Abstract
Autophagy, a major degradative pathway for proteins and organelles, is essential for survival of mature neurons. Extensive autophagic-lysosomal pathology in Alzheimer’s disease brain contributes to Alzheimer’s disease pathogenesis, although the underlying mechanisms are not well understood. Here, we identified and characterized marked intraneuronal amyloid-β peptide/amyloid and lysosomal system pathology in the Alzheimer’s disease mouse model TgCRND8 similar to that previously described in Alzheimer’s disease brains. We further establish that the basis for these pathologies involves defective proteolytic clearance of neuronal autophagic substrates including amyloid-β peptide. To establish the pathogenic significance of these abnormalities, we enhanced lysosomal cathepsin activities and rates of autophagic protein turnover in TgCRND8 mice by genetically deleting cystatin B, an endogenous inhibitor of lysosomal cysteine proteases. Cystatin B deletion rescued autophagic-lysosomal pathology, reduced abnormal accumulations of amyloid-β peptide, ubiquitinated proteins and other autophagic substrates within autolysosomes/lysosomes and reduced intraneuronal amyloid-β peptide. The amelioration of lysosomal function in TgCRND8 markedly decreased extracellular amyloid deposition and total brain amyloid-β peptide 40 and 42 levels, and prevented the development of deficits of learning and memory in fear conditioning and olfactory habituation tests. Our findings support the pathogenic significance of autophagic-lysosomal dysfunction in Alzheimer’s disease and indicate the potential value of restoring normal autophagy as an innovative therapeutic strategy for Alzheimer’s disease.
Keywords: autophagy, lysosome, cystatin B, cathepsin, Alzheimer’s disease
Introduction
Lysosomes (DeDuve and Wattiaux, 1966) are acidified vesicular organelles containing over 60 proteases and other acid hydrolases (Turk et al., 2003) that degrade macromolecules delivered by two converging pathways, endocytosis and autophagy. The endocytic pathway transports extracellular materials and certain plasma membrane proteins to lysosomes through early endosomes and late endosomes/multivesicular bodies (Piper and Luzio, 2001). The autophagic pathway delivers intracellular constituents to lysosomes by several processes, which include chaperone-mediated autophagy, microautophagy and macroautophagy (Mizushima et al., 2008). Chaperone-mediated autophagy, shown to be defective in several neurodegenerative diseases, involves the facilitated direct entry into lysosomes of selected cytosolic proteins containing a chaperone-mediated autophagy-targeting motif (Cuervo, 2010). Macroautophagy mediates the bulk degradation of cytoplasm and is the principal mechanism for the turnover of organelles. Macroautophagy is also a vital pathway for degrading abnormal and aggregated proteins, particularly under stress or injury conditions (Rubinsztein, 2006; Levine and Kroemer, 2008) including neurodegenerative protein misfolding diseases.
During macroautophagy (hereafter referred to by the general term ‘autophagy’), an elongated membrane structure envelopes cytoplasmic constituents and encloses to form an autophagosome, which subsequently fuses with lysosomes to effect degradation of substrates (see DeDuve and Wattiaux, 1966; Nixon, 2006 for glossary). Late endosomes/multivesicular bodies containing endocytosed cargoes destined for degradation may also fuse with autophagosomes before lysosomal clearance (Fader and Colombo, 2009). Both autophagy and the endocytic pathway are considered to be major pathways for amyloid precursor protein (APP) processing and amyloid-β peptide (Aβ) generation (Nixon, 2007; Pickford et al., 2008), particularly under Alzheimer’s disease-related conditions. Although Aβ may be generated at several sites in cells, the Aβ generated in endosomes and autophagic vacuoles and delivered to lysosomes is mainly cleared through lysosomal proteolysis under normal conditions (Nixon, 2007).
Dysfunction of the endosomal–lysosomal pathway causes the earliest known neuronal pathology in Alzheimer’s disease and is promoted by genetic factors that cause early onset Alzheimer’s disease or increase Alzheimer’s disease risk (for reviews, see Nixon and Cataldo, 2006; Nixon et al., 2008). Preceding the appearance of neurofibrillary tangles and neuritic plaques, neuronal endosomes enlarge, reflecting an abnormal acceleration of endocytosis (Cataldo et al., 2008; Jiang et al., 2010). Lysosome proliferation in affected neurons, indicating the early mobilization of the lysosomal system, accompanies the initial appearance of extracellular β-amyloid (Cataldo et al., 1996) and is followed by development of striking pathology of the autophagic-lysosomal pathway, including robust, relatively selective accumulation of autophagic vacuoles and lysosomal dense bodies in dystrophic neurites throughout the Alzheimer’s disease brain (Nixon et al., 2005; Yu et al., 2005). The autophagy pathology in Alzheimer’s disease brain resembles that induced by knocking out specific cathepsins or by administering lysosomal protease inhibitors (Felbor et al., 2002; Koike et al., 2005; Boland et al., 2008). These observations and others showing that Aβ and other autophagic substrates, such as the autophagosome marker microtubule-associated protein 1 light chain 3 (LC3), accumulate intraneuronally in vesicular compartments in Alzheimer’s disease brain have suggested that autolysosomal proteolysis is markedly impaired in Alzheimer’s disease (Boland et al., 2008; Nixon et al., 2008). This conclusion is underscored by the recent observation that presenilin 1 is essential for lysosomal acidification and autophagy and that mutations of presenilin 1 in early-onset familial Alzheimer’s disease cause marked loss of these functions (Lee et al., 2010).
