Abstract
Etiologic risk factors for hepatocellular carcinoma can be involved in the transformation process by directly targeting intracellular signaling pathways or by indirectly stimulating chronic cycles of hepatocyte destruction and regeneration. However, the contribution of these two routes to hepatocarcinogenesis has not been determined, partly because of the difficulty in distinguishing damaged and regenerated hepatocytes. Here we report that induced deletion of the damaged DNA binding protein 1 (DDB1) abrogates the self-renewing capacity of hepatocytes, resulting in compensatory proliferation of DDB1-expressing hepatocytes. Constitutive stimulation of this regeneration process leads to development of hepatocellular carcinoma, which surprisingly contains no disruption of the DDB1 gene, indicating a cell-nonautonomous role of DDB1 inactivation in tumor initiation. Our results suggest that viruses and hepatoxins may cause liver tumors by simply driving hepatocyte turnover without directly targeting cancer cells.
Keywords: ubiquitin ligase, mouse model, gene knockout
Hepatocellular carcinoma (HCC), representing 80% to 90% of all primary liver cancers, is the fifth most common cancer and the third leading cause of cancer death worldwide (1). HCC development follows a multistep progression from hepatic dysplasia to neoplasia, with accumulation of somatic mutations and chromosomal alterations (2). Conventionally, these genetic alterations are identified in human liver tumor samples by gene expression profiling, genomic and proteomic analysis, and direct sequencing. Candidate genes are subsequently validated in cell cultures and animals. This methodology has led to the identification of oncogenes and tumor suppressor genes important for liver tumor development, such as those regulating Wnt/β-catenin, p53, EGFR receptor tyrosine kinases, and MET/HGF pathways (3).
Alternatively, interactions between host factors and HCC-associated pathogens such as hepatitis B virus (HBV) and hepatitis C virus (HCV) can be investigated to understand early molecular and cellular alterations leading to HCC initiation. HBV encodes a nonstructural regulatory protein, HBx, that is required for HBV replication and implicated in HBV-associated HCC (4). How HBx contributes to hepatocarcinogenesis is unclear, but is believed to involve its myriad target host proteins, one of which is the damaged DNA binding protein 1 (DDB1) (5). DDB1 is the linker protein for the Cullin 4 (Cul4) E3 ubiquitin ligase (6), which regulates ubiquitination and proteasomal degradation of proteins essential for nucleotide excision repair [DDB2 (7) and CSB (8)], cell cycle progression [p21 (9, 10), p27 (11), and Myc (12)], DNA replication [Cdt1 (13) and POLH-1 (14)], and cell growth [c-Jun (15) and TSC2 (16)]. Other substrates of the E3 ligase, including XPC (7), histones H3 and H4 (17), and histone H2A (18), are modified without degradation to facilitate DNA damage repair. Several viral regulatory proteins such as the HIV Vpr protein (19) and the simian virus 5 V protein (20) can hijack the E3 ligase to target undesired host factors and benefit the respective viral life cycle.
In this study, we addressed the role of DDB1 in mouse liver development and pathogenesis. By using mice with inducible or constitutive deletion of DDB1 in hepatocytes, we show that DDB1 loss prevents hepatocytes to replicate DNA, induces compensatory proliferation of DDB1-expressing hepatocytes, and eventually leads to development of HCC that surprisingly contains the intact DDB1 gene. Our genetic experiments suggest that cancer-initiating events for HCC may not necessarily directly target cancer-initiating cells, but can stimulate cell transformation by altering tissue homeostasis. Our results have significant implication for designing therapies against HCC in human patients.
Results
DDB1 Is Required for Hepatocyte Proliferation.
