of stably silenced elements provided information about the maintenance of MuDR silencing, but we also wanted to examine the initiation of silencing in the first generation of exposure of MuDR to Muk. To do so, we examined TIRA methylation in a number of tissues in F1 plants that carried both Muk and MuDR. Developing leaves were collected at a similar age (leaf plastochron index) to minimize potential differences in DNA methylation associated with growth variation among these leaves, and leaves from multiple genotyped individuals were pooled to minimize plant-to-plant variation. Consistent with previous observations (13), we found that TIRA was increasingly methylated in the first few leaves, with nearly every cytosine in all sequence contexts methylated by leaves 3 and 4 (Fig. 1E and Figs. S2 and S3). Surprisingly, however, TIRA methylation dropped dramatically in leaves 5, 6, and 7 but was substantially restored in subsequent leaves. Because sampled leaves were pooled from F1 plants derived from crosses using Muk as both male (two families) and female (two families), the observed effect is not attributable to a maternal effect caused by the presence of Muk in the female lineage. We also found that, even in individual leaves, the absence of TIRA methylation is transient; although TIRA was not methylated in immature leaf 6, it was more methylated in mature leaf 6 but less methylated than in juvenile or adult leaves (Fig. 1D and Fig. S2).
Interestingly, leaf 5 to leaf 7 represent a transitional stage from juvenile to adult phases of growth in maize (14). Transition leaves are those leaves that have mixtures of juvenile and adult characteristics. In our line, the first leaf with visible signs of both adult and juvenile characteristics is leaf 6. Our data indicate that the progression of transposon silencing is not simply a cumulative process as the maize shoot develops but, rather, appears to be a programmatic process that is sensitive to signals associated with vegetative phase change.
A transient reduction of TIRA methylation was also observed in progeny of F1 plants that carried only silenced MuDR elements in the absence of Muk (Fig. S4A). However, the degree of reduced TIRA methylation in transition leaves of these F2 plants was less dramatic than was observed in the first generation of silencing. Nevertheless, these data suggest that both the initiation (in F1 plants) and maintenance (in F2 plants) of MuDR silencing are sensitive to changes that occur during vegetative phase change. They also show that cytosine methylation in all three sequence contexts is maintained and even added to in the generation immediately after the loss of the silencing trigger (Muk).
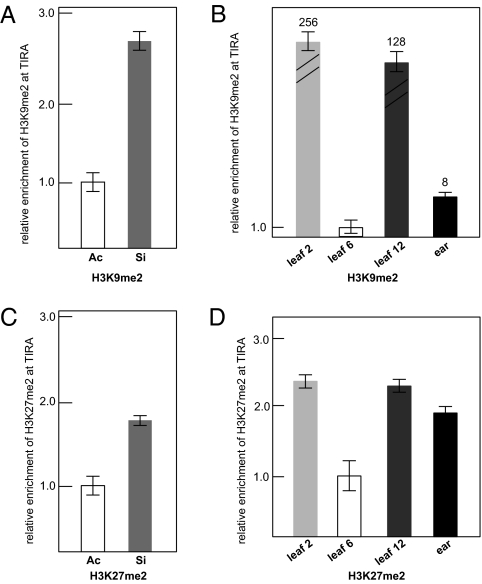
Changes in histone modification mirrored changes in TIRA methylation. H3K9 and H3K27 dimethylation are often associated with epigenetic silencing of transposons in plants (15–17). We found that dimethylation of both of these histone tails corresponded well with DNA methylation in active, silenced, and F1 plants. Both modifications were enriched in stably silenced TIRA and in all tissues of F1 plants, with the notable exception of the transition leaves (Fig. 2). This enrichment did not extend beyond TIRA into the flanking sequence, indicating that they were a direct result of the interaction of Muk with TIRA of MuDR and did not spread from or to the flanking sequences (Fig. S5B).
Fig. 2.
ChIP analysis of enrichment of histone marks at TIRA. (A) Relative enrichment of H3K9me2 in leaf 6 of stably silenced MuDR compared with an active MuDR element as determined by quantitative PCR. ChIP results were normalized to copia and then to the value of active MuDR. (B) Quantitative PCR analysis of relative enrichment of H3K9 dimethylation at TIRA in different tissues of F1 plants. ChIP results were normalized first to copia and then to the value of active MuDR. (C) Quantitative PCR analysis of H3K27 dimethylation in TIRA of active and stably silenced MuDR elements in young leaf 6. (D) Quantitative PCR of relative enrichment of H3K27 dimethylation in TIRA of various tissues of F1 plants. The value for leaf 6 was arbitrarily set at 1.
