Abstract
Plant germ cells develop in specialized haploid structures, termed gametophytes. The female gametophyte patterns of flowering plants are diverse, with often unknown adaptive value. Here we present the Arabidopsis fiona mutant, which forms a female gametophyte that is structurally and functionally reminiscent of a phylogenetic distant female gametophyte. The respective changes include a modified reproductive behavior of one of the female germ cells (central cell) and an extended lifespan of three adjacent accessory cells (antipodals). FIONA encodes the cysteinyl t-RNA synthetase SYCO ARATH (SYCO), which is expressed and required in the central cell but not in the antipodals, suggesting that antipodal lifespan is controlled by the adjacent gamete. SYCO localizes to the mitochondria, and ultrastructural analysis of mutant central cells revealed that the protein is necessary for mitochondrial cristae integrity. Furthermore, a dominant ATP/ADP translocator caused mitochondrial cristae degeneration and extended antipodal lifespan when expressed in the central cell of wild-type plants. Notably, this construct did not affect antipodal lifespan when expressed in antipodals. Our results thus identify an unexpected noncell autonomous role for mitochondria in the regulation of cellular lifespan and provide a basis for the coordinated development of gametic and nongametic cells.
Keywords: cell–cell communication, gametes, programmed cell death
In angiosperms, gametes form in few-celled haploid structures, termed gametophytes. The female gametophyte of most flowering plants originates from a single haploid spore through three syncytial division cycles. Subsequent cellularization generates two synergids, three antipodal cells, and two types of female gametes, an egg and a central cell. The different cell types have distinct functions in the reproductive process. Synergids mediate short-range pollen tube attraction and direct the subsequent release of the two sperm cells (1). The fertilized egg gives rise to an embryo, and the fusion of the second sperm cell with the central cell initiates the formation of endosperm, which nurtures the developing embryo. The central cell initially comprises two haploid polar nuclei, which, in many flowering plant species, fuse before fertilization, generating a diploid secondary nucleus (2). The diploid status of the central cell translates into triploid endosperm with a maternal/paternal ratio of 2:1. This ratio has been shown to critically impact on seed size as, for example, a relative decrease in the maternal contribution results in bigger seeds (3). Antipodals, the accessory cells that lie adjacent to the central cell, might also play a nutritive role by transferring nutrients from the maternal sporophyte to the female gametophyte (4). In several grass species, like wheat and maize, antipodal cells proliferate (5). By contrast, in most higher eudicots antipodal cells do not persist but undergo programmed cell death (PCD) (4) (Fig. 1 A–C). The adaptive value of this derived developmental program (2) is unclear. Recent studies have demonstrated a remarkable developmental plasticity of antipodal cells: in lachesis and clotho mutants, which are defective for the putative splicing factors PRP4 and Snu114, respectively, antipodals can adopt a central cell fate (6, 7). Furthermore, overexpression of an auxin biosynthesis gene can activate egg cell marker expression in antipodal cells (8). The gametic potential of antipodal cells has raised the hypothesis that antipodal cells act as a backup in case of gametic failure (6). However, to this end it is not known how PCD in antipodal cells is activated or bypassed. In the present work we show that PCD in antipodals is suppressed after mitochondrial dysfunction in the central cell. Our results thus provide a mechanistic basis for the coordinated development of both cell types and reveal a unique noncell autonomous mechanism for the regulation of cellular lifespan.
Fig. 1.
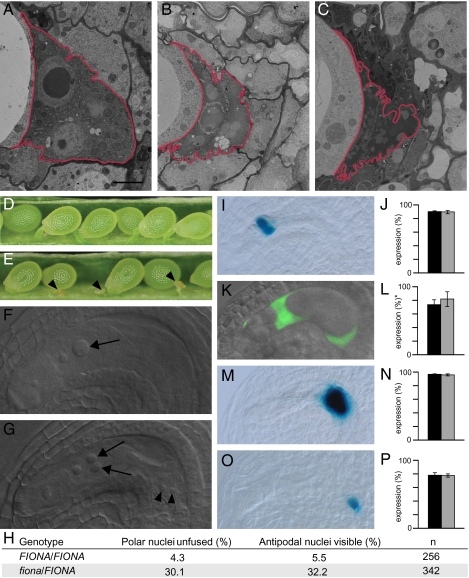
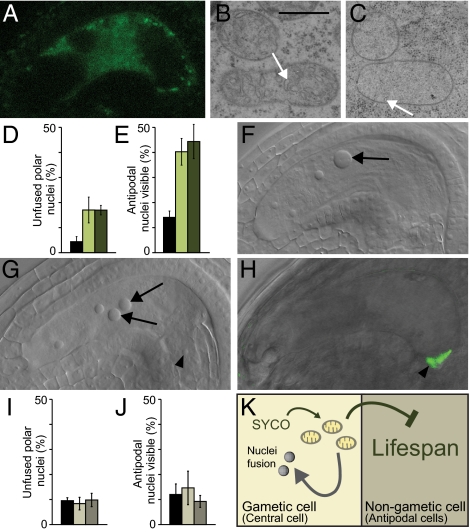
