Abstract
Transposons are mobile genetic elements that are an important source of genetic variation and are useful tools for genome engineering, mutagenesis screens, and vectors for transgenesis including gene therapy. We have used second-generation sequencing to analyze ≈2 × 105 unique de novo transposon insertion sites of the transposon Hermes in the Saccharomyces cerevisiae genome from both in vitro transposition reactions by using purified yeast genomic DNA, to better characterize intrinsic sequence specificity, and sites recovered from in vivo transposition events, to characterize the effect of intracellular factors such as chromatin on target site selection. We find that Hermes transposon targeting in vivo is profoundly affected by chromatin structure: The subset of genome-wide target sites used in vivo is strongly associated (P < 2e-16 by Fisher's exact test) with nucleosome-free chromatin. Our characterization of the insertion site preferences of Hermes not only assists in the future use of this transposon as a molecular biology tool but also establishes methods to more fully determine targeting mechanisms of other transposons. We have also discovered a long-range sequence motif that defines S. cerevisiae nucleosome-free regions.
Keywords: target site preference, integration, hAT element, next gen sequencing
Virtually all known genomes harbor transposable elements. Transposon integration site selection is of interest not only to expand our understanding of a transposon's behavior but also to facilitate use of that transposon as a molecular biology tool, or even as a clinical tool (e.g., in gene therapy; ref. 1).
The distribution of elements within an extant genome reflects the interplay between element insertion and deletion; these two phenomena can be difficult to separate, because the deleted elements are not usually observed. Thus, understanding how transposons choose target sites can provide insight into genome evolution.
Most transposable elements that have been studied do not choose their integration sites at random but rather use preferred integration sites, or “hotspots.” Hotspots for integration can be defined by particular sequence preferences (2), by interactions with host proteins (3–6), or by multiple mechanisms using different proteins (7–9). Additionally, in the eukaryotic cell, the accessibility of DNA to transposase and other proteins is considerably affected by chromatin structure (the 3D nucleoprotein complex of DNA wrapped around nucleosomes). Transposons can also be deleted. Because insertion of a transposable element into an essential gene could be lethal to a unicellular host and would variably impact a multicellular eukaryote depending on the timing of the insertion, transposable elements are generally highly regulated via mechanisms as diverse as methylation and RNAi (10–12).
Here, we have studied target site selection by the DNA cut-and-paste transposon Hermes in the genome of the baker's yeast Saccharomyces cerevisiae. Using next gen sequencing, we have analyzed large numbers of de novo insertions generated in vitro using purified transposase and naked genomic DNA as a target and in vivo using chromatin as a target to define Hermes target site selectivity. We find that the packing of DNA into nucleosomes precludes insertions into many sites that are targets for insertion in vitro.
Our high-throughput approach reveals the influence of local DNA sequence in determining target site choices and demonstrates that the major determinant of Hermes target site choice in vivo is accessibility of target DNA.
Results
Experimental Overview.
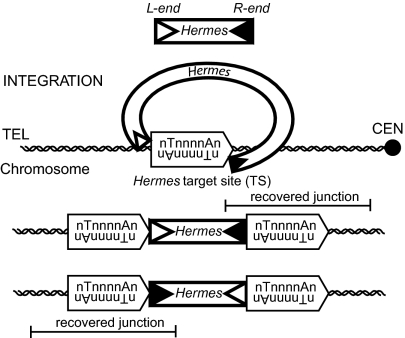
We have established an in vitro system for Hermes transposase by using naked DNA as a target (13). As described here, we have established a genetic selection protocol (SI Materials and Methods) to recover Hermes insertions in vivo into the yeast genome by using the NatMX gene driven by the TEF promoter as an antibiotic selection marker that can be detected even in regions of heterochromatin (14). A hyperactive version of the Hermes transposase was used to isolate a large number of integrants. We have used massively parallel sequencing methods to specifically and sensitively map de novo integration sites generated in both in vitro and in vivo. Because Hermes can insert in either orientation once it recognizes an 8-bp nTnnnnAn target site (Fig. 1), our pipeline is based on the positions of target sites recognized, not the orientations of the recovered insertion sites.