We previously reported that DDB1 is essential for survival of proliferating cells in mouse brain and skin, but dispensable for nondividing cells such as neurons (21, 22). Although highly differentiated, hepatocytes can reenter cell cycle and self-renew in response to loss of liver mass. To analyze the role of DDB1 in hepatocyte proliferation, we generated DDB1F/F;Mx1-Cre+/− mice, which harbor homozygous floxed DDB1 alleles (DDB1F) (21) and an Mx1-Cre transgene (23). We confirmed a very efficient deletion of DDB1 in DDB1F/F;Mx1-Cre+/− mouse liver after one i.p. injection of polyinosinic:polycytidylic acid [poly(I:C)], which induces an IFN response to activate the expression of Cre recombinase. The floxed DDB1 alleles were almost completely deleted 1 d after injection, shown by PCR analysis of liver genomic DNA (Fig. 1A), and the DDB1 protein was undetectable 5 d after injection, as shown by Western analysis of total liver lysates (Fig. 1B). Mx1-Cre is also activated by poly(I:C) in other cell types such as hematopoietic cells, but we detected only a small reduction of DDB1 levels in spleen lysates, suggesting that our induction protocol mainly targets the liver.
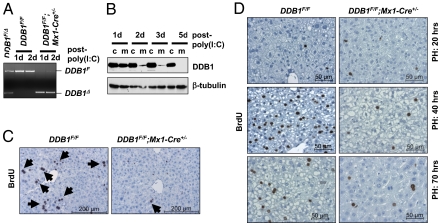
Fig. 1.
Induced deletion of DDB1 deprived hepatocytes of replicative capacity during liver development and regeneration. (A) PCR analysis of floxed (DDB1F) and deleted DDB1 (DDB1Δ) alleles in liver genomic DNA prepared from adult mice with indicated genotypes 1 or 2 d after poly(I:C) injection. Tail DNA from heterozygous DDB1F/Δ mice was used as a quantitative control for the two alleles. (B) Western analysis of DDB1 protein levels in whole liver lysates from control (c, DDB1F/F) or mutant (m, DDB1F/F;Mx1-Cre+/−) mice 1, 2, 3, or 5 d after poly(I:C) injection. (C) IHC staining for incorporated BrdU on liver sections from 1-mo-old mice 3 d after injection. Arrows indicate positive hepatocytes. (D) BrdU IHC staining on liver sections from 2-mo-old mice 20, 40, or 70 h after PH surgery. Surgery was performed on mice 3 d after injection with poly(I:C).
We performed two experiments to determine the effect of DDB1 loss on hepatocyte proliferation. First, we injected poly(I:C) to young mice (1 mo old) in which hepatocytes are actively proliferating. Loss of DDB1 in these mice resulted in an abolishment of proliferating hepatocytes, shown by their lack of BrdU incorporation (Fig. 1C and Fig. S1A). Second, we deleted DDB1 in adult mice (≥2 mo old) in which hepatocytes are mostly quiescent, and then we performed partial hepatectomy (PH) surgeries to stimulate hepatocytes to reenter the cell cycle. In contrast to massive DNA replication (Fig. 1D) and mitotic figures (Fig. S1B) found in control mouse liver 40 h after PH, DDB1-deficient liver exhibited few cycling hepatocytes (Fig. S1 C and D). To definitively prove that DDB1-deficient hepatocytes failed to replicate DNA, we isolated hepatocytes from poly(I:C)-treated mice at various times after PH, labeled hepatocyte DNA with propidium iodide (PI) in a hypotonic buffer, and analyzed DNA contents by flow cytometry. Consistent with PH-induced hepatocyte reentry into cell cycle, hepatocytes from control mice exhibited a reduction of diploid cells and concurrent increase of polyploid cells following DNA replication (Fig. S1E). By contrast, DNA contents in hepatocytes from DDB1-deficient liver exhibited little fluctuation within 100 h after PH (Fig. S1E). Based on these results, we conclude that DDB1 is required for hepatocyte proliferation in vivo.
P21 Accumulates in DDB1-Deficient Liver After PH.
Failure of hepatocyte proliferation usually results in defective liver regeneration and animal death (24). We found that six of eight mice with DDB1-deficient liver died within 24 d after PH (Fig. S2A). Liver from these mice contained variable degrees of DDB1-expressing ductile cells amid inflammatory infiltrates around portal areas, but very few DDB1-expressing hepatocytes (Fig. S2B). To understand the underlying molecular changes, we compared the protein levels of cell cycle regulators that are known to be targeted by the DDB1-Cul4A ubiquitin ligase (6) in DDB1-deficient and control livers after PH. Whereas levels of Cdt1 and p27 showed no change and c-Jun was constitutively up-regulated following PH, p21 accumulated dramatically 100 h after PH, a time when regenerating control liver expressed no p21 (Fig. S2C). Abundant p21 protein was also found in all dead animals with failed regeneration of DDB1-deficient liver, consistent with an inhibitory role of p21 in hepatocyte cell cycle progression and liver regeneration (25). These results suggest that DDB1-deficient hepatocytes fail to proliferate, possibly as a result of accumulation of p21 during liver regeneration.