Transcriptional activity of mudrA correlated well with both DNA and histone H3 methylation. F1 plants only showed significant levels of mudrA expression in tissues that lacked TIRA methylation and that exhibited reduced H3K9me2 and H3K27me2 levels (Fig. 3 A and B). No such mudrA expression was observed in transition leaves of F2 plants (Fig. S4B). Importantly, the differential expression pattern of mudrA in F1 plants was not attributable to differential expression of Muk, which was expressed in the shoot apex, embryo, and immature ear but not in any of the leaves or the immature tassel (Fig. 3C). These data demonstrate that Muk transcript is not required in the leaves to cause silencing in that tissue, which suggests that the previously reported enrichment of small RNAs in leaf 2 of F1 plants (10) are the result of an amplification step in these leaves that occurs in the absence of Muk transcript. Given that Muk does express in the shoot apex and young embryo, we suggest that this process may involve transport of a signal (generated by Muk) from the shoot apex to the developing leaves, a process reminiscent of systemic silencing of viruses in plants (18). Although it is also possible that methylation is simply propagated in leaves from methylation established in the meristem, we suggest that such propagation of methylation is unlikely, at least in transition leaves. When young, these leaves lack TIRA methylation, but once they are fully grown they gain it (Fig. 1D). In this case, the methylation cannot be caused by propagation; it must have been added de novo in the leaf, presumably from a signal produced in the meristem, where Muk is expressed.
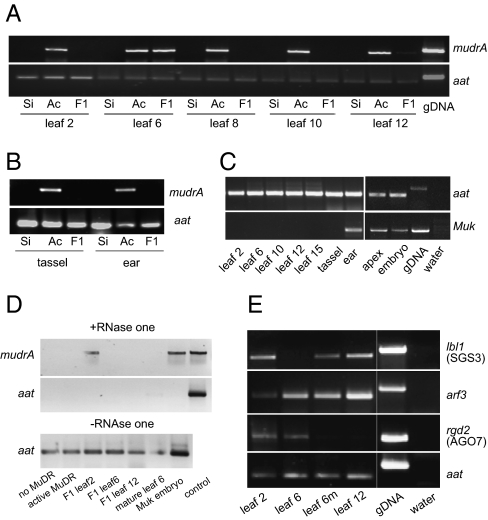
Fig. 3.
Analysis of tissue-specific changes in gene expression. (A) RT-PCR analysis of mRNA levels of mudrA in different leaves of plants carrying silenced (Si), active (Ac), and F1 MuDR elements. (B) RT-PCR analysis of mRNA levels of mudrA in the tassel and ear of plants carrying silenced, active, and F1 MuDR elements. (C) RT-PCR analysis of Muk in various tissues. (D) RT-PCR detection of dsRNA in various tissues of F1 plants. (E) RT-PCR analysis of maize homologs of SGS3 (lbl1), ARF3, and AGO7 (rgd2) in different leaves of F1 plants. Leaf 6m represents a mature and fully expanded leaf 6; all other leaves were developing young leaves. gDNA, genomic DNA.
Previous work in our laboratory had demonstrated that dsRNA can be detected in tissues in which Muk is expressed (10), consistent with the proposed hairpin structure of the Muk transcript. Because leaf 2 does not express Muk (Fig. 3C), it would not be expected to contain the hairpin. siRNAs are, however, detected in leaf 2 (10). We hypothesized that these siRNAs are the result of an RNA-dependent RNA polymerase-dependent amplification step in the leaves, which would produce dsRNA that could be processed by a dicer into siRNAs. To test this hypothesis, we assayed for the presence of dsRNA in F1 leaves that exhibit (leaves 2 and 12) or lack (leaf 6) TIRA methylation. The results showed that the dsRNA was detectable in leaf 2 but not in leaf 6 (Fig. 3D). However, it was also absent in leaf 12, which did exhibit substantial levels of TIRA methylation (Fig. 1E and Fig. S2), suggesting that silencing in these later leaves is not associated with the production of dsRNA.