Fig. 1.
Hermes transposition mechanism. Hermes insertions have 8-bp target site duplications. Our experimental method specifically retrieves adjoining genomic DNA sequence at the right end of Hermes to retain both position and orientation information.
Table 1 gives the numbers of in vitro and in vivo target sites recovered in this study, broken down by the number of experiments in which each target site was found. Full details of the analysis pathway for each system are given in Fig. S1 A and B). Note that among transposon insertion sites in chromatin, in vivo we found that an unexpectedly large number of target sites are recovered in multiple experiments and that this is significantly different from what is expected when integration occurs randomly over all possible sites (P < 10−5 by simulation; SI Materials and Methods).
Table 1.
Target sites recovered in vitro and in vivo
| Recovery in fraction of experiments | Target sites with bidirectional insertions | Target sites with only one insertion | Total |
| In vivo | |||
| 6/6 | 844 | 133 | 977 |
| 5/6 | 1,729 | 356 | 2,085 |
| 4/6 | 3,477 | 985 | 4,462 |
| 3/6 | 7,492 | 3,294 | 10,786 |
| 2/6 | 14,969 | 15,161 | 30,130 |
| 1/6 | 10,430 | 116,730 | 127,160 |
| In vitro | |||
| 1/1 | 35,633 | 142,974 | 178,607 |
Rows indicate the number of times targets sites harboring insertions were mapped in the single in vitro experiment and in the six independent in vivo experiments. In the in vivo experiments, sites that contained bidirectional insertions and those containing insertion in a single orientation are counted separately.
Matched Random Control (MRC) Set.
To characterize Hermes target site selectivity, it was critical to create an adequate control distribution for comparison. We defined an 8-bp target site only by the nTnnnnAn sequence and then simulated the constraints of the experimental design by disallowing insertions too close to cleavage sites of the restriction enzyme used, because these insertions would not be recovered. We created the same number of total insertion sites as were observed in the initial results for each experiment, then carried out the same processing to generate a set of control target sites.
Analysis of Hermes Transposase Insertion Site Selection in Vitro.
Target site selection on naked DNA in vitro by Hermes transposase reveals the intrinsic target sequence specificity of the transposase in the absence of histones or other DNA-associated proteins. Moreover, any insertion can be recovered even if it would be lethal to living yeast.
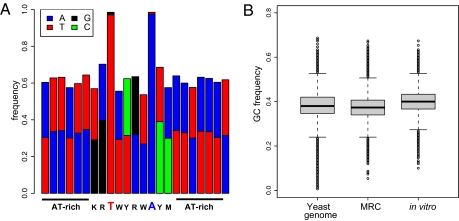
We analyzed 178,607 different target sites recovered in a single in vitro reaction by using isolated yeast DNA as a target (Table 1). Although we accepted every recovered in vivo read as a valid insertion event, for in vitro insertions, we considered as target sites those positions in which insertions were recovered in both directions; this way, we were confident that we were analyzing completed insertion events. When the in vitro target sites are aligned (Fig. 2A), we observe the previously described nTnnnnAn target site duplication (13), as well as a longer, subtle consensus. To determine the possible effects of using the longer consensus, at every position in the genome we generated a log likelihood of that base being the starting point for a motif that agreed with the consensus. The sites that emerged as the most favorable targets were nearly exactly the sites created in the MRC, so we continue to use this as the baseline.
Fig. 2.
Sequences of in vitro target sites. (A) Composition plot of target sites from the in vitro experiment. IUBMB nomenclature for bases is used. R, purine; Y, pyrimidine; W, A/T; M, A/C; and K, G/T. (B) Sequence composition.
Target Sites Used in Vitro Are GC-Rich.