DDB1-Expressing Hepatocytes Replace DDB1-Deficient Hepatocytes During Regeneration.
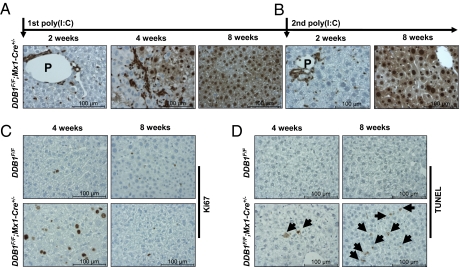
Interestingly, the two mice with DDB1-deficient liver that survived after PH had their livers completely restored with DDB1-expressing hepatocytes (Fig. S1F). In fact, all DDB1F/F;Mx1-Cre+/− mice showed regenerated livers with DDB1-expressing hepatocytes 8 wk after poly(I:C) injection, following a defined time course of gradual replacement of DDB1-depleted hepatocytes by DDB1-expressing hepatocytes (Fig. 2A). This regeneration progress is independent of mouse sex (male or female), mouse age at the time of injection (1 mo, 2 mo, or 5 mo tested), or injection frequency (one injection or two consecutive injections). In livers with mixed population of DDB1-positive and DDB1-negative hepatocytes, hepatocyte proliferation was very prominent, indicated by their positive staining for proliferation marker Ki67 (Fig. 2C). Only DDB1-expressing hepatocytes were in the cell cycle (Fig. S1G), consistent with a proliferation defect of DDB1-deficient hepatocytes. TUNEL-positive apoptotic cells were detected in regenerating livers but became more abundant in livers repopulated with DDB1-expressing hepatocytes (Fig. 2D), suggesting that DDB1-depleted hepatocytes, unlike DDB1-depleted neuronal and epidermal progenitor cells (21, 22), do not undergo immediate apoptotic death but can survive for a very long time, presumably to maintain liver structural integrity and metabolic functions. No tissue necrosis (on histologic examination) or cell senescence (based on senescent β-galactosidase staining) were found during the regenerative process. Based on these results, we conclude that the growth defect of DDB1-depleted hepatocytes induces compensatory proliferation of DDB1-expressing hepatocytes to regenerate a new liver.
Fig. 2.
Complete liver regeneration with DDB1-expressing hepatocytes after induced DDB1 deletion. DDB1 staining on liver sections from (A) adult DDB1F/F;Mx1-Cre+/− mice 2, 4, or 8 wk after poly(I:C) injection or (B) mice 2 or 8 wk after the second poly(I:C) injection. (C) Ki67 IHC staining and (D) TUNEL analysis on liver sections from mice 4 or 8 wk after poly(I:C) injection. Arrows indicate TUNEL-positive cells.
Strikingly but not surprisingly, the newly regenerated DDB1-expressing hepatocytes can be depleted of DDB1 again by a second injection of poly(I:C) into the DDB1F/F;Mx1-Cre+/− mice with freshly regenerated liver, and these mice once more recovered completely with DDB1-expressing hepatocytes 8 wk later (Fig. 2B). This observation not only exemplifies the seemingly unlimited regenerative capacity of liver, but also stimulated consideration of a continuous deletion of DDB1 in regenerating hepatocytes by Alb-Cre.
Continuous Deletion of DDB1 Causes Chronic Hepatocyte Turnover and Mild Liver Damage.