In the present study, we investigated the possibility that improving neuronal autophagic-lysosomal proteolytic function in a mouse model of Alzheimer’s disease pathology would ameliorate brain pathology and prevent neuronal dysfunction and memory decline. TgCRND8 mice, overexpressing a version of APP695 including Swe and Ind mutations and producing more Aβ42 than Aβ40 (Chishti et al., 2001), develop lysosomal system pathology, accumulate intraneuronal Aβ and robustly deposit β-amyloid extracellularly in neuritic plaques, leading to marked memory deficits (Janus et al., 2000). We sought to increase cathepsin activities in autolysosomes and lysosomes of TgCRND8 mice by deleting the gene for cystatin B (or stefin B) (Pennacchio et al., 1998) to relieve inhibition of multiple cathepsins. The cystatin B knockout (CBKO) mice that were crossed with the TgCRND8 mice were generated as a model of the recessively inherited neurodegenerative disorder, myoclonus epilepsy of type 1, in which the loss of cystatin B leads to loss of cerebellar granule cells, progressive cerebellar atrophy and loss of motor coordination (Pennacchio et al., 1998). Despite these deficits in this model, cystatin B deletion represented a strategy to selectively enhance the activities of cysteine proteases and test the hypothesis that improving lysosomal proteolytic function in TgCRND8 mice would ameliorate Alzheimer’s disease-related pathologies. Cystatin B, an endogenous inhibitor of cysteine proteases, is a member of the stefin family (Type 1 cystatins) (Turk et al., 2008) within the cystatin superfamily (Tizon and Levy, 2006; Turk et al., 2008). Although cystatin B is reported to be present in several intracellular sites (Riccio et al., 2001; Alakurtti et al., 2005), we show here that it is prominently localized to compartments of the lysosome system, and that reducing cystatin B levels enhances lysosomal enzyme activities thereby stimulating lysosomal protein turnover. We further demonstrate that enhancing lysosomal proteolysis exerts significant therapeutic effects in TgCRND8 mice, including an improved clearance of autophagy substrates resulting in reduced levels of intracellular and extracellular Aβ and reversal of multiple cognitive deficits. Our findings underscore the pathogenic importance of lysosomal system dysfunction in Alzheimer’s disease and establish proof of concept that enhancing lysosome function in Alzheimer’s disease models and, by extension in Alzheimer’s disease, may ameliorate neuropathology and cognitive deficits.
Materials and methods
Animals and tissue preparation
All animal procedures were performed following the National Institutes of Health Guidelines for the Humane Treatment of Animals, with approval from the Institutional Animal Care and Use Committee at the Nathan Kline Institute for Psychiatric Research. Animals of both sexes were used in this study. CBKO mice were initially from Dr Richard M. Myers at Stanford University School of Medicine (Pennacchio et al., 1998), and were on a 129X1/SvJ (129X1) strain background. TgCRND8 mice, which express mutant human APP, Swedish (K670N/M671L) plus Indiana (V717F) mutations, were created on a 129S6/SvEvTac (129S6) strain background by Dr David Westaway at the University of Toronto (Chishti et al., 2001). The CBKO mice were bred to the TgCRND8 mice to generate CBKO+/−/APP+/− double heterozygotes that were bred with CBKO to generate CBKO−/−/APP+/−. Subsequently, CBKO−/−/APP+/− were crossed with CBKO to accelerate the generation of CBKO−/−/APP+/−. The breeding process generated four main types of experimental animals: (i) wild type; (ii) CBKO; (iii) TgCRND8; and (iv) CBKO/TgCRND8 (i.e. the CBKO−/−/APP+/−)—all on a 129X1 × 129S6 strain background. All mice were genotyped by polymerase chain reaction. The mice were studied at 6.0 ± 0.2 months of age, which is simply described as ‘6-month-old’ or ‘6 months of age’ hereafter, except where otherwise indicated.