fiona mutants exhibit defects in polar nuclei fusion and an extended antipodal lifespan. (A–C) Antipodals in wild-type (A) 1 d, (B) 2 d, and (C) 3 d after emasculation. The antipodals are marked by a red border. (Scale bar, 2 μm.) (D) Wild-type silique showing full seed set. (E) Silique of a selfed fiona/FIONA plant containing normal and aborted seeds (arrowheads). (F and G) Whole-mount clearings of gametophytes 2 d after emasculation. (F) Wild-type gametophyte containing a secondary nucleus in the central cell (arrow). (G) fiona gametophyte containing unfused polar nuclei (arrows) and persisting antipodal cells (arrowheads). (H) Frequencies of unfused polar nuclei and persistent antipodal cells in wild-type and fiona/FIONA plants. (I–P) Expression of central cell and antipodal cell marker genes. Black bars represent wild-type, gray bars represent fiona/FIONA plants. (I) Expression of the central cell marker pMEA::NLS_GUS (7). (J) Frequencies of ovules expressing pMEA::NLS_GUS; n = 534 for wild type, n = 499 for fiona/FIONA. (K) Expression of the central cell marker DD22::GFP (35). (L) Frequencies of ovules expressing DD22::GFP; n = 176 for wild type, n = 151 for fiona/FIONA. *For ease of comparison the scores from DD22::GFP/- plants were adjusted to a homozygous marker situation. (M) Expression of the antipodal cell marker pAt1g36340::GUS (16). (N) Frequencies of ovules expressing pAt1g36340::GUS; n = 395 for wild type, n = 549 for fiona/FIONA. (O) Expression of the antipodal cell marker GT3733. (P) Frequencies of ovules expressing GT3733; n = 275 for wild type, n = 262 for fiona/FIONA. Gametophytes were analyzed 2 d after emasculation. Error bars, mean ± SEM.
Results and Discussion
FIONA Is Necessary for the Fusion of Central Cell Nuclei.
We have isolated the fiona mutant in an ethyl methanesulfonate (EMS)-induced screen for female gametophytic mutants. Heterozygous fiona plants exhibit 24.6% infertile seeds (Fig. 1 D and E; n = 1045), suggesting that homozygous fiona mutants are embryo lethal. Light microscopy inspection revealed that the infertile seeds correspond to early embryo and endosperm arrests, revealing an essential role of FIONA during seed development (Fig. S1). Because we could not recover homozygous plants, we performed all of the analyses on heterozygous mutants, which segregate 50% wild type and 50% mutant gametophytes. In wild type, the central cell initially comprises two haploid polar nuclei, which mainly fused before fertilization, resulting in the formation of a large secondary nucleus (Fig. 1 F and H). By contrast, in most fiona female gametophytes, polar nuclei size was slightly reduced and the nuclei failed to fuse (Fig. 1 G and H). Defects in polar nuclei fusion have been reported for several female gametophytic mutants (9–11). In lachesis and clotho mutants, the fusion defect is a likely consequence of a misspecified central cell (6, 7). To test whether fiona central cells were similarly affected, we introduced two central cell markers. Marker expression in fiona/FIONA was comparable to wild type (Fig. 1 I–L), suggesting that the central cell identity was not broadly perturbed.
FIONA Regulates PCD of Antipodal Cells.
In addition to the alterations in central cell development, we frequently observed persistent antipodal cells in fiona mutant gametophytes (Fig. 1 F–H), indicating that the developmental program that ultimately results in antipodal cell death requires FIONA activity. In lachesis and clotho mutants, defects in antipodal cell death correlate with the progressive conversion of antipodal cells into central cells (6, 7). By contrast, fiona antipodals did not seem to switch cell fate, as evidenced by the correct expression of two antipodal cell markers (Fig. 1 M–P) and a lack of pMEA::NLS_GUS central cell marker expression in antipodal cells (n = 499). These results indicated that FIONA interferes with a process downstream of antipodal cell commitment but upstream of the PCD pathway. The overall morphology of the egg cell and synergids in fiona mutants was normal, and egg cell marker expression was unaltered (Fig. S2 A and B). Light-microscopy inspection revealed an occasional increase in synergid nucleolus size (5%, n = 342), and expression of the synergid marker ET2634 (6) was slightly reduced (Fig. S2 C–F). This phenotype, however, did not affect the pollen tube attracting activity of the synergids (Fig. S2G).
Nuclear Fusion in fiona Central Cells Relies on Paternal Cues.
A common feature of previously described polar nuclei fusion mutants is their reduced fertility (6, 7, 9, 11). Most of these mutants fail to proceed beyond the haploid phase. The recently described bip1-4 bip2-1 double mutant does get fertilized despite an apparent polar nuclei fusion defect; however, the resulting seeds abort (12). In bip1-4 bip2-1, central cell proliferation seems to be initiated prematurely after fusion of the male sperm nucleus to only one of the polar nuclei, resulting in the formation of diploid instead of triploid endosperm.