The predicted target sites are an accurate model of the in vitro sites, as evidenced by the fact that 89% of the in vitro target sites overlap with predicted sites. However, the in vitro site choice appears to be much more specific than the model, as only 25% of the predicted sites are actually found in the in vitro data. Taking 150-bp windows centered at the midpoint of each predicted target site, the set of MRC sites has a composition very much like the average yeast genomic composition, at 38.4% GC. By contrast, the target sites recovered in the in vitro reaction are much more GC-rich, at 40.2% GC (Fig. 2B).
Intergenic Bias of Hermes Insertion in Vivo.
To probe the effect of chromatin on Hermes insertions, we generated six independent sets of insertion sites in vivo, three in haploids and three in diploids to yield 175,600 sites (Table 1). Alignment of the in vivo target site duplication sequences revealed the same overall sequence pattern observed in vitro, nTnnnnAn.
We compared the distribution of target sites in intergenic regions (IGRs) to the distribution of target sites in ORFs (Table 2). Approximately 70% of the yeast genome is occupied by ORFs; however, the in vivo target site distribution in both haploid (40.8% in ORFs and 59.2% in IGRs) and diploid (45.4% in ORFs and 54.6% in IGRs) experiments suggests that Hermes preferentially targets intergenic regions. Although the haploid cells may be less likely to survive disruption to ORFs (although we still see insertions in 758 of the 1,211 Saccharomyces Genome Database genes annotated as essential) (Table S1), most diploid cells can presumably tolerate an insertion that completely disrupts one allele of an essential gene; in both cases, the counts of Hermes insertions recovered from ORFs are much lower than expected (P < 0.001).
Table 2.
Target sites in ORFs and classes of IGRs
| Haploid, % | Diploid, % | in Vitro, % | MRC, % | Yeast genome, % | |
| Genome | |||||
| ORF | 40.8* | 45.4* | 75.3* | 69.8 | 70.6 |
| IGR | 59.2* | 54.6* | 24.7* | 30.2 | 29.4 |
| Breakdown of intergenic regions | |||||
| Tandem | 51.0 | 51.0 | 49.2 | 49.8 | 51.5 |
| Convergent | 11.1 | 10.12 | 15.3 | 17.08 | 15.5 |
| Divergent | 37.4 | 38.64 | 33.74 | 32.19 | 33.2 |
Number of Hermes target sites in ORFs and IGRs in in vivo haploid and diploid yeast datasets and the in vitro dataset is contrasted with a matched random control (MRC) as well as the ratios from the yeast genome. Also shown is the distribution of target sites in classes of intergenic regions: IGRs flanked by two 5′ gene boundaries (divergent), IGRs flanked by two 3′ gene boundaries (convergent), and other orientations (tandem).
*P < 0.001, Fisher's exact test.
Intergenic regions come in three varieties: those between two 5′ ends of genes (divergent), those between two 3′ ends of genes (convergent), and those flanked by genes that are on the same strand (tandem). As shown in Table 2, the target site counts in the convergent and divergent categories are significantly different from the simulated data (P < 2.2e-16, Fisher's exact test) and suggest a propensity for Hermes to insert near the 5′ end of genes, because insertions into divergent regions are overrepresented and insertions into convergent regions are less common.
ORF Boundaries Are Targeted by Hermes in Vivo.
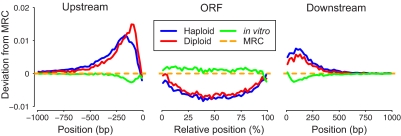
We plotted the genomic distribution of target sites in both haploids and diploids across all genes in the yeast genome (Fig. 3). Strikingly, we found that insertions generated in vivo were much more common near gene boundaries, which are AT-rich, generally falling just outside ORFs (P << 2.2e-16). In contrast, in vitro Hermes target sites are underrepresented just outside of ORFs, consistent with the preference for GC-rich regions described above.
Fig. 3.
Target site distribution in the neighborhood of all yeast ORFs. In haploid (blue line), diploid (red line), and in vitro (green line) datasets, the insertion site frequency is represented as a deviation from the MRC (yellow dotted line). Data upstream and downstream of each gene are presented as positions relative to the transcription start site or transcription termination site. Insertions are plotted in percentage intervals along the gene.