To obtain tissues for experiments, animals were anaesthetized with a mixture (0.01 ml/g body weight, intraperitoneally) of ketamine (10 mg/ml) and xylazine (1 mg/ml). Mice for light microscopic analyses were usually fixed by cardiac perfusion using 4% paraformaldehyde in 0.1 M sodium cacodylate buffer [pH 7.4, Electron Microscopy Sciences (EMS), Hatfield, PA, USA]. Following perfusion fixation, the brains were immersion-fixed in the same fixative overnight at 4°C. For conventional transmission electron microscopic study, 4% paraformaldehyde was supplemented with 2% glutaraldehyde (EMS). For biochemical analyses, including western blotting, enzymatic activity assays and enzyme-linked immunosorbent assay, the brains were quickly frozen on dry ice and stored at –70°C. In some cases, when both morphological and biochemical analyses were planned on the same mouse, the brain was removed after brief perfusion with saline. One hemisphere was frozen at –70°C and the other half was immersion-fixed in 4% paraformaldehyde for three days at 4°C.
Subcellular fractionation for isolation of autophagic vacuoles and lysosomes from brain tissue
For each mouse genotype, cerebral cortices from five or more brains were pooled. Using a protocol modified from Marzella et al. (1982), the samples were homogenized and subjected to differential centrifugation to separate a fraction enriched in autophagic vacuole, lysosomes and mitochondria as described previously (Cuervo et al., 1995; Singh et al., 2009). The different organelles in this fraction were isolated by floatation in a discontinuous gradient of metrizamide (50, 26, 24, 20 and 10%). A fraction containing mainly autophagosomes (AV10) was recovered in the 10–20% interface, a fraction enriched in autophagolysosomes (AV20) was isolated at the 20–24% interface, and lysosome-enriched fraction was recovered in the 24–16% interface. These fractions were washed by centrifugation in 0.25 M sucrose and pellets were immersed in a cacodylate fixation buffer for electron microscopic analysis or analysed directly by western blot or enzyme assay. A fraction enriched in endoplasmic reticulum resealed vesicles (microsomes) and the cytosolic fraction was obtained in the pellet and supernatant, respectively, after centrifugation of the supernatant at 100 000 g for 1 h.
Antibodies for immunocytochemistry, immunofluorescence, western blotting and enzyme-linked immunosorbent assay
The following antibodies were used in this study: RU2, rabbit polyclonal antibody directed against mouse cathepsin D (made in-house, diluted 1:5000–10 000 for immunofluorescence and immunocytochemistry and 1:5000 for western blotting); D-2-3, sheep polyclonal antibody directed against human cathepsin D (made in-house, diluted 1:1000 for immunofluorescence); cathepsin B rabbit polyclonal antibody directed against human cathepsin B (Cortex Biochem, CR6009RP, diluted 1:400 for immunocytochemistry); cathepsin L rabbit polyclonal antibody directed against human cathepsin L (Athens Research and Technology, 01-12-030112, diluted 1:400 for immunocytochemistry); cystatin B rabbit polyclonal antibody directed against rat cystatin B (made in-house, diluted 1:100 for western blotting and 1:10 for immunocytochemistry); rat monoclonal antibody directed against mouse lysosomal membrane associated protein 2 (Developmental Studies Hybridoma Bank, University of Iowa, ABL-93, diluted 1:100 for immunofluorescence); LC3 rabbit polyclonal antibody directed against rat LC3 (microtubule associated protein 1 light chain 3) (made in-house, diluted 1:2000 for western blotting); LC3 rabbit polyclonal antibody directed against an N-terminal portion of human LC3 (Novus Biologicals, NB100-2200, diluted 1:1000 for western blotting); LC3 polyclonal antibody directed against rat LC3 (diluted 1:250 for immunofluorescence, a gift from Dr Y. Uchiyama, Juntendo University Graduate School of Medicine, Japan); lysobisphosphatidic acid monoclonal antibody directed against rat lysobisphosphatidic acid (a gift from Dr Jean Gruenberg, University of Geneva, Switzerland); ubiquitin mouse monoclonal antibody directed against bovine ubiquitin (Chemicon, MAB1510, diluted 1:300 for immunofluorescence); ubiquitin polyclonal antibody directed against human ubiquitin (Dako, Z 0458, diluted 1:1500 for western blotting); glial fibrillary acidic protein mouse monoclonal antibody directed against pig glial fibrillary acidic protein (Sigma, G3893, diluted 1:400 for immunofluorescence); Iba-1 rabbit polyclonal antibody directed against the C-terminus of the microglia/macrophage-specific protein Iba-1 (Waco Chemicals, 019-19741, diluted 1:500 for immunofluorescence); neuronal nuclei mouse monoclonal antibody directed against neuronal nuclei protein from mouse brain (Millipore, MAB377MI, diluted 1:50). Antibodies directed against APP, Aβ and/or other APP proteolytic species included: 22C11 mouse monoclonal antibody directed against the N-terminus (amino acids 66–81) of APP695 (Millipore, MAB348, diluted 1:100 for immunocytochemistry); 6E10 (mouse monoclonal antibody specific to Aβ1-16, diluted 1:1000 for western blotting) and 4G8 (mouse monoclonal antibody specific to Aβ17-24, diluted 1:250 for immunocytochemistry), both from Covance (Emeryville, CA,USA; catalogue No: SIG-39320 and SIG-39220, respectively); R226 (polyclonal antibody specific to Aβ42, diluted 1:500 for immunocytochemistry), the kind gift of Dr Pankaj D. Mehta (Institute for Basic Research in Developmental Disabilities, Staten Island, NY, USA); C1/6.1 monoclonal antibody against the C-terminal 20 residues of APP (made in-house, diluted 1:400 for immunocytochemistry) (Mathews et al., 2002); and additional mouse monoclonal antibodies—the generous gift of Dr Marc Mercken (Janssen Pharmaceutica/Johnson & Johnson, Belgium) (Mathews et al., 2002): JRF/AβN/25 (specific to Aβ1-7, diluted 1:500 for immunocytochemistry), JRF/Aβtot/17 (specific to Aβ1-16, diluted 1:500 for immunocytochemistry and 1:1000 for western blotting), JRF/cAβ40/10 (specific to Aβ40, diluted 1:500 for immunocytochemistry), JRF/cAβ42/26 (specific to Aβ42, diluted 1:500 for immunocytochemistry).