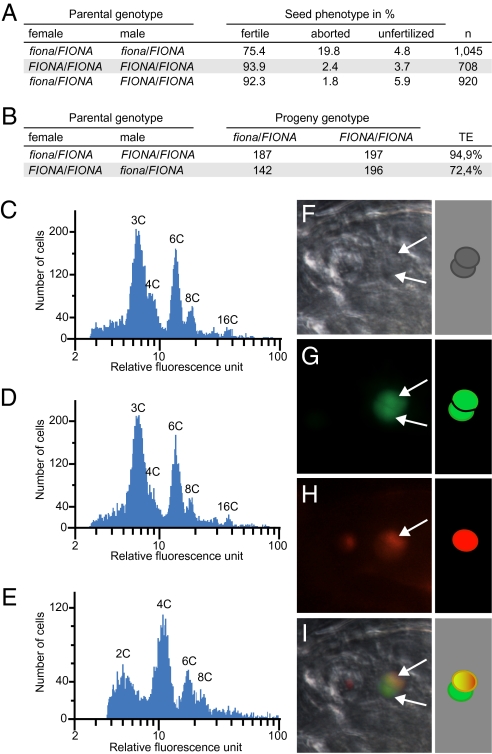
To our surprise, fiona gametophytes were fully fertile when pollinated with wild-type pollen (Fig. 2A), and the fiona mutation was transmitted through the female without significant decrement (Fig. 2B). Importantly, the frequency of unfused polar nuclei in fiona female gametophytes remained constant even when the flowers were kept artificially unfertilized over a period of 4 d (n = 140). This contrasts markedly with the situation in, for example, slow walker1 and slow walker2 mutants, in which unfused polar nuclei result from a developmental delay of the female gametophyte (13, 14). We therefore reasoned that the fertility of mutant gametophytes was likely to be due to a fertilization-triggered polar nuclei fusion. Alternatively, we could not exclude the possibility that in fiona, similar to the situation in bip1-4 bip2-1 double mutants, diploid endosperm is formed, resulting from fusion of the male sperm nucleus to one of the haploid polar nuclei. The latter scenario would consequently affect the maternal/paternal ratio of the endosperm. We, therefore, measured seed size and determined endosperm ploidy as an indirect readout of polar nuclei fusion. Seeds derived from fiona/FIONA plants were comparable in size to wild-type seeds (wild type 429 μm, n = 533; fiona/FIONA 432 μm, n = 484). Furthermore, the endosperm nuclei in fiona/FIONA seeds were uniform in size and did not differ from wild type (n = 156 for wild type; n = 154 for fiona/FIONA), which contrasts with the situation in bip1-4 bip2-1 double mutants, which exhibit irregular nuclei size in the endosperm (12).
Fig. 2.
fiona mutant female gametophytes give rise to viable seeds. (A) Analysis of siliques resulting from crosses of fiona/FIONA and FIONA/FIONA plants. (B) Segregation analysis of fiona, as determined by reciprocal crosses between fiona/FIONA and FIONA/FIONA plants and PCR-based genotyping of the progeny. TE, transmission efficiency. (C) Flow cytometry ploidy analysis of seeds derived from wild-type plants. (D) Flow cytometry ploidy analysis of seeds derived from fiona/FIONA plants. (E) Flow cytometry ploidy analysis of seeds derived from emasculated fie/FIE flowers. (F–I) Fertilization of a fiona mutant gametophyte expressing pAt5g40260::NLS_GFP with sperm cells expressing HTR10-mRFP1. Arrows point at the polar nuclei. (F) Bright-field image. (G) pAT5g40260::NLS_GFP. (H) HTR10-mRFP1. (I) Overlay.
Additionally, flow cytometry analysis of fiona/FIONA seeds detected a 3C peak, as in wild type (Fig. 2 C and D). This profile contrasted with the one generated for fie/FIE mutants (Fig. 2E), which form autonomous diploid endosperm in the absence of fertilization (15). To trace the fusion of male and female gametes during fertilization in fiona gametophytes, we generated fiona/FIONA plants expressing a nuclear-localized GFP under control of the gametophyte specific promoter pAt5g40260 (16). We then pollinated fiona/FIONA, pAt5g40260::NLS_GFP plants with pollen expressing the sperm cell-specific reporter gene HTR10-mRFP1 (17). In fiona gametophytes, we observed unfused polar nuclei, of which one had fused with a sperm nucleus (Fig. 2 F–I), confirming that central cell nuclei in fiona can undergo karyogamy (n = 9).
Intriguingly, the female gametophyte of fiona mutants shares morphological and mechanistic similarities with the female gametophyte of several grass species, such as wheat. In wheat, nuclei fusion in the central cell is initiated after fertilization by heterotypic fusion between the male sperm nucleus and one of the polar nuclei (18). Furthermore, antipodal cells do not undergo PCD but persist. In view of the similarities between the fully fertile fiona gametophytes and the gametophyte pattern of wheat, it is tempting to speculate whether a FIONA-related mechanism has been used during evolution, accounting for part of the diversity found among flowering plant female gametophytes.