Hermes Inserts in 5′ and 3′ Nucleosome-Free Regions (NFRs) at Yeast ORFs.
Like the regions of preferred Hermes insertion in vivo, NFRs (also often termed nucleosome-depleted regions) are present upstream and downstream of genes in vivo (15) and, indeed, we find that Hermes inserts in vivo preferentially into NFRs (Fig. 4).
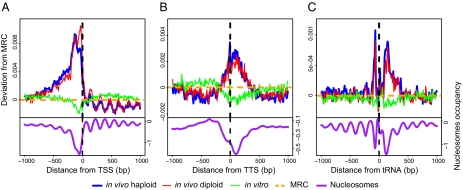
Fig. 4.
Hermes target NFRs near the borders of yeast ORFs. (A) Hermes insertions are strongly correlated with regions of lower nucleosome occupancy upstream of transcription start sites (TSS). All TSS in the yeast genome are aligned, and Hermes target sites in haploid (blue), diploid (red), in vitro (green), and MRC (dashed, orange) datasets are plotted (after normalization) with respect to each neighboring TSS (Upper). Nucleosome occupancy (purple) as in Lee et al. (15) (Lower). (B) Hermes insertions are correlated with regions of lower nucleosome occupancy at transcription termination sites (TTS). Colors are as in A. (C) Insertion site distribution at yeast tRNAs Hermes target sites in haploid (blue), diploid (red), in vitro (green), and MRC (dashed, orange) datasets are plotted (after normalization) with respect to each tRNA TSS (Upper). Nucleosome occupancy (purple) as in Lee et al. (15) (Lower).
We plotted the genomic environment of every Hermes target site that fell within 1 kb of a transcription start site (TSS) (Fig. 4A) and transcription termination site (TTS) (Fig. 4B) of any yeast gene (TSS and TTS from ref. 16). Strikingly, these Hermes target sites, especially in the case of the diploid dataset, coincide with regions of low nucleosome occupancy, which was determined based on published data (Fig. S2). Notably, in 5′ ends of genes, apparently phased peaks of Hermes insertion are observed, corresponding to phased patterns of nucleosome occupancy (and thus NFRs).
A similar picture emerges when all Hermes insertion sites are plotted across all tRNA genes (Fig. 4C). Here, the preference for NFRs is quite distinct, because the peaks of target site insertions follow the well-characterized valleys in nucleosome occupancy that coincide with the reported binding regions of the TFIIIB and TFIIIC transcription factor complexes (15) across the tRNA genes.
We suggest that these patterns result from preferential insertion into regions in which DNA is most physically accessible to Hermes. The target sites recovered the in vivo experiment are valid sites but are a different subset of the MRC than the in vitro sites; in particular, they are less GC-rich than the sites preferred in vitro. The different propensity of Hermes to insert near genes in the two systems reflects this preference.
Genome-Wide Bias of Hermes for Low Nucleosome-Occupancy Regions.
To determine whether nucleosome occlusion of target DNA is the primary determinant of target site choice or simply a coincident finding (because gene boundaries and NFRs are intrinsically closely related), the distribution of Hermes target sites was compared with nucleosome occupancy. We used published data (15) to define NFRs, regions of intermediate nucleosome occupancy (IORs), and nuclesome-occupied regions (NORs) (SI Materials and Methods and Fig. S2).
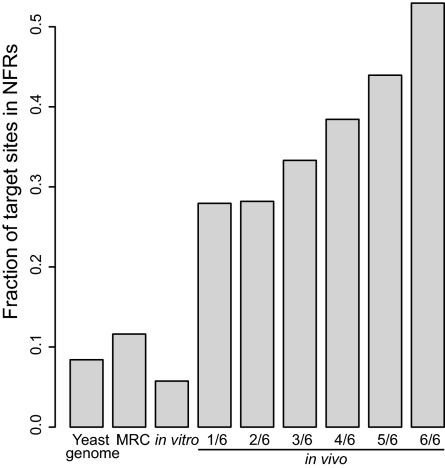
The target sites that are used most frequently, or hotspots, (those that were recovered in all six experiments) are most strongly biased toward NFRs (P < 2.2e-16, t test) (Table 1, Fig. 5, and Table S2). These hotspots do not result from preferential insertion due to targeting of a particular 8-bp target site duplication because their sequences are basically the same (Fig. S3). Strikingly, the target sites recovered from the highest number of experiments were most likely to be in NFRs (Fig. 5), suggesting that NFRs very strongly influence the selection of target sites by the Hermes transposase. In contrast, the in vitro target sites are generally underrepresented in NFRs (NFRs are AT-rich compared with the in vitro target sites recovered).