Ultrastructural analyses
For electron microscopy, vibratome brain sections were cut and post-fixed in 1% osmium tetroxide. Following alcohol dehydration, sections were embedded in EPON® resin (EMS) and ultrathin sections prepared and stained with uranyl acetate and lead citrate. Material was viewed with a Philips CM 10 electron microscope equipped with a digital camera (Hamamatsu, model C4742-95) aided by AMT Image Capture Engine software (version 5.42.443a). For light microscopic examination, 1 μm sections were stained with toluidine blue.
Analysis of proteolysis
Total protein degradation in cultured primary fibroblasts or neurons was measured by metabolic labelling (Auteri et al., 1983; Cuervo et al., 2004) with a 48 h pulse with [3H]-leucine (2 µCi/ml) to preferentially label long-lived proteins. During the chase, aliquots of the medium taken at different time points were precipitated in trichloroacetic acid and proteolysis was measured as the percentage of the initial acid-precipitable radioactivity (protein) converted to acid-soluble radioactivity (amino acids and small peptides) over time.
Enzymatic activity assays for cathepsins
Cathepsin D activity was assayed as pepstatin-inhibitable activity toward 14C-labelled methemoglobin, and cathepsin B and cathepsin L activities were assayed by measuring aminomethyl coumarine (AMC) released from Z-Arg-Arg-AMC (specific for cathepsin B) and Z-Phe-Arg-AMC (specific for cathepsin B and L). Detailed protocols were described previously (Yang et al., 2009) and are also provided in the online Supplementary Methods.
Contextual fear conditioning
Protocols were the same as those described previously (Ohno et al., 2001; Kimura and Ohno, 2009). The training session for contextual fear conditioning consisted of a 180 s exploration period followed by two conditioned stimulus-unconditioned stimulus pairings separated by 1 min interval (foot shock intensity, 1.0 mA; duration, 2 s). After the last shock, the mice (wild-type n = 10, CBKO n = 10, TgCRND8 n = 9, and CBKO/TgCRND8 n = 7; all 6-months-old) were allowed to stay in the chamber for another 30 s and were then returned to their home cages. Contextual fear memory was measured by scoring freezing behaviour (the absence of all but respiratory movement) for 180 s with a FreezeFrame automated scoring system (Coulbourn Instruments, Allentown, PA, USA) when the mice were placed back into the same conditioning chamber 24 h after training (Supplementary Methods).
Odour habituation test
The same set of mice used for contextual fear conditioning (wild-type n = 10, CBKO n = 9, TgCRND8 n = 8 and CBKO/TgCRND8 n = 7) were individually housed for at least 72 h and tested in their home cages for olfactory investigation behaviour in an odour habituation test (Sundberg et al., 1982) using previously described methods (Wesson et al., 2010) (Supplementary Methods). Measures of odour investigation behaviour (n = 7 odours) were recorded over two daily sessions. Normalized investigation data were analysed with ANOVA followed by Fisher’s protected least significant difference.
Protocols for primary cell cultures, immunolabeling, immunoblotting, immunoprecipitation, enzyme-linked immunosorbent assay and generation of an anti-cystatin B antibody can be found in the online Supplementary Methods.