By using DDB1F/F;Mx1-Cre+/− mice, we have established that hepatocytes without DDB1 expression failed to proliferate, resulting in their displacement by DDB1-expressing hepatocytes. As this regeneration process can be repeatedly induced, we generated DDB1F/F;Alb-Cre+/− mice to induce continuous hepatocyte-specific deletion of DDB1 and, as a consequence, continuous liver regeneration. These young mice (1–3 mo) contained mixed amounts of deleted and nondeleted DDB1 alleles in their hepatocytes, determined by PCR and Southern blot analysis of hepatocyte genomic DNA, and reduced DDB1 protein levels in hepatocyte lysates compared with those from control mice (Fig. 3A). These changes reflected the mixed population of DDB1-expressing and DDB1-deficient hepatocytes in DDB1F/F;Alb-Cre+/− mouse liver (Fig. 3B), in which hepatocyte proliferation and death were very active (Fig. S3A; quantified in Fig. 3C) and only DDB1-expressing hepatocytes were dividing (Fig. S3B). Closer examination of the regenerative DDB1-positive nodules revealed many hepatocytes weakly stained for DDB1 in nuclei only (Fig. 3B Right), which turned out to be hepatocytes that were freshly deleted of the DDB1 gene (Fig. S1H). These results indicate that constitutive expression of Cre in hepatocytes leads to continuous replenishment of DDB1-deficient hepatocytes with DDB1-expressing hepatocytes.
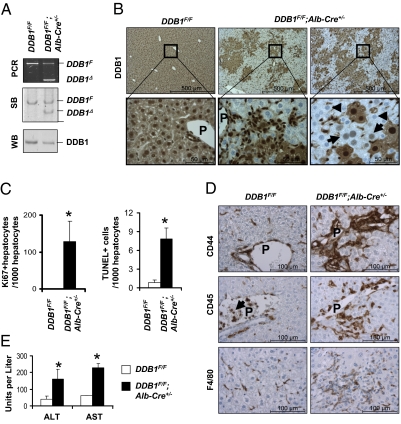
Fig. 3.
DDB1F/F;Alb-Cre+/− mice exhibited continuous hepatocyte turnover and chronic liver damage. (A) PCR and Southern blot (SB) analysis for DDB1F and DDB1Δ alleles in genomic DNA, and Western blot (WB) analysis of DDB1 protein levels from hepatocytes isolated from adult mice. (B) DDB1 IHC staining on liver sections from 6-wk-old mice. Arrows indicate hepatocytes freshly inactivated of the DDB1 gene, and arrowheads indicate hepatocytes completely depleted of DDB1 protein. (C) Quantification of Ki67-positive hepatocytes and TUNEL-positive cells on liver sections from three 6-wk-old mice. Values shown as mean ± SEM (*P < 0.01). (D) IHC staining for CD44, CD45, and F4/80. Abundant positive cells are found around portal areas. Arrows indicate CD45-positive blood cells in portal vein. (E) Measurement of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels. Values shown as means ± SEM; n = 3 (*P < 0.01).
The regenerating liver from DDB1F/F;Alb-Cre+/− mice exhibited increased infiltration with liver parenchyma immune cells, including activated CD44+ T cells, CD45+ leukocytes, and F4/80+ macrophages (Fig. 3D). Areas of liver steatosis and fibrosis were frequently found (Fig. S3C). Polyploid hepatocytes with 8N and 16N chromosomes were dramatically increased (Fig. S3D). Serum levels of alanine aminotransferase and aspartate aminotransferase were elevated compared with controls (Fig. 3E), indicating that the regenerating liver is undergoing chronic damage.
In DDB1F/F;Alb-Cre+/− mice 4 mo or older, Cre-induced DDB1 deletion was significantly reduced (Fig. S4A). Resistance of many hepatocytes to Cre excision is likely caused by an age-dependent decline of Cre expression in these mice (Fig. S4B). Reexpression of Cre by intrasplenic delivery of Ade-Cre expression constructs efficiently induced DDB1 deletion in those resistant hepatocytes (Fig. S4C). Nevertheless, hepatocyte turnover in these adult mutant mice was still prominent (Fig. S4D) compared with age-matched DDB1F/F controls, which showed barely any proliferation and death. Together these results indicate that constitutive expression of Cre in hepatocytes leads to continuous replenishment of DDB1-deficient hepatocytes with DDB1-expressing hepatocytes.
Aged DDB1F/F;Alb-Cre+/− Mice Develop Liver Tumors.