Fig. 5.
Favored target sites are more common in NFRs. The percentages of target sites in each of the categories of the in vivo data that fall into NFRs are shown, along with the percent of in vitro and matched random control target sites in NFRs and the percentage of the yeast genome that is occupied by NFRs.
Preferred Hermes Target Sites Are the Centers of Long-Range DNA Composition Biases.
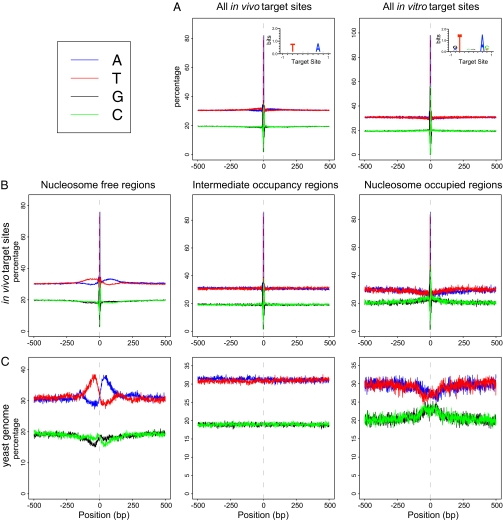
We also explored the sequence environment of each 8-bp target site duplication by plotting the nucleotide frequency at each position within a 1-kb window centered on each target site (Fig. 6A). This extended sequence context reveals a pattern that is striking as a whole, yet not detectable in any single sequence. These regions have a distinct T-rich segment on the 5′ of the target site midpoint and an A-rich region on the 3′ end. This pattern is detectable to almost 200 bp on each side of target sites and is symmetric, in keeping with the propensity of Hermes to insert in either orientation in a target site. Strikingly, this long-range composition bias is not seen in the in vitro data (Fig. 6A), again indicating that Hermes is choosing its target sites in vivo based on a feature of DNA in vivo that is not present in vitro.
Fig. 6.
Extended target site motifs by nucleosome occupancy. (A) Nucleotide frequency distributions in 1-kb regions surrounding all in vivo and all in vitro target sites recovered in this study. (B) Nucleotide frequency distribution in the 500 bp flanking the midpoint of all Hermes target sites found in NFRs (Left), regions of intermediate nucleosome occupancy (IOR) (Center), and nucleosome occupied regions (NOR) (Right) as defined in Fig. S2. (C) Corresponding nucleotide frequency distribution of 500 bp defining NFRs, IORs, and NORs in the yeast genome.
The T/A asymmetry indicates that long-range sequence and/or structural mechanisms participate in determining Hermes targeting. Because Hermes target sites are overrepresented in NFRs in vivo, we broke the dataset into the target sites that fell in NFRs, IORs, and NORs, and plotted the composite nucleotide frequency of the 1-kb windows surrounding the target sites in each category (Fig. 6B). The symmetric variation in nucleotide frequency is quite prominent in the set of target sites in NFRs but not in the other two subsets. Then we aligned all yeast NFRs, centered on their midpoints, and constructed a composition plot (Fig. 6C). The unusual long-range T/A bias is even stronger in this plot than when only Hermes target sites are considered, suggesting that it is peculiar to the yeast NFRs and not just to Hermes target sites. Because Hermes targets the center of the yeast NFRs, the midpoint for both patterns is the same.