Results
TgCRND8 mice exhibit marked autophagic-lysosomal dysfunction
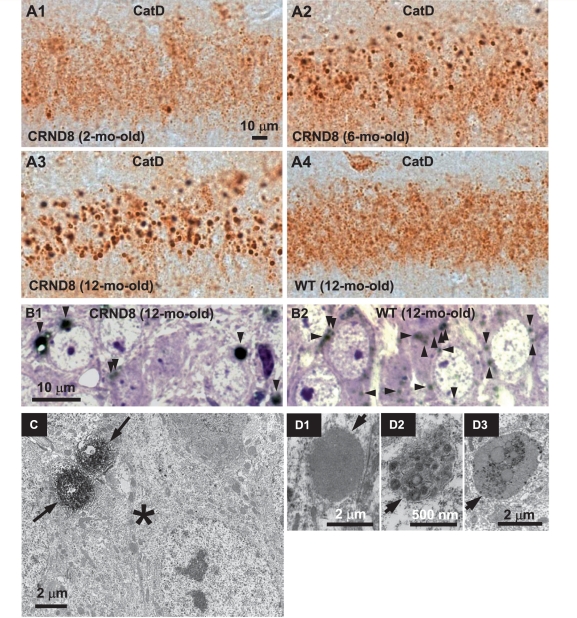
Neurons in affected brain regions of 6-month-old TgCRND8 mice, immunolabelled with an antibody (RU2) against mouse cathepsin D, exhibited strikingly enlarged cathepsin D-positive lysosomal compartments with reduced numbers of normal-sized lysosomes (Fig. 1A2). Although only rarely seen in 2-month-old TgCRND8 mice (Fig. 1A1), these giant compartments progressively increased in number and size to at least 12 months of age (Fig. 1A3) and were absent in age-matched wild-type mice (Fig. 1A4). Giant cathepsin D-positive compartments were most abundant in the hippocampal CA1 sector, including the pyramidal cell layer (Fig. 1A1–3) and neuronal processes within the stratum radiatum (e.g. Fig. 6A3, arrowheads) and, at a lower density, in the CA3 sector and cerebral cortex (data not shown). Antibodies against cathepsin B and cathepsin L revealed similar patterns (data not shown).
Figure 1.
Abnormal lysosome-related compartments in the brains of TgCRND8. Sections from the hippocampal CA1 sector of 2- (A1), 6- (A2) and 12-month-old (A3) TgCRND8 (strain 129S6) and 12-month-old wild-type (WT) (A4) mice (n = 3–6 mice per age per genotype) immunostained with an antibody against mouse cathepsin D (RU2). Representative microphotograph images depict abundant abnormally enlarged lysosome-related granules in CA1 neurons of TgCRND8 at 6- and 12-months of age. (B) Images of the CA1 sector from toluidine blue-stained 1 µm thick sections that were derived from vibratome sections of 12-month-old TgCRND8 (B1) and wild-type (B2) immunostained with RU2. Toluidine blue-stained structures are in blue/purple and RU2-stained granules are in black and denoted by arrowheads. (C) Vibratome brain sections from 6-month-old TgCRND8 were immunostained with the anti-cathepsin D antibody RU2 and then processed for electron microscopy embedding. Areas of the hippocampal CA1 sector containing RU2-positive giant granules (as those seen in Fig. 1A2) were selected for ultrathin sectioning. The electron micrograph shows that a neuronal soma (asterisk) contains two giant RU2-positive granules decorated by strong diaminobenzidine products (arrows). (D) Electron micrographs of the CA1 hippocampal area in 6-month-old TgCRND8 mice depict giant granules (arrows) with single-limiting membranes, containing undigested materials. Scale bars: 10 µm (A and B); 2 µm (C, D1 and D3); 500 nm (D2). CatD = cathepsin D; CRND8 = TgCRND8.
Figure 6.
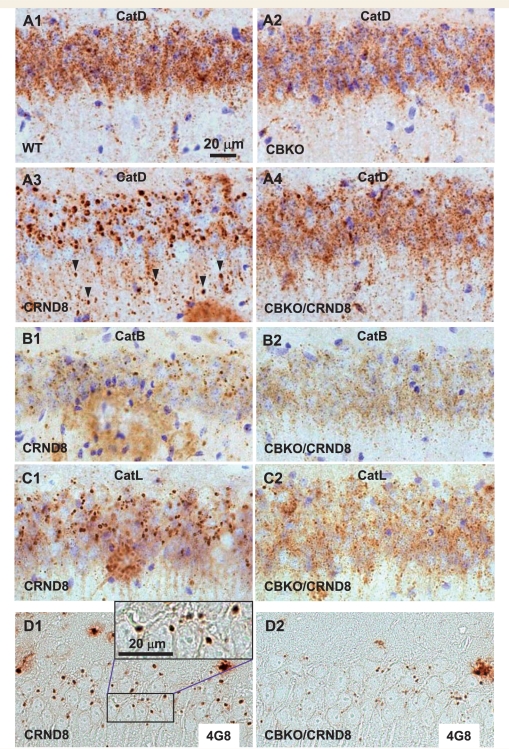
Elimination of giant autolysosomes in the brains of CBKO/TgCRND8. Vibratome (A–C) or paraffin (D) brain sections from a group of wild-type, CBKO, TgCRND8 and CBKO/TgCRND8 mice (strains: 129S6 × 129X1 for the four genotypes) at 6 months of age were processed with antibodies to cathepsin D (RU2) (A), cathepsin B (B), cathepsin L (C) or Aβ (4G8) (D). All images are from the hippocampal CA1 sector showing reduction of giant autolysosomes—revealed by antibodies to either cathepsins or Aβ—in the CBKO/TgCRND8 compared to TgCRND8. The images in (A) are representative of the results from 9–13 mice per genotype, while those in (B–D) are from 3–4 mice per genotype. Arrowheads (A3) depict giant autolysosomes within the stratum radiatum. Scale bars: 20 µm (A–D). CatB = cathepsin B; CatD = cathespin D; CatL = cathepsin L; CRND8 = TgCRND8.