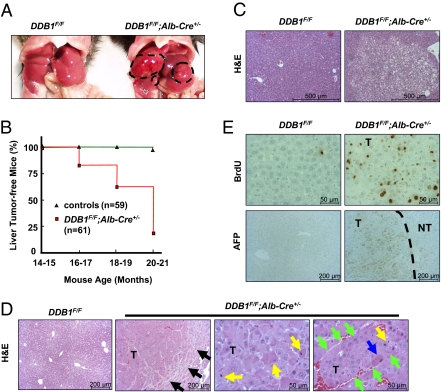
DDB1F/F;Alb-Cre+/− mice developed normally and exhibited no signs of obvious sickness until they developed liver tumors (Fig. 4A), starting at approximately 17 mo and affecting more than 80% of the animals 20 mo or older (Fig. 4B and Table S1). Histological examination of livers from 12- to 17-mo-old DDB1F/F;Alb-Cre+/− mice showed frequent premalignant nodules featuring clear-cell dysplasia (Fig. 4C). Approximately the same ratio of male and female mutant mice had tumors. These tumors were characteristically HCC based on histological features such as trabecular growth with multiple hepatocyte layers in one plate, increased cellularity, lack of portal tracts, and increased mitotic figures and apoptotic cells (Fig. 4D). Cancer cells replicated substantially and expressed biomarkers such as α-fetoprotein and cytokeratin 8 (Fig. 4E and Fig. S5A). Some tumors, but not all, were surrounded by fibrotic tissues (Fig. S5B). Examination of livers from DDB1F/F;Alb-Cre+/− mice before tumorigenesis did not reveal significant fibrosis, suggesting that HCC development is independent of fibrosis in this animal model. As in some human HCCs, E-cadherin was down-regulated and β-catenin up-regulated in these tumors, compared with their neighboring nontumor tissues (Fig. S5C). Levels of several other proteins implicated in HCC development, including c-Myc, p38, JNK, IκBα, and IκBβ, did not show significant changes in the tumor lysates compared with lysates from nontumor tissue and WT livers (Fig. S5D).
Fig. 4.
Aged DDB1F/F;Alb-Cre+/− mice developed HCC. (A) Representative photograph of liver tumors (outlined with dashed white lines) in a 19-mo-old DDB1F/F;Alb-Cre+/− mouse. (B) Age-dependent increase of liver tumor incidence in DDB1F/F;Alb-Cre+/− mice. Each age group has at least 10 animals of each genotype. n, total number of mice. (C) H&E staining of premalignant nodules shows clear cell dysplasia in a 15-mo-old DDB1F/F;Alb-Cre+/− mice. (D) Histological analysis of the liver tumor (T). Black arrows indicate the tumor edge, yellow arrows mitotic figures, blue arrows apoptotic cells, and green arrows rims of a trabecular growth of multiple layers of malignant cancer cells. (E) IHC analysis of the tumor. AFP, α-fetoprotein. Dashed line separates nontumor (NT) from tumor part.
Liver Cancer Cells Contain No Deletion of DDB1.
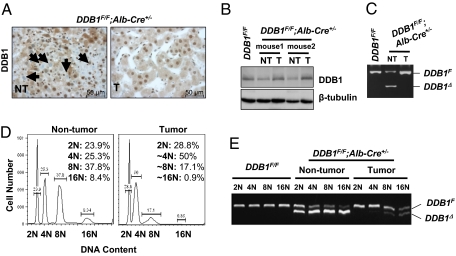
In cultured cells (26) and developing embryos (22), loss of DDB1 causes genomic instability, which is associated with cancer disposition. Surprisingly, cancer cells in the liver tumors from DDB1F/F;Alb-Cre+/− mice apparently contained the parental floxed DDB1 allele rather than the deleted allele, judged from four experimental observations. First, nearly all cells within HCCs were positive for DDB1 immunostaining, in contrast to a mixed population of DDB1-positive and DDB1-negative hepatocytes in the neighboring nontumor tissues (Fig. 5A). Second, Western analysis demonstrated that levels of DDB1 protein in the tumors were higher than those in the neighboring nontumor tissues, and similar to those in WT livers (Fig. 5B). Third, PCR genotyping for the floxed and deleted DDB1 alleles in isolated hepatocytes demonstrated a predominant population of cancer cells with no deletion of the floxed DDB1 allele (Fig. 5C). Finally, unlike highly polyploid hepatocytes in nontumor tissues, cancer cells isolated from tumors were mostly aneuploid (Fig. 5D). To exclude the contamination of nonparenchymal cells (containing 2N chromosomes and no DDB1 deletion) in total hepatocytes, aneuploid cancer cells with more than 2N chromosomes were isolated by FACS and genotyped by PCR. Whereas hepatocytes from nontumor tissues always contained the deleted DDB1 allele, cancer cells contained almost exclusively the floxed allele (Fig. 5E). Given the clonal nature of tumor growth, we conclude that HCCs in DDB1F/F;Alb-Cre+/− mice originate from DDB1-expressing cells undergoing continuous regeneration but apparently develop ways to circumvent Cre-driven deletion of DDB1.