The Gametic Central Cell Triggers PCD of Adjacent Antipodal Cells.
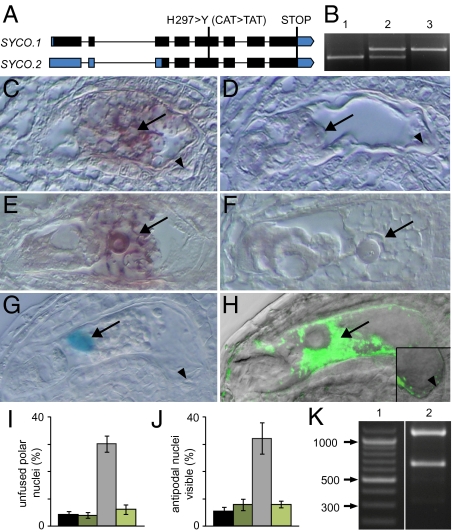
We mapped the fiona mutation to the At2g31170 locus, which codes for SYCO ARATH, one of three cysteinyl-tRNA synthetases encoded by the Arabidopsis genome (19). In the following we will refer to the gene as SYCO and to fiona as syco-1. In syco-1, a single base pair mutation causes an amino acid exchange from histidine to tyrosine on position 297 of a total of 563 amino acids (Fig. 3A). This histidine is one of five conserved histidine residues at the base of the catalytic center and is important for protein function in Escherichia coli (20). Expression of SYCO genomic DNA, driven by a 1.25-kb upstream promoter fragment, was able to rescue the mutant phenotype, suggesting that syco-1 is a loss-of-function mutation (Fig. 3B). RT-PCR detected SYCO expression throughout the plant (Fig. S3A). To determine the temporal and spatial expression pattern of SYCO in the female gametophyte, we generated promoter reporter constructs and determined SYCO mRNA distribution using in situ hybridization. In mature female gametophytes, a clear hybridization signal was found in the central cell (Fig. 3 C–F). In agreement with this, GUS expression in the female gametophyte of pSYCO::NLS_GUS plants was exclusively detected in the central cell (Fig. 3G). We did not detect any pSYCO::NLS_GUS expression before cellularization (Fig. S3 B and C; n = 250). These results suggest that the extended lifespan of syco-1 antipodal cells is an indirect consequence of the modified developmental program of the central cell. To test whether central cell expression of SYCO was sufficient to complement the mutant defects, we expressed a SYCO_GFP fusion protein under the control of the central cell specific MEDEA (MEA) promoter (Fig. 3H), which did not confer expression before cellularization in syco-1/SYCO pMEA::SYCO_GFP plants (n = 115).
Fig. 3.
The cysteinyl-tRNA synthetase SYCO is expressed and required in the central cell but not in antipodals. (A) Gene structure of SYCO. Exons are depicted in bars, introns in lines. UTRs are indicated in blue, coding exons in black. The change in DNA and amino acid sequence in syco-1 is indicated. (B) PCR-based genotyping of the SYCO locus reveals that the genomic SYCO DNA driven by a 1.25-kb upstream promoter allows the generation of homozygous mutants. 1, SYCO/SYCO; 2, syco-1/SYCO; 3, syco-1/syco-1, pSYCO::SYCO/-. (C–F), In situ hybridization of SYCO mRNA in the female gametophyte before (C and D) and after (E and F) antipodal degeneration. (C and E) Antisense probe. (D and F) Sense probe. Arrow and arrowhead point at central cell and antipodals, respectively. (G) NLS_GUS expression driven by a 1.25-kb promoter fragment upstream of SYCO is restricted to the central cell nucleus (arrow). (H) Localization of pMEA::SYCO_GFP in the central cell (arrow) of the female gametophyte 2 d after emasculation (overlay of GFP fluorescence with differential interference contrast image). Inset: Magnification of the proximal end of the central cell and of the antipodals (arrowhead). (I and J) pMEA::SYCO_GFP complements the polar nuclei fusion (I) and antipodal PCD (J) defects in syco-1/SYCO plants. Black bar, wild type, n = 256; dark green bar, pMEA::SYCO_GFP, n = 355; gray bar, syco-1/SYCO, n = 342; light green bar, syco-1/SYCO, pMEA::SYCO_GFP, n = 426. Gametophytes were analyzed 2 d after emasculation. Error bars, mean ± SEM. (K) Digestion of PCR-amplified SYCO_GFP with BspMI discriminates between SYCO.1 and SYCO.2 cDNA. SYCO.1 is digested to 1,162 bp and 666 bp, SYCO.2 to 1,162 bp, 338 bp, and 332 bp. 1, ladder; 2, pMEA::SYCO_GFP.