The enlarged size of cathepsin D-positive compartments in TgCRND8 mice (Fig. 1B1) compared to wild-type mice (Fig. 1B2) was more evident in vibratome sections of hippocampus immunostained with cathepsin D antibodies, which were then embedded in EPON, cut into 1 µm sections and stained with toluidine blue (Fig. 1B). At the ultrastructural level, the cathepsin D-stained granules were abnormally large and located within neuronal soma (Fig. 1C) and processes (data not shown). Given that the highly electron dense diaminobenzidine (DAB) product, reflecting strong cathepsin D immunoreactivity, in the immunoelectron microsocopy samples (e.g. Fig. 1C) prevented a clear viewing of the contents within these granules, we performed additional ultrastructural analysis. In conventionally processed electron microscopy samples, these giant lysosomal compartments in the cell bodies and neurites of CA1 neurons appeared as electron-dense, single-membrane-limited structures of 1.5–3.0 µm diameter containing incompletely digested materials, including membranous structures and a minor lipopigment component (Fig. 1D1–D3 and Supplementary Fig. 1), and therefore could be classified as autolysosomes. To determine whether or not these compartments existed in glial cells, we performed double immunofluorescent labelling with an antibody to cathepsin D and an antibody against a marker for either astrocytes (glial fibrillary acidic protein) or microglia (Iba-1) (Supplementary Fig. 2) and found no cathepsin D-labelled giant autolysosomes in cells with either the glial fibrillary acidic protein or the Iba-1 signal. However, most, if not all, of these giant autolysosomes exist within neuronal nuclei-stained neurons (Supplementary Fig. 2), consistent with the neuronal localization of these giant autolysosomes as shown in Fig. 1B–D and Supplementary Fig. 1.
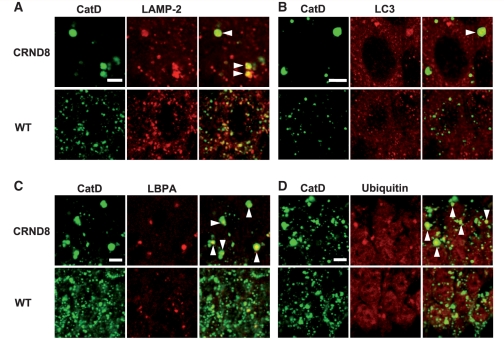
In double immunofluorescence studies supporting the identification of these giant granules as autolysosomes, the cathepsin D-positive giant compartments in TgCRND8 (Fig. 2A–C) were found to be co-immunolabelled by markers for lysosomes (lysosomal membrane associated protein 2, Fig. 2A), autophagosomes/autolysosomes (LC3, Fig. 2B) or late endosomes (lysobisphosphatidic acid, Fig. 2C), consistent with the convergence of both the autophagic and endocytic pathways on these compartments. Interestingly, cathepsin D-positive giant autolysosomes were also labelled by anti-ubiquitin antibody (Fig. 2D) that, together with the data shown in Fig. 8, indicates accumulation of ubiquitinated proteins within these compartments, which are incompletely degraded.
Figure 2.
Markers/substrates of the endocytic-autophagic-lysosomal pathways presenting in the giant autolysosomes. Double immunofluorescent labelling of brain sections from 12-month-old TgCRND8 and wild-type (WT) mice (n = 3 mice per genotype) with an antibody against cathepsin D (CatD) (rabbit polyclonal antibody RU2 for A, C and D, sheep polyclonal antibody D-2-3 for B) and an antibody directed against either a lysosomal marker, lysosomal membrane associated protein 2 (LAMP-2) (A), an autophagosomes/autolysosomes marker, LC3 (B), a late endosome marker, lysobisphosphatidic acid (LBPA) (C), or ubiquitin (D). The arrowheads in the overlays depict those giant granules which display double-immunostaining. Images shown are all from the hippocampal CA1 sector. Scale bars: 5 µm (A–D). CRND8 = TgCRND8.