Fig. 5.
Liver cancer cells from DDB1F/F;Alb-Cre+/− mice had no deletion of the DDB1 gene. (A) IHC staining for DDB1 on a tumor and its neighboring nontumor liver tissues. Arrows indicate DDB1-negative hepatocytes in nontumor tissue. (B) Western analysis of the levels of DDB1 protein in nontumor and tumor lysates. (C) PCR genotyping of DDB1F and DDB1Δ alleles in genomic DNA from hepatocytes isolated from nontumor and tumor tissues. (D) Flow cytometry analysis of DNA contents in nontumor and tumor hepatocytes. Percentage of cells with different chromosomes is indicated. (E) PCR genotyping of DDB1 alleles in hepatocytes and cancer cells sorted by FACS according to their DNA contents.
Discussion
DDB1F/F;Alb-Cre+/− mice recapitulate many key features of human HCC pathogenesis, including chronic inflammation and hepatocyte turnover (3). The unique genetics behind this mouse model is that parenchymal hepatocytes are deprived of their regenerative capacity by Cre-driven DDB1 deletion, leading to compensatory proliferation of DDB1-expressing hepatocytes. We propose that substantial proliferation of these cells might expose them to damages from the inflammatory environment and allow for accumulation of mutations that eventually lead to their transformation. This mechanism might be shared by other genetic models such as albumin-plasminogen activator transgenic mice (27).
DDB1 is ubiquitously expressed in noncancerous hepatocytes and HCC cells from patients (Fig. S6), suggesting that deregulation of DDB1 expression may not be a principal mechanism underlying human hepatocarcinogenesis. Instead, the rationale behind our genetic studies of DDB1 inactivation in liver is that DDB1 is a well-established target protein of HBx, a viral protein implicated in HBV-associated liver tumor pathogenesis (4). The direct binding of HBx to DDB1 is required for nearly all biological functions of HBx, including arrest of cell growth, stimulation of HBV replication, and transcription activation (28). The role of DDB1 in the HBx tumor model is being investigated.
Our HCC model offers insights into HCC pathogenesis. Unlike many other solid tumors, liver tumors have no clear genetic predisposition, but usually arise from chronic attacks by viruses such as HBV and HCV or chemical carcinogens (3). Hepatoxins are metabolized in hepatocytes, leading to toxicity and death of targeted hepatocytes rather than their transformation. HBV and HCV infection clearly contributes to liver cancer development by inducing immune-mediated inflammation, and whether these viruses directly cause cell transformation is still an area of intense studies (29). Our work demonstrates that cancer-initiating mutations or damages do not necessarily reside in cancer-initiating cells. Genetic alterations, such as DDB1 inactivation, can initiate HCC indirectly through cell–cell communication. Our work argues that viruses, toxins, and somatic mutations may cause HCC mainly by inducing microenvironmental changes, such as a local immune response (30), rather than directly transforming their target cells. Therefore, caution has to be exercised when deciphering the exact roles of targeted genes in HCC development in KO mice. Furthermore, identifying cancer-driving mutations from tumor samples, as illustrated in many large-scale sequencing projects (31), may have missed some early cancer-initiating genetic events, particularly for HCC.
Materials and Methods
Mice.