As expected, expression of pMEA::SYCO_GFP in a syco-1 mutant background reconstituted the original central cell program, resulting in polar nuclei fusion (Fig. 3I). Importantly, PCD of antipodals was also restored to wild-type levels (Fig. 3J), indicating that antipodal cell death is triggered by the central cell. The competence of the antipodals to adopt a gametic cell fate has led to the hypothesis that antipodals might serve as a backup for the central cells (6). Our results imply that central cell integrity is a critical parameter determining antipodal lifespan, which is in agreement with this theory.
Mitochondrial Dysfunction in the Central Cell Extends Antipodal Lifespan.
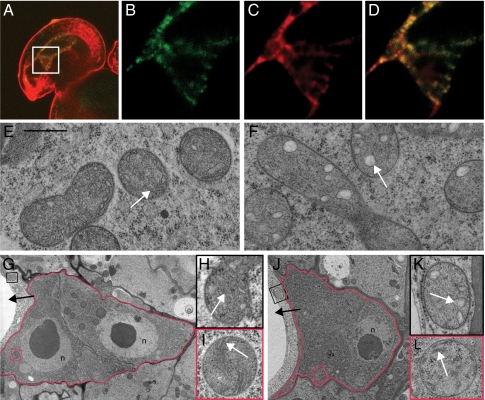
SYCO possesses two splice variants (Fig. 3A). In contrast to SYCO.2, SYCO.1 contains a unique N-terminal domain, which, in transient expression studies, was shown to target GFP to the mitochondria and chloroplasts (21). To determine which splice variant is generated in the female gametophyte, we amplified cDNA from plants expressing SYCO_GFP in the central cell, using primers that discriminated against endogenous SYCO cDNA. By this means, we mainly detected SYCO.1, indicating that the predominant variant generated in the central cell contains the dual targeting sequence (Fig. 3K). To determine the subcellular localization of SYCO in planta, we analyzed the GFP-tagged version of the protein in the central cell. pMEA:: SYCO_GFP accumulated in patches and colocalized with the mitochondrion-specific dye MitoTracker Orange (Fig. 4 A–D).
Fig. 4.
SYCO localizes to mitochondria and is necessary for cristae integrity in the central cell. (A–D) Subcellular localization of SYCO_GFP in the central cell. (A) Entire ovule (overlay). (B–D) Magnification of box in A. (B) pMEA::SYCO_GFP. (C) MitoTrackerOrange. (D) Overlay. (E and F) Mitochondria of wild-type (E) and syco-1 (F) central cells 2 d after emasculation. (G) Antipodal cells (red border) of a wild-type gametophyte 0 d after emasculation. (H and I) Magnification of boxes in G: central cell mitochondrium (H) and antipodal mitochondrium (I). (J) Antipodal cells (red border) of a syco-1 gametophyte 2 d after emasculation. (K and L) Magnification of boxes in J: central cell mitochondrium (K) and antipodal cell mitochondria (L). Black arrow, central cell; white arrow, cristae; n, nucleus. (Scale bar, 500 nm in E, F, H, I, K, and L.)
The subcellular SYCO distribution prompted us to investigate the mitochondria in the central cell and antipodal cells of syco-1 gametophytes. Ultrastructural analysis on sections of wild-type gametophytes detected mitochondria containing numerous clearly visible cristae (Fig. 4 E and G–I). Corresponding sections of syco-1 gametophytes revealed a regular morphology in the mitochondria of antipodal cells (Fig. 4 J and L). By contrast, the mitochondria of the central cell lacked regular cristae (Fig. 4 F and K and Fig. S4), indicating that SYCO is necessary for the integrity of central cell mitochondria.
To determine whether there is a causal relationship between the integrity of central cell mitochondria and the lifespan of antipodals, we aimed to specifically impair the mitochondrial function in the central cells of wild-type plants. The mitochondrial ATP/ADP translocator AAC is a key enzyme in the inner membrane of mitochondria, and the ADP/ATP catalyzing function of AAC2 is conserved between yeast and plants (22). In yeast, a dominant mutation is described which impairs the electron transport chain, resulting in membrane uncoupling (23). We introduced an analogous mutation into Arabidopsis AAC2 (Fig. S5) and expressed this dominant version from a central cell promoter. As expected, pMEA::aac2A199D_GFP hemizygous plants segregated 50% gametophytes which expressed the construct specifically in the central cell (Fig. 5A). Ultrastructural analysis revealed that the respective central cell mitochondria lack regular cristae (Fig. 5 B and C), confirming the conserved role of AAC2 for mitochondrial membrane integrity. We next analyzed the morphology of pMEA::aac2A199D_GFP plants by performing cleared whole mounts. In two independent transgenic lines we observed unfused polar nuclei (Fig. 5 D, F, and G), indicating that nuclei fusion in the central cell is indeed a mitochondria-dependent process. Importantly, this construct additionally repressed PCD of antipodals (Fig. 5 E–G). By contrast, expression of aac2A199D in antipodals, making use of the promoter pHSFa2 (Fig. 5H), did not significantly affect antipodal PCD, as evidenced by the analysis of independent transgenic lines (Fig. 5 I and J). Our data thus establish an essential role of central cell mitochondria for antipodal PCD.