The production of dsRNA in leaf 2 is consistent with the amplification of siRNAs, presumably because of the activity of an RNA-dependent RNA polymerase in this tissue. Although RNA-DEPENDENT RNA POLYMERASE2 (RDR2) is often involved in RNA-directed DNA methylation (19), Muk-induced silencing of MuDR and the production of siRNAs in leaf 2 occurs in its absence, making RDR2 an unlikely candidate for being the responsible polymerase (20). Another potential candidate is RDR6, which, in conjunction with SUPPRESSOR OF GENE SILENCING3 (SGS3), produces dsRNA from a variety of templates, including tasiRNA precursors, viral RNA, and aberrant transgene RNA (21, 22). SGS3 is thought to stabilize precursor RNAs, allowing them to be targets of RDR6 activity (23). Among other things, RDR6/SGS3 activity is required for virus-induced gene silencing–directed DNA methylation and long-range systemic silencing (22, 24). It is also essential for the tasiRNA pathway, which bears distinct similarities to Muk-induced transacting silencing of MuDR (25).
To see whether the changes in methylation of TIRA were caused by changes in expression of genes required for the production of dsRNA, we examined expression levels of candidate genes by RT-PCR. We did not see a change in expression of a maize homolog of RDR6 (Fig. S6A). However, in leaves where TIRA methylation was lost, expression of leafbladeless1 (lbl1) [the maize homolog of SGS3 (26)] was dramatically reduced (Fig. 3E). A reduction in lbl1 expression was also observed in plants that carried only active MuDR (Fig. S6B), suggesting that the loss of lbl1 in these leaves is unlikely to be a consequence of the interaction between Muk and MuDR in F1 plants. Further, in mature leaf 6, in which TIRA methylation was partially restored, lbl1 expression was also partially restored. Based on these results, we suggest that amplification of the small RNAs in leaf 2 results from lbl1-dependent production of dsRNA, and loss of lbl1 in the transition leaves results in an absence of methylation of TIRA in these leaves. We did not see variation in expression of the maize homolog of METHYL TRANSEFERASE1 (MET1) or of a homolog of REPRESSOR OF SILENCING1 (ROS1), which are required for maintenance methylation and active dimethylation, respectively (12) (Fig. S6A). The lack of variation in expression of MET1 or ROS1 suggests that the absence of methylation we observe in transition leaves is likely a consequence of the loss of lbl1 rather than a passive loss or active elimination of DNA methylation (27).
In both maize and Arabidopsis, SGS3/lbl1 is required to produce a tasiRNA that targets and negatively regulates AUXIN RESPONSE FACTOR3 (ARF3) (28). Down-regulation of ARF3 is required for maintenance of the juvenile stage in Arabidopsis; if SGS3 is mutated or ARF3 is overexpressed, plants enter the adult phase prematurely (25, 29). We found that in the same tissue in which lbl1 expression was lost, expression of a maize homolog of ARF3, arf3a, was increased (Fig. 3E and Fig. S6C ). It should be noted that there were some tissues in which high levels of lbl1 were not associated with a reduction of arf3a expression. However, in these tissues, expression of ragged seedling2 (rgd2), the maize homolog of AGO7 (30), was not detected (Fig. 3E). In Arabidopsis, AGO7 is required for the production of tasiRNAs in this pathway (29, 31), and arf3a in maize overexpresses in maize rgd2 mutants (30). Thus, we suggest that although lbl1 is expressed in adult leaves, rgd2 is not available for the effective down-regulation of arf3a. Together, these data suggest that the transient loss of lbl1 expression in growing transition leaves causes coordinate changes in both MuDR silencing and the tasiRNA silencing pathway.
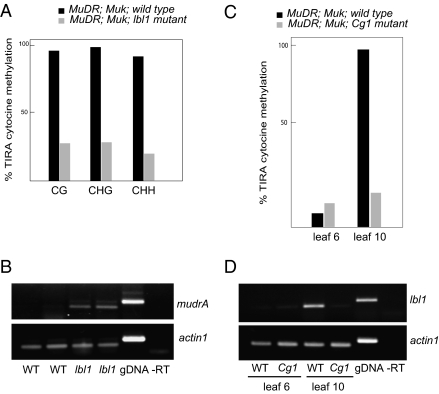
Given that the transient loss of lbl1 expression in transition leaves was associated with the loss of MuDR TIRA methylation, we wanted to know if mutations in lbl1 would cause a loss of methylation in leaves in which TIRA normally becomes methylated. To test this hypothesis, we crossed lbl1 homozygous mutants to plants carrying MuDR and to plants that were homozygous for Muk. lbl1 heterozygotes carrying MuDR were then crossed to lbl1 heterozygotes carrying Muk to generate MuDR;Muk F1 sibling plants that were phenotypically mutant or wild type for lbl1. Because TIRA is invariably methylated in leaf 3 of F1 plants (Fig. 1E), tissue from this leaf from mutant and wild-type siblings was examined for evidence of TIRA methylation. TIRA methylation was substantially reduced in F1 mutant leaf 3. In contrast, sibling plants that carried a wild-type copy of lbl1 were nearly completely methylated (Fig. 4A). Consistent with the methylation data, we also found that mudrA transcript was absent in the wild-type siblings but present in the mutants (Fig. 4B). These data demonstrate that TIRA methylation of mudrA induced by Muk depends on lbl1, and they strongly suggest that the loss of lbl1 expression in transition leaves is the cause of the transient loss of TIRA transcriptional gene silencing that we observed in F1 transition leaves. In our mixed genetic background, the lbl1 mutants exhibited severe polarity defects in older leaves, making it difficult to determine the timing of phase transition. However, leaves 3 and 4 of the mutants were entirely juvenile in appearance, suggesting that the loss of methylation we observed in leaf 3 was not caused by a shift in the timing of phase change in these plants.