A recent publication (17) reported short poly(T) and poly(A) tracts in NFRs. We see these sequences at a higher than expected rate, but the longer-range motif is not solely a consequence of those short patterns; instead, it is a consequence of both the summation of the shorter patterns and a larger scale sequence motif.
Notably, a quite distinct nucleotide composition pattern is seen at target sites in NORs and in NORs themselves (Fig. 6C), with an unusually GC-rich area at the center of the NORs. Analysis of the sequence composition of NORs, using 5-base sequence fragments, reveals that the central portion of NORs has an unusually high number of GC-rich 5-mers.
Hermes Target Sites Not in Hotspots Cluster Spatially.
So far we have considered the Hermes transposase targeting simply as an aggregate of the individual target sites recovered in the various experiments. A second analysis focused on spatial clustering of insertions across the yeast genome (Fig. 7A). Whereas hotspots may measure a high propensity for insertion into a very specific site, clusters should occur in areas that are generally predisposed to an above average number of insertions but that do not necessarily conform precisely to the conditions that create insertion hotspots. Given that hotspots were found disproportionately in NFRs, we expected that clusters might be found near promoter regions and other biologically active areas of DNA that have more dynamic accessibility.
Fig. 7.
Clusters of insertions. (A) Hotspots and clusters. (B) Distribution of target site clusters in NORs, IORs, and NFRs. (C) Sequence composition of 150-bp windows. Windows are centered at the midpoint of randomly chosen fragments of the yeast genome, MRC dataset, in vitro or in vivo cluster, and hotspots.
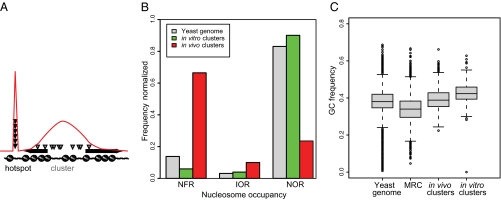
Target sites from all six in vivo experiments were pooled, hotspots were removed from the dataset, and, as discussed in Materials and Methods, a kernel smoothing algorithm was applied to define boundaries of clusters and to determine whether a large number of target sites were actually closer in genomic space than would be expected at random. This analysis yielded 1,807 clusters, ranging in size from 4 to 1,650 bp (average 255; very short clusters can result from insertions into small sequences that have high densities of T's and A's). An example of clusters from this analysis is shown in Fig. S4. Fig. S5 gives a bird's-eye view of the clusters on all chromosomes from the in vivo experiments. Positions of target sites are given in Dataset S1. We also identified in vitro clusters by using the same strategy. The target site sequence in both in vivo and in vitro clusters, nTnnnnAn, was the same as for hotspots, nTnnnnAn.
In vivo clusters occur predominately in NFRs (Fig. 7B). By contrast, the very few in vitro clusters observed were overrepresented in NORs, again supporting the idea that the in vitro targeting reflects sequence preferences, whereas targeting in vivo reflects the nucleosome status of the target DNA. Additionally, the genomic context of in vivo clusters had significantly higher GC content than the genome, and higher than that of NFRs (Fig. 7C).
Discussion
This study has used de novo generation and analysis of large numbers of both in vitro and in vivo target sites to define transposable element target site specificity.
We analyzed 178,607 in vitro target sites and 175,600 in vivo target sites; each set of target sites was derived from millions of sequencing reads. The mean density of insertions underlying the compiled target sites was between 121/kb (haploid and in vitro experiments) and 24/kb (diploid experiments).
In vitro and in vivo analyses both confirm that the 8-bp target site duplication nTnnnnAn is the key sequence element driving target site selection. However, we found that insertions in vivo were not randomly distributed throughout the genome and occurred predominantly in intergenic regions, especially at gene borders. We suggest that this pattern reflects preferential Hermes targeting to NFRs (thus using the most accessible DNA as a target) even though our in vitro studies revealed that Hermes preferentially inserts into GC-rich DNA and NFRs on average are AT-rich in comparison with the genome as a whole (66.7% vs. 60.7% AT).