Fig. 5.
aac2A199D causes mitochondrial cristae degradation and extends antipodal lifespan when expressed in the central cell of wild-type plants. (A) Localization of pMEA::aac2A199D_GFP in the central cell of the female gametophyte. (B and C) Central cell mitochondria in wild type (B) and pMEA::aac2A199D_GFP (C). White arrows indicate cristae. (D and E) Frequencies of unfused polar nuclei (D) and persistent antipodal cells (E) in wild-type (black bar) and two independent pMEA::aac2A199D_GFP transgenic lines (green bars); n = 361 for wild type, n = 452 for line 1 (pMEA::aac2A199D_GFP/-; light green), n = 212 for line 2 (pMEA::aac2A199D_GFP/ pMEA::aac2A199D_GFP; dark green). Gametophytes were analyzed 2 d after emasculation. (F and G) Whole-mount clearing of a wild-type (F) and pMEA::aac2A199D_GFP (G) gametophyte 2 d after emasculation. Arrows indicate the fused secondary nucleus and the unfused polar nuclei, respectively. Arrowheads indicate antipodals. (H) Localization of pHSFa2::aac2A199D_GFP in antipodals (arrowhead) of the female gametophyte (overlay of GFP fluorescence with differential interference contrast image). (I and J) Frequencies of unfused polar nuclei (I) and persistent antipodal cells (J) in wild-type (black bar) and two independent pHSFa2::aac2A199D_GFP/pHSFa2::aac2A199D_GFP transgenic lines (gray bars); n = 241 for wild type, n = 239 for line 1 (light gray), n = 173 for line 2 (dark gray). (K) Model for SYCO function. SYCO-dependent mitochondria integrity is necessary for nuclei fusion in the central cell and negatively regulates antipodal lifespan in a noncell autonomous manner. Error bars, mean ± SEM. (Scale bar, 500 nm in B and C.)
Conclusions
Mitochondria are the principal energy source of the cells. In plants, they have also been implicated in polar nuclei fusion, which is prohibited after mutations in the mitochondrial ribosomal subunit L21 (11) or a mitochondria-localized DnaJ protein (9). Here we have shown that cristae degeneration induced by expression of the dominant aac2A199D allele prevents polar nuclei fusion, confirming the proposed role of mitochondria during nuclear fusion (11) (Fig. 5K). Mitochondria have, in addition, long been recognized to be associated with PCD in both plants and animals (24, 25), and their dysfunction has been shown to correlate with cell and organismal aging in yeast, invertebrates, and mammals (26). Seemingly paradox, the reduction of mitochondria function can also extend lifespan as shown for Caenorhabditis elegans and Drosophila (27, 28). The mechanisms that underlie lifespan extension are elusive; however, one theory is that reactive oxygen species, which accumulate as a byproduct of a functional electron transport chain, enhance aging through their damaging effects on mitochondrial DNA (26). Our results suggest that the function of mitochondria in lifespan regulation is conserved in plants. However, the spatial discrepancy between the mitochondrial defect in the central cell and the PCD defect in antipodals is surprising and reveals that mitochondria dysfunction can affect cellular lifespan in a noncell autonomous manner (Fig. 5K). The data presented here extend the current view of mitochondria-associated lifespan regulation and draw the attention away from the dying cell and toward adjacent nondying cells as a potential cause for PCD.
syco-1 provides a fascinating example of how small-scale molecular changes can affect complex reproductive traits. The similarity between the female gametophytes of syco-1 and grasses, together with our finding that SYCO is required for mitochondrial cristae integrity, might suggest that differences in the metabolic profile between Arabidopsis and wheat central cells account for the distinct female gametophyte patterns of these phylogenetic distant plant species. This theory will remain a topic for future investigation.
Materials and Methods
Plant Material and Growth Conditions.
Plants were grown on soil in growth chambers under long-day conditions (16 h light/8 h dark) at 20 °C. The syco-1 allele was isolated from plants containing markers ET1119 and GT3733 (6) in the Landsberg erecta (Ler) accession after mutagenesis; seeds were mutagenized by incubation in 0.15% EMS for 10 h.
Molecular Cloning and Complementation.
Mapping of syco-1 was performed as described previously (29) using polymorphisms annotated by CEREON (30). The Columbia accession (Col-0) was used as the crossing partner. syco-1 was located to chromosome 2 in an area of 20 kb between polymorphisms CER442808 and CER442815 on BAC T16B12/F16D14. We sequenced all ORFs in this interval and identified a single heterozygous locus in At2g31170.