Fig. 4.
(A) Analysis of percentage of methylated cytosines in TIRA in MuDR;Muk F1 lbl1 mutants and wild-type siblings. Data are presented for cytosines methylated in each of the three cytosine contexts: CG, CHG, and CHH. (B) Expression of mudrA in MuDR;Muk F1 lbl1 mutant and wild-type leaf 3. (C) Analysis of percentage of total cytosine methylation in MuDR;Muk F1 Cg1 mutant and wild-type leaves 6 and 10. (D) Expression of lbl1 in MuDR;Muk F1 Cg1 mutant and wild-type leaves 6 and 10.
In maize, five to six leaves normally form in embryonic development (32). Because we observed a loss of TIRA methylation in the last two leaves made in the embryo, it was possible that our observations reflected changes associated with the transition from embryonic to postembryonic development rather than with phase transition per se. To test this hypothesis, we examined the effects of a mutation that dramatically alters the timing of phase transition (14, 33). Corngrass1 (Cg1) causes a delay in phase change because of overexpression of the miR156 precursor, an important regulator of phase change in both maize and Arabidopsis (34). The severity of the phenotype of Cg1 varies depending on the genetic background (35). In our mixed genetic background, Cg1 resulted in the prolonged production of transition leaves, with both adult and juvenile traits throughout much of plant development. F1 MuDR;Muk siblings that were mutant or wild type for Cg1 were compared for TIRA methylation in leaves 6 and 10. As expected, the wild-type plants exhibited low levels of methylation in the transition leaf (leaf 6) and much higher levels in an adult leaf (leaf 10) (Fig. 4C). In contrast, Cg1 plants showed low levels of TIRA methylation in both leaf 6 and leaf 10, consistent with the prolonged transition phenotype we observed in this plant. The shift in TIRA methylation in Cg1 mutants indicates that epigenetic modification at TIRA is altered by changes in timing of vegetative phase transition. Further, we found that, although expression of lbl1 was low in leaf 6 and increased in leaf 10 in wild-type siblings, it remained low in both leaves in the Cg1 mutants, supporting the hypothesis that changes in lbl1 expression cause changes in TIRA methylation and suggesting that Cg1 is epistatic to lbl (Fig. 4D).
As described above, RT-PCR indicated that mudrA transcript is produced in immature leaf 6 of F1 plants, suggesting that functional MURA protein may be produced in that tissue. Work over a number of years has demonstrated that active transposase is invariably associated with hypomethylation of methyl-sensitive HinfI sites within the TIRs of nonautonomous Mu1 elements, which share TIR sequence homology with MuDR but contain distinct internal sequences (9). To assess transposase activity in various plant tissues, the same DNA that was used for bisulfite analysis of TIRA methylation was digested with HinfI, blotted, and probed with a fragment of Mu1. Surprisingly, we found that Mu1 was methylated in all of the tissues we examined (Fig. S7). Thus, although TIRA is hypomethylated and mudrA is expressed in immature leaf 6, it does not appear that a functional MURA protein is produced. This result suggests that there are additional levels of posttranscriptional inhibition of MURA activity in this tissue.