Figure 8.
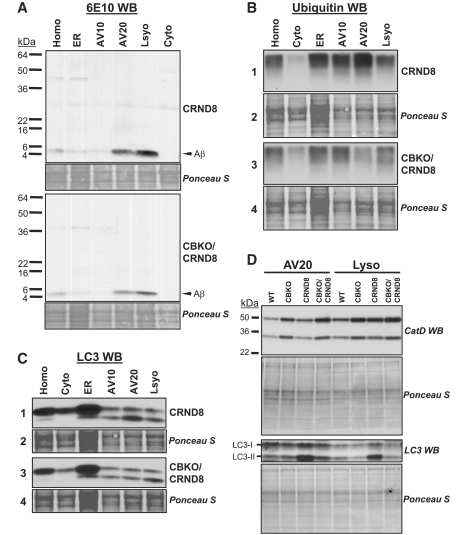
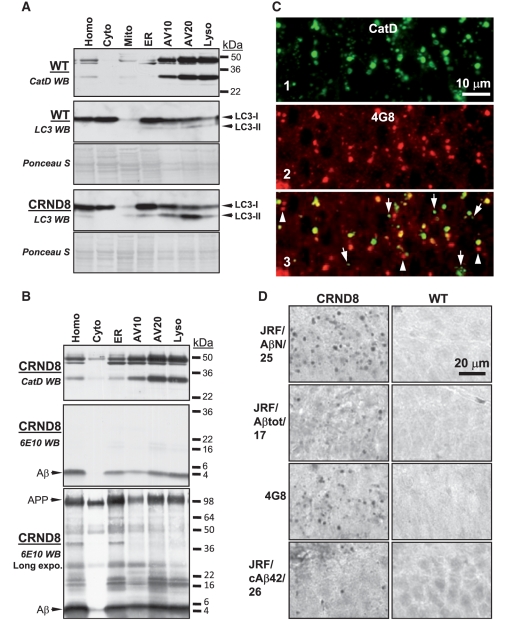
Cystatin B deletion in TgCRND8 promoting clearance of Aβ, ubiquitinated proteins and LC3-II in isolated autophagic-lysosomal compartments. Subcellular fractions containing the autophagic-lysosomal compartments AV10, AV20 and lysosomes were isolated from brains of 6-month-old TgCRND8 and CBKO/TgCRND8 (n = 8 per genotype). Equal amounts of proteins from each fraction were subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis and processed for western blotting with an antibody directed against Aβ (A), ubiquitin (B) or LC3 (C), and representative blots are shown. (D) AV20 and Lyso fractions from all four genotypes were loaded onto one gel and processed with an anti-cathepsin D (RU2) or anti-LC3 antibody. Corresponding ponceau S-stained blots are shown below the antibody-probed blots as loading controls (A–D). AV10 = early autophagic vacuole/autophagosome; AV20 = late autophagic vacuole/autolysosome; CRND8 = TgCRND8; Cyto = cytosol; ER = endoplasmic reticulum-enriched; Homo = homogenate; Lyso = lysosome; WB = western blotting; WT = wild-type.
To investigate whether or not autophagic proteolysis may be impaired, we studied LC3-II, a membrane-associated form of LC3 that first appears on newly formed autophagosomes and is then normally degraded quickly by lysosomal enzymes after autophagosome/lysosome fusion (Mizushima and Yoshimori, 2007). Using subcellular fractionation on metrizamide density gradients, we isolated lysosomes and autophagic vacuoles of relatively lower and higher density from the brains of 12-month-old TgCRND8 and wild-type mice, and confirmed their identities based on their characteristic ultrastructural features as previously described (Cuervo et al., 1995; Cao et al., 2006), and the presence of LC3-II (Fig. 3A) and cathepsin D immunoreactivity (e.g. Fig. 3A, B, top) on immunoblot analyses. Consistent with our previous immunocytochemical studies, immunoblot analysis showed that the two autophagic vacuole fractions and lysosome fractions from TgCRND8 mice contained abnormally high levels of LC3, including very high proportions of LC3-II (Fig. 3A). In contrast, LC3-II was much lower in the corresponding wild-type fractions relative to LC3-I/total LC3 (Fig. 3A), indicating efficient autolysosome turnover of LC3-II in wild-type mice but impaired LC3-II turnover in TgCRND8 mice (also see Fig. 8D).
Figure 3.