To generate the pSYCO::SYCO complementation construct, a 1,253-bp promoter fragment upstream of the start codon was PCR-amplified from genomic DNA using primers 5′-AGTGAGGCGCGCCTTTTGATTTTGATTTTGTGTTGTTTTTGTCGTTAAGAC-3′ and 5′-AGTGATGATCATGGTGTGGAACGATAAGCTCCTTC-3′. The annotated SYCO ORF was amplified from Ler genomic DNA using primers 5′-AGTGATTAATTAAACTAGTATGGCTTCTTCTGTCCTTAATCTCTTC-3′ and 5′-GAAAAATTATTACGTATCTCATACC-3′. For the translational fusion construct, genomic SYCO was amplified using 5′-ACTGACAATTGAGGTGTAGTAGTTACAGGTTCTTG-3′ as antisense primer, released PacI/MfeI and cloned in front of an eGFP lacking the start codon. AAC2 was amplified from Ler cDNA using the primers 5′-GTTAATTAATGGTTGAACAGACTCAGCACC-3′ and 5′-ACAATTGGGCACCTCCAGATCCATACTTC-3′. The point mutation A199D was inserted by site-directed mutagenesis (31) using the primer 5′- P-AGGCACCAtCAGCACCTC-3′. The mutated aac2A199D was cloned PacI/MfeI in front of an eGFP lacking the start codon. To generate the pSYCO::NLS_GUS construct, the same promoter fragment as used for complementation was cloned into binary vector pGIIBar-pLIS::NLS_GUS in place of the LIS promoter (6). For amplification of the previously published MEA promoter (6) primers 5′-AGTGAGGCGCGCCAATAGGTCGAGAAAATGCTGTGAATCG-3′ and 5′-AGTGACGATCGTAACCACTCGCCTCTTCTTTTTTTCTC-3′ were used. For amplification of pAT5g40260 (16), primers 5′-AGTGAGGCGCGCCTTTCTGTACTTTTGAAAATA-3′ and 5′-AGTGATTAATTAATAAAATCGCCGTTTACAAA-3′ were used.
PCR-based Genotyping.
Plants were genotyped for the syco-1 allele using primers 5′-CCAATTCAGATTCTTGATAATGGC-3′ and 5′-AAGTCTCAGAGCAAGAGGATG-3′. The resulting 661-bp fragment was digested with HphI, yielding two fragments of 133 bp and 528 bp from the product of the syco-1 allele and three fragments of 133 bp, 260 bp, and 268 bp from the product of the wild-type allele. To discriminate against the transgene in plants containing pSYCO::SYCO or pMEA::SYCO_GFP, primers 5′-CATTGAATACTTGTTTACTCATTTAAGTC-3′ and 5′-ATCACTAACAAAGTATAACCATTAAGC-3′ were used. The resulting 2,248-bp fragment was digested with HphI, yielding 192-bp and 2,056-bp fragments in wild type.
Reverse Transcription–Polymerase Chain Reaction.
RNA isolation, reverse transcription, and RT-PCR were performed as previously described (6). SYCO cDNA was amplified using primers 5′-AATGGTTTTGTTACTGTTGACTCC-3′ and 5′-AAGTCTCAGAGCAAGAGGATG-3′. To discriminate between the two splice variants in female gametophytes, cDNA from gynoecia (2 d after emasculation) of pMEA::SYCO_GFP plants was amplified using primers 5′-CTTCTTCTGTCCTTAATCTCTTC-3′ and 5′-AACTTCAGGGTCAGCTTGC-3′ and digested with BspMI.
Flow Cytometry.
For flow cytometry analysis, seeds containing heart-stage embryos were crushed in a 2-mL test tube with nuclear extraction buffer (CyStain UV-precise kit; Partec) using a pistil. For analysis of autonomous endosperm formation in fie, seeds from emasculated flowers were harvested. The cells were filtered through a 50-μm CellTrics Filter (Partec) and stained with nuclear staining buffer (CyStain UV-precise kit). Flow cytometry was performed on a Partec Ploidy Analyzer (32). Extraction from young leaves was taken as a reference value.
Histology and Microscopy.
For analysis of mature gametophytes, the oldest closed flower bud of a given inflorescence was emasculated and harvested 2 or 4 d later. For counting endosperm nuclei in syco-1/SYCO and wild type, whole-mount clearings of 3-d-old crosses with wild-type pollen were analyzed. Cytochemical staining for GUS activity and whole-mount clearings were performed as described previously (6). The GFP constructs and MitoTracker Orange (Molecular Probes) were observed using a Leica TCS-SP2 confocal scanning microscope. Confocal images were obtained with a 63× water-immersion objective lens and via Leica Confocal software. For analysis of the nuclear fusion event, emasculated flowers were pollinated with HTR10-mRFP1 (17) and analyzed under a Leica DMI6000B 10–14 h after pollination.
In situ hybridization on emasculated flowers was performed as described previously (33). The antisense riboprobe was amplified using primers 5′-CTTCTTCTGTCCTTAATCTCTTC-3′ and 5′-TAAGCTTTAATACGACTCACTATAGGGAGACTCAAGAGTACATCGAAAGTGAC-3′, the sense probe was amplified using primers 5′-TAAGCTTTAATACGACTCACTATAGGGAGACTTCTTCTGTCCTTAATCTCTTC-3′ and 5′-CTCAAGAGTACATCGAAAGTGAC-3′.