We have also found that NFRs have a distinct pattern of a T-rich region on the top strand 5′ of the target site midpoint and an A-rich region on the top strand at the 3′ end. This pattern is symmetric and detectable to almost 200 bp on each side of the nucleosome-free target sites. Although it has been long known that poly(dA:dT) segments tend to exclude nucleosomes because they are resistant to bending, such a distinctive sequence signature has not been recognized (18).
We also analyzed the spatial distribution of insertion sites, finding a number of clusters of insertion sites both in vitro and in vivo. These regions are GC-rich, consistent with the in vitro results. Clusters in vivo could result from targeting to a region with favorable sequence composition but varying nucleosome occupancy, so that all insertion events do not happen at exactly the same site. This occlusion model would predict changes in transposon insertion pattern at genomic locations whose nucleosome occupancy is governed by the action of histone modifying enzymes and chromatin remodelers that may in turn be modulated by physiological stimuli; occupancy of this region may be more dynamic than the rest of the genome. More limited studies using restriction enzyme digestion and the binding of sequence-specific DNA binding proteins have also shown that DNA is more accessible in NFRs (18, 19).
Other transposons such as the Drosophila P element, the maize Mu transposon, and the retrotransposon Tf1 in Schizosaccharomyces pombe show a preference for insertion into the 5′ ends of genes and may reflect the preferential use of target sites in NFRs (20–24). This preference is not a general attribute, however; the integrase of some elements such as HIV actually exploits nucleosome-induced bending of DNA to identify target sites throughout the genome, both in vitro and in vivo (25–27). The fungal LTR elements Ty1 and Ty3 insert preferentially into DNA on the nucleosome surface upstream of pol III genes, and Ty5 inserts preferentially into heterochromatin (reviewed in refs. 28 and 29).
The Hermes element also appears to be especially sensitive to DNA composition, preferring GC-rich regions, even when its choice of target sites is constrained, as in vivo, to only a subset of the genome. This property makes Hermes an excellent sensor of the local DNA environment, even beyond its target site, and raises the possibility of the Hermes target sites observed being part of larger regions of DNA structure and composition that are not easily apparent when examined one by one.
Thus, transposons can be sensitive and useful probes of chromatin structure. We suggest that Hermes is a useful probe for NFRs, which are related to gene expression and other DNA transactions. Probing gene activity in a way other than relying on RNA as readout may be a powerful approach. Two recent publications report correlations of replication origins and NFRs (30, 31), and MacAlpine et al. also reports targeting of the origin replication complex (which targets nucleosome-free regions) to active promoters, suggesting that the Hermes transposon may be a useful tool in this realm as well.
Materials and Methods
Hermes Insertion and Recovery in Vivo in S. cerevisiae.
To recover in vivo integrations, we first constructed a yeast ARS CEN plasmid, pSG36, (SI Materials and Methods) containing a URA3 marker, a Hermes-NatMX transposon, and a GALs promoter-regulated Hermes transposase gene. Cells containing a chromosomal transposition event were recovered by selecting for resistance to the antibiotic, ClonNAT, and 5′-fluoroorotic acid (5-FOA). ClonNAT resistance resulting from expression of the NAT1 gene driven by the TEF promoter can de detected even in regions of heterochromatin (14). Cell harboring plasmids with unexcised transposons were counter selected by using 5-FOA. We prepared amplicon libraries of transposition insertion sites by LMPCR (SI Materials and Methods) using Mse1-digested genomic DNA isolated from haploid and diploid strains of yeast in which transposition was induced for ≈80 generations in liquid synthetic complete medium containing galactose as in the scheme outlined in ref. 32. We measured transposition frequencies (SI Materials and Methods), and at least one in 100 induced cells had an integration event. Oligos used in this study are listed in Table S3.
We also analyzed Hermes insertions in vitro into deproteinized yeast genomic DNA by using a modified version of our previously described in vitro system to ascertain the extent of the influence of nucleosomes on the pattern of insertions in vivo and to uncover any intrinsic sequence preference that the Hermes transposase may have (SI Materials and Methods and ref. 13. Hermes R-end-genomic DNA junctions, recovered by LM-PCR, were also sequenced by using Illumina instruments, by the core facility at the University of California, Riverside.