Discussion
Our observations suggest a link between phase change in maize and the initiation of epigenetic silencing of a transposon. Both processes are associated with a reduction of lbl1 expression during plant development, which, in turn, is associated with an increase in the levels of the tasiRNA target arf3a as well as dramatic changes in epigenetic modification of a MuDR element that is undergoing silencing. The relationship between arf3 regulation and MuDR modification suggests that lbl1 regulation may act to coordinate vegetative phase transition with transposon silencing. These data are consistent with a role for changes in lbl1 and arf3a transcription levels in promoting phase change in maize, a hypothesis supported by the observation that mutations in SGS3 or ectopic expression of ARF3 can both cause premature phase change in Arabidopsis (36).
Expression of Muk, which is driven by an ectopic flanking promoter (10), exhibits tissue specificity, making it possible to examine the ways that expression of a trigger in one tissue can affect silencing in a different tissue. Muk is expressed in the shoot apex and immature ear but not in leaves. However, we have found that target sequences can be efficiently methylated in juvenile leaves of F1 plants. This observation is similar to that made in Arabidopsis demonstrating that naturally occurring small RNAs derived from inverted repeats can be transported through the vasculature and cause transcriptional gene silencing in a target tissue (37). In maize, our data suggest that this process requires lbl1 activity. All detectable forms of transcriptional silencing of MuDR in the F1 transition leaves, including cytosine methylation in all sequence contexts as well as H3K9 and H3K27 dimethylation, are lost when lbl1 expression is reduced. Mutants in lbl1 exhibit substantial decreases in TIRA methylation and an increase in mudrA expression, even in juvenile leaves. Further a mutation that extends the period of transitional growth, Cg1, also extends the period of lbl1 down-regulation and TIRA hypomethylation.
The transient release of MuDR expression in transition leaves was initially puzzling: expression of the transposase gene would be expected to be deleterious to the host organism. However, this transcriptional activity is not associated with the production of functional transposase, suggesting that the transposon is being permitted to express a transcript but not to make a functional transposase. In addition, it occurs in a tissue that does not produce the germ line in maize but that is adjacent to one that will.
One intriguing possibility is that this period, the phase change, represents an opportunity for the plant genome to “unmask” potentially dangerous transposable elements. Progressive changes in methylation of transposons over the developmental course have lead to the suggestions that developmental time and shifts in epigenetic regulation of transposons may be functionally related (38); this idea is supported by our observations. In maize, the transition leaves represent a shift from nonreproductive to reproductive growth. Recent work in the vegetative nucleus in pollen, the developing ovule, and possibly the endosperm suggests that transposon expression is up-regulated in those tissues to permit the host to identify potentially dangerous, invasive DNA in a tissue that will not contribute to the next generation (39–43). Our data suggest that leaves during vegetative phase transition may represent an additional tissue in which transposons (and perhaps viruses, given the role that SGS3 plays in systemic silencing of these pathogens) are permitted to express transcript to enhance the process of transposon recognition and silencing. Such a system of silencing reinforcement would imply systemic trafficking of information in the form of small RNAs derived from inverted repeats, a process that has recently been documented in plants (44, 45).
It is also intriguing that both the timing of phase change and transcriptional release of mudrA may both be coordinately regulated, at least in part, by changes in lbl1 expression. The theme that is emerging is one in which germ-line differentiation requires effective transposon regulation, and therefore the two processes are often coordinated. We suggest that vegetative phase transition may represent an additional example of this process, adapted to the unique requirements of organisms that lack a sequestered germ line.
Materials and Methods
Plant Materials.
Generation of plants carrying MuDR, Muk, and derivatives was as previously described (8, 10). For mutant analysis, plants homozygous for lbl1 (46) or heterozygous for Cg1 (34) were crossed to plants carrying MuDR and to plants carrying Muk. The resulting progeny were intercrossed to generate sibling plants carrying both MuDR and Muk in mutant and wild-type genetic backgrounds.
Tissue Collection.
Leaves were collected when they were ≈6 cm long, as they emerged from the whirl. Immature ears were harvested once they were ≈6 cm long. Shoot apex tissue included the shoot apical meristem along with the youngest leaf primordia surrounding the shoot apical meristem from 2-wk-old seedlings.
Chromatin Immunoprecipitation (ChIP).
ChIP was carried out as described previously (47, 48) with modifications as described in SI Materials and Methods and with the primers provided in Table S1.
Real-Time PCR Analysis.
Quantitative PCR was performed by using FastStart Universal SYBR Green Master (ROX) (Roche) in a 25-μl PCR according to the manufacturer's instructions and using primers specific to each gene (described in SI Materials and Methods and Table S1). Relative fold change was determined by using the comparative CT method (49) normalized to control sequences.
Genomic Bisulfite Sequencing.