Mutant mice used in this study were in a mixed BL6/129 genetic background and generated by breeding the conditional DDB1 mice (21) with Alb-Cre transgenic (32) and Mx1-Cre transgenic (23) mice, both acquired from Jackson Laboratory. All animals were maintained in pathogen-free facilities and all experiments followed regulations of the authors’ institutional animal care and use committee and the Burnham Institute. Deletion of DDB1 in DDB1F/F;Mx1-Cre+/− mice was achieved by one i.p. injection of poly(I:C) (GE Healthcare) at 13 mg/kg body weight.
Immunostaining and Imaging.
Mouse liver tissues or dissected tumors were collected and processed for histological, immunohistochemical (IHC), immunofluorescence (IF), Southern blot, Western blot, or PCR genotyping analysis, essentially as described previously (21). Five-micrometer paraffin tissue sections were used for all analysis except a few performed on frozen sections as indicated. The following antibodies were used: rabbit polyclonal antibodies against human DDB1 (Bethyl Laboratories) and human AFP (Santa Cruz Biotechnology) and monoclonal antibodies against mouse F4/80 and CD45 (eBioscience) and mouse CD44 (Endogen). Additional antibodies used in Western blot analysis are rabbit polyclonal antibodies against human p21 (Santa Cruz Biotechnology) and Cdt1 (gift of N. Hideo, University of Hyogo, Kobe, Japan). For BrdU incorporation assays, mice were injected intraperitoneally with 100 mg/kg BrdU and killed 1 h later. For steatosis assays, frozen liver sections were incubated in 60% isopropanol for 2 min and then in oil red solution for 20 min. All IHC pictures were taken with a Zeiss Axio Imager LED microscope, and IF pictures were taken with a Nikon inverted TE300 fluorescent microscope.
Hepatocyte Isolation.
Hepatocytes were isolated following a standard three-step protocol involving collagenase perfusion of the whole liver and low-speed centrifugation of detached cells. Isolated hepatocytes were then analyzed without being cultured.
Liver Function Assays.
Blood samples (150 μL) collected from mice via retro-orbital bleeding were analyzed by using a mammalian liver enzyme profile rotor on a VetScan VS2 analyzer. At least three animals of matching ages from each genotype were assayed.
PH Surgery.
Two-thirds PH surgery was performed as described (33). Briefly, adult mice (2–3 mo old) were injected with poly(I:C) to induce DDB1 deletion in the liver. After 3 d, mice were anesthetized by isoflurane inhalation, and resection of left lateral and median lobes or sham operation was performed.
Supplementary Material
Acknowledgments
We thank R. Oshima, Z. Ronai, S. Courtneidge, and G. S. Feng for reading the manuscript and providing helpful suggestions. We thank R. Cardiff for help with interpreting liver pathology; D. Brenner, P. Zhou, and M. Karin for discussions; H. Nishitani (University of Hyogo, Hyogo, Japan) for Cdt1 antibody; the histology core facilities at Sanford-Burnham Medical Research Institute and Columbia University for support; and L. Slater, K. de Los Santos, Y. Altman, and E. Monosov for technical assistance. S.P.G is an investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health grants (to S.P.G. and Y.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015793108/-/DCSupplemental.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 3.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson S, Xiong Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 8.Groisman R, et al. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbas T, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda-Carboni GA, et al. A functional link between Wnt signaling and SKP2-independent p27 turnover in mammary tumors. Genes Dev. 2008;22:3121–3134. doi: 10.1101/gad.1692808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertz IE, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 16.Hu J, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Kapetanaki MG, et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröfelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc Natl Acad Sci USA. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 21.Cang Y, et al. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell. 2006;127:929–940. doi: 10.1016/j.cell.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 22.Cang Y, et al. DDB1 is essential for genomic stability in developing epidermis. Proc Natl Acad Sci USA. 2007;104:2733–2737. doi: 10.1073/pnas.0611311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 24.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 25.Wu H, et al. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 26.Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol Cell Biol. 2006;26:7977–7990. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandgren EP, Palmiter RD, Heckel JL, Brinster RL, Degen JL. DNA rearrangement causes hepatocarcinogenesis in albumin-plasminogen activator transgenic mice. Proc Natl Acad Sci USA. 1992;89:11523–11527. doi: 10.1073/pnas.89.23.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 30.Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.