Strategy for Sequence Analysis.
Our processing pipeline (Fig. S1 A and B) was stringent; any read that did not exactly contain the expected 10 bp of the substrate Hermes transposon Rend (allowing a single ambiguous base, i.e., one “N,” in the sequencing read) was excluded. The remaining reads were trimmed and aligned to the yeast genome by using the Bowtie short read alignment program (33). At this point, we required an exact match to the yeast genome and excluded any read that aligned more than once to the yeast genome, because that junction could not be placed unambiguously. To remove bias in the read counts due to PCR or other amplification biases, we simply collapsed all repeated insertion reads to create a set of nonredundant insertion sites, giving each insertion site equal weight regardless of how many times it appeared in the raw sequencing data.
Target Sites.
Each sequencing read comes from a junction between the Hermes transposon and the yeast genome (Fig. 1). To define the insertion sites in a biologically relevant way and to create a consistent framework for analysis, we identified the target site for each insertion and our analyses always refer to the center of that site, not to the coordinates of the junctions between Hermes and the yeast genome. This is particularly important for the in vitro experiment, in which we were concerned that for technical reasons we might recover some junctions representing incomplete translocation products; here we did not consider a target site unless we recovered insertions in both orientations in that site, indicating that it was specifically targeted at least twice.
MRC.
We created a MRC set of transposon insertions by randomly choosing nTnnnnAn sites that are >10 bp away from an MseI site (mimicking the experimental protocol). We chose the same number of sites as the number of sequencing reads that aligned to the yeast genome and processed them as in Fig. S1A. This set of simulated data were used throughout as a random model that is more realistic than considering every genomic position a potential insertion site.
Supplementary Material
Acknowledgments
We thank Rupak Mitra, Jef Boeke, Rafael Irizarry, Henry Levin, and members of the N.L.C. Laboratory for valuable discussions. We thank Peter Atkinson (University of California, Riverside, California) for Hermes plasmids and Larisa Mitkina (Johns Hopkins School of Medicine, Baltimore) for providing the hyperactive Hermes mutant. Funding for this work was provided by National Institutes of Health Grants GM076425 (to N.L.C.) and CA09139. N.L.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016382107/-/DCSupplemental.
References
- 1.VandenDriessche T, Chuah MK. Moving gene therapy forward with mobile DNA. Hum Gene Ther. 2009;20:1559–1561. doi: 10.1089/hum.2009.1109. [DOI] [PubMed] [Google Scholar]
- 2.Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci USA. 1992;89:7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 4.Chalker DL, Sandmeyer SB. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126:837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalker DL, Sandmeyer SB. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Kuduvalli PN, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waddell CS, Craig NL. Tn7 transposition: Recognition of the attTn7 target sequence. Proc Natl Acad Sci USA. 1989;86:3958–3962. doi: 10.1073/pnas.86.11.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parks AR, et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell KA, Burns KH, Boeke JD. A descent into the nuage: the maelstrom of transposon control. Dev Cell. 2008;15:179–181. doi: 10.1016/j.devcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovsin N, Peterlin BM. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann N Y Acad Sci. 2009;1178:268–275. doi: 10.1111/j.1749-6632.2009.05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 14.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 16.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu R, Li H. Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters. Genome Res. 2010;20:473–484. doi: 10.1101/gr.103226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellen HJ, et al. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spradling AC, et al. Gene disruptions using P transposable elements: An integral component of the Drosophila genome project. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y, Levin HL. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010;20:239–248. doi: 10.1101/gr.099648.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 27.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushman FD. Targeting survival: Integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 29.Yieh L, Kassavetis G, Geiduschek EP, Sandmeyer SB. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J Biol Chem. 2000;275:29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 30.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JM, Evertts AG, Levin HL. The Hermes transposon of Musca domestica and its use as a mutagen of Schizosaccharomyces pombe. Methods. 2009;49:243–247. doi: 10.1016/j.ymeth.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.