Genomic DNA was isolated as previously described (6). Bisulfite conversion was performed with an EpiTect Bisulfite kit (Qiagen). PCR fragments from TIRA were amplified by using TIRAmF6 and TIRAR3 (Table S1). PCR product was purified and cloned with a CloneJET PCR Cloning Kit (Fermentas), and 10 independent clones were sequenced from each sample. The resulting sequences were analyzed with kismeth (http://katahdin.mssm.edu/kismeth/revpage.pl) (50).
dsRNA Assay.
dsRNA analysis was performed as described previously (10).Detailed methods and associated references are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Margaret Woodhouse and Diane Burgess for critical reading of the manuscript and R. Keith Slotkin and Christopher Hail for initial characterization of Muk expression patterns. This work was supported by the National Science Foundation's Plant Genome Research Program (DBI-0820828 to D.L. and DBI-0337083 to M.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016884108/-/DCSupplemental.
References
- 1.Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- 2.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 3.Lau NC. Small RNAs in the animal gonad: Guarding genomes and guiding development. Int J Biochem Cell Biol. 2010;42:1334–1347. doi: 10.1016/j.biocel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosher RA, Melnyk CW. SiRNAs and DNA methylation: Seedy epigenetics. Trends Plant Sci. 2010;15:4204–4210. doi: 10.1016/j.tplants.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 6.Lisch D, Chomet P, Freeling M. Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu elements in a minimal line. Genetics. 1995;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walbot V, Evans MMS. Unique features of the plant life cycle and their consequences. Nat Rev Genet. 2003;4:369–379. doi: 10.1038/nrg1064. [DOI] [PubMed] [Google Scholar]
- 8.Slotkin RK, Freeling M, Lisch D. Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics. 2003;165:781–797. doi: 10.1093/genetics/165.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisch D. Mutator transposons. Trends Plant Sci. 2002;7:498–504. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- 10.Slotkin RK, Freeling M, Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- 11.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martienssen R, Baron A. Coordinate suppression of mutations caused by Robertson's mutator transposons in maize. Genetics. 1994;136:1157–1170. doi: 10.1093/genetics/136.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bongard-Pierce DK, Evans MMS, Poethig RS. Heteroblastic features of leaf anatomy in maize and their genetic regulation. Int J Plant Sci. 1996;157:331–340. [Google Scholar]
- 15.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y, et al. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007;19:9–22. doi: 10.1105/tpc.106.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Q, Guo HS. Systemic antiviral silencing in plants. Virus Res. 2006;118:1–6. doi: 10.1016/j.virusres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodhouse MR, Freeling M, Lisch D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 2006;4:e339. doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 22.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 23.Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalantidis K, Schumacher HT, Alexiadis T, Helm JM. RNA silencing movement in plants. Biol Cell. 2008;100:13–26. doi: 10.1042/BC20070079. [DOI] [PubMed] [Google Scholar]
- 25.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmermans MCP. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, et al. Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol Cells. 2008;26:611–615. [PMC free article] [PubMed] [Google Scholar]
- 28.Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 29.Fahlgren N, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 30.Douglas RN, et al. ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell. 2010;22:1441–1451. doi: 10.1105/tpc.109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 32.Sheridan WF. Maize developmental genetics: Genes of morphogenesis. Annu Rev Genet. 1988;22:353–385. doi: 10.1146/annurev.ge.22.120188.002033. [DOI] [PubMed] [Google Scholar]
- 33.Whaley WG, Leech JH. The developmental morphology of the mutant “corn grass”. Bull Torrey Bot Club. 1950;77:274–286. [Google Scholar]
- 34.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 35.Poethig RS. Heterochronic mutations affecting shoot development in maize. Genetics. 1988;119:959–973. doi: 10.1093/genetics/119.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter C, et al. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 38.Poethig RS. Phase change and the regulation of shoot morphogenesis in plants. Science. 1990;250:923–930. doi: 10.1126/science.250.4983.923. [DOI] [PubMed] [Google Scholar]
- 39.Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol. 2009;19:1677–1681. doi: 10.1016/j.cub.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 40.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olmedo-Monfil V, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 45.Dunoyer P, et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Timmermans MCP, Schultes NP, Jankovsky JP, Nelson T. Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development. 1998;125:2813–2823. doi: 10.1242/dev.125.15.2813. [DOI] [PubMed] [Google Scholar]
- 47.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 48.Haring M, et al. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 50.Gruntman E, et al. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics. 2008;9:371. doi: 10.1186/1471-2105-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.