Ultrastructural analysis of high-pressure frozen, freeze-substituted, and resin-embedded ovules was carried out as previously described (34).
Statistical Analysis.
Mutant transmission efficiency was analyzed using a χ2 test.
Supplementary Material
Acknowledgments
We thank F. Berger (Temasek LifeSciences Laboratory, National University of Singapore, Singapore), G. N. Drews (Department of Biology, University of Utah, Salt Lake City), R. Fischer (Plant and Microbial Biology Department, University of California, Berkeley, CA), U. Grossniklaus (Institute of Plant Biology and Zürich Basel Plant Science Center, University of Zürich, Zürich, Switzerland), and V. Sundaresan (Department of Plant Biology, University of California, Davis, CA) for seeds; D. Ripper for technical support; M. Nowack and A. Schnittger for help with ploidy analysis; and F. de Courcy, W. Friedman, P. Kahle, T. Laux, D. Weijers, and members of the R.G.-H. laboratory for helpful discussions and comments on the manuscript. This work was supported by the University of Tübingen, Deutsche Forschungsgemeinschaft Grant SFB446 (to R.G.-H.), and a stipend from the Landesgraduiertenförderung Baden-Württemberg (to C.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012795108/-/DCSupplemental.
References
- 1.Dresselhaus T, Márton ML. Micropylar pollen tube guidance and burst: Adapted from defense mechanisms? Curr Opin Plant Biol. 2009;12:773–780. doi: 10.1016/j.pbi.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Williams JH, Friedman WE. The four-celled female gametophyte of Illicium (Illiciaceae; Austrobaileyales): Implications for understanding the origin and early evolution of monocots, eumagnoliids, and eudicots. Am J Bot. 2004;91:332–351. doi: 10.3732/ajb.91.3.332. [DOI] [PubMed] [Google Scholar]
- 3.Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- 4.Holloway SJ, Friedman WE. Embryological features of Tofieldia glutinosa and their bearing on the early diversification of monocotyledonous plants. Ann Bot (Lond) 2008;102:167–182. doi: 10.1093/aob/mcn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghavan V. Molecular Embryology of Flowering Plants. Cambridge, UK: Cambridge Univ Press; 1997. [Google Scholar]
- 6.Gross-Hardt R, et al. LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 2007;5:e47. doi: 10.1371/journal.pbio.0050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moll C, et al. CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 2008;56:913–921. doi: 10.1111/j.1365-313X.2008.03650.x. [DOI] [PubMed] [Google Scholar]
- 8.Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science. 2009;324:1684–1689. doi: 10.1126/science.1167324. [DOI] [PubMed] [Google Scholar]
- 9.Christensen CA, et al. Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell. 2002;14:2215–2232. doi: 10.1105/tpc.002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagnussat GC, et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- 11.Portereiko MF, et al. NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006;141:957–965. doi: 10.1104/pp.106.079319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruyama D, Endo T, Nishikawa S. BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:1684–1689. doi: 10.1073/pnas.0905795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N, et al. SLOW WALKER2, a NOC1/MAK21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in Arabidopsis. Plant Physiol. 2009;151:1486–1497. doi: 10.1104/pp.109.142414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi DQ, et al. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell. 2005;17:2340–2354. doi: 10.1105/tpc.105.033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohad N, et al. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu HJ, Hogan P, Sundaresan V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 2005;139:1853–1869. doi: 10.1104/pp.105.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol. 2007;17:1032–1037. doi: 10.1016/j.cub.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Hoshikawa K. Cytological studies of double fertilization in wheat (Triticum aestivum L.) Proc Crop Sci Soc Jpn. 1959;28:142–146. [Google Scholar]
- 19.Duchêne AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang CM, Perona JJ, Hou YM. Amino acid discrimination by a highly differentiated metal center of an aminoacyl-tRNA synthetase. Biochemistry. 2003;42:10931–10937. doi: 10.1021/bi034812u. [DOI] [PubMed] [Google Scholar]
- 21.Peeters NM, et al. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- 22.Haferkamp I, Hackstein JH, Voncken FG, Schmit G, Tjaden J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur J Biochem. 2002;269:3172–3181. doi: 10.1046/j.1432-1033.2002.02991.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Salinas K, Zuo X, Kucejova B, Chen XJ. Dominant membrane uncoupling by mutant adenine nucleotide translocase in mitochondrial diseases. Hum Mol Genet. 2008;17:4036–4044. doi: 10.1093/hmg/ddn306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 25.Reape TJ, McCabe PF. Apoptotic-like programmed cell death in plants. New Phytol. 2008;180:13–26. doi: 10.1111/j.1469-8137.2008.02549.x. [DOI] [PubMed] [Google Scholar]
- 26.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copeland JM, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 29.Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jander G, et al. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowack MK, et al. Bypassing genomic imprinting allows seed development. Nature. 2007;447:312–315. doi: 10.1038/nature05770. [DOI] [PubMed] [Google Scholar]
- 33.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 34.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffen JG, Kang IH, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007;51:281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.