Delayed degradation and accumulation of LC3-II and Aβ within autophagic-lysosomal compartments in the brains of TgCRND8. Subcellular fractions containing the autophagic-lysosomal compartments AV10, AV20 and lysosomes were isolated from brains of 12-month-old TgCRND8 and wild-type (WT) (n = 6 per genotype), and equal amounts of protein from each fraction were subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis and processed for western blotting (WB) with an antibody directed against cathepsin D (CatD) (A, B, top panels), LC3 (A) or 6E10 (B). Immunoblot for cathepsin D showing that the AV10, AV20 and lysosome fractions exhibit strongest cathepsin D signals, confirming the autophagic-lysosomal origin of these fractions. Corresponding ponceau S stained blots are shown below LC3 antibody-probed blots as loading controls (A). (C) Double immunofluorescent labelling showing that most giant autolysosomes and some smaller lysosomes were co-immunolabelled (C3) by RU2 (C1) and 4G8 (C2) in the CA1 sector of a 6-month-old TgCRND8. Granules displaying only RU2 signal or 4G8 signal are labelled with arrows and arrowheads, respectively. (D) Detection of giant autolysosomes in the CA1 sector by additional antibodies directed against various regions of the Aβ domain, including JRF/AβN/25, JRF/Aβtot/17, 4G8 and JRF/cAβ42/26. The images are representative of the results from three 12-month-old mice per genotype. Scale bars: 10 µm (C); 20 µm (D). AV10 = early autophagic vacuole/autophagosome; AV20 = late autophagic vacuole/autolysosome; CRND8 = TgCRND8; Cyto = cytosol; ER = endoplasmic reticulum-enriched; Homo = homogenate; Lyso = lysosome; Mito = mitochondria.
Further, immunoblot analyses with monoclonal antibody 6E10 on subcellular fractions of TgCRND8 brains revealed accumulation of Aβ within autophagic-lysosomal compartments (Fig. 3B, middle and bottom). Double immunofluorescence labelling of tissue sections using monoclonal antibody 4G8 to detect APP-related metabolites, including Aβ, revealed abundant 4G8 signal in most cathepsin D-positive giant autolysosomes and in some smaller lysosomes in CA1 neurons of TgCRND8 mice (Fig. 3C) but no 4G8 immunolabelling in wild-type mice (not shown, but see Figs 3D and 4G8: TgCRND8 versus wild-type). Additional immunocytochemistry with antibodies against a panel of APP epitopes exhibited immunoreactivity in granular compartments of TgCRND8 mice when we used antibodies against sequences within Aβ, such as JRF/AβN/25, JRF/Aβtot/17, and JRF/cAβ42/26 (Fig. 3D), but not antibodies directed against sequences within the N-terminus or the extreme C-terminal portion of APP (22C11 and C1/6.1, respectively) (data not shown). These analyses demonstrated that Aβ is not efficiently degraded by lysosomal hydrolases and therefore, accumulates in autophagic vacuoles and lysosomes.
Deletion of cystatin B enhances lysosomal activities and accelerates protein turnover
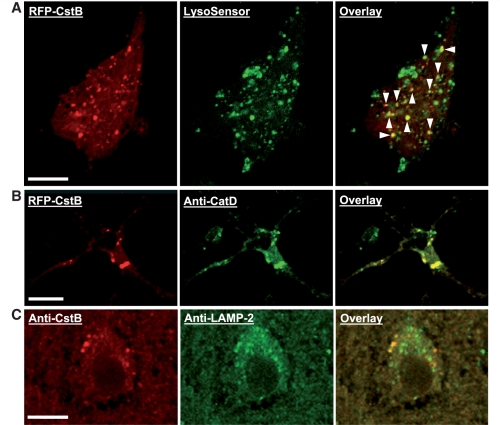
We observed no detectable differences in brain protein levels of cystatin B between wild-type and TgCRND8 mice (Supplementary Fig. 3). Therefore, it is unlikely that cystatin B elevation contributes to the lysosomal pathology in these mice. We evaluated cystatin B deletion in this study, however, as a strategy to enhance cathepsin activities and further assess lysosomal proteolysis enhancement as a possible therapeutic approach in this Alzheimer’s disease mouse model. Previous immunocytochemical studies have yielded conflicting results regarding the subcellular locations of cystatin B, which have included localization to nucleus, cytosol and punctate structures within the cytoplasm (for review, see Joensuu et al., 2008). In the present study, primary cortical neurons from wild-type mice transfected with red fluorescent protein-tagged mouse cystatin B DNA plasmid exhibited a predominantly punctate staining pattern with less intense signal in the cytoplasm. The majority of cystatin B-positive granular compartments were also labelled by the lysosomal marker, LysoSensor (Fig. 4A), and with a specific anti-mouse cathepsin D antibody (Fig. 4B). Immunocytochemical studies performed on mouse brain sections, using an affinity purified polyclonal antibody that detects no protein band at the molecular weight of cystatin B in CBKO mice (Supplementary Fig. 3), showed that vesicular compartments in cortical neurons were prominently labelled by cystatin B antibody, many of which were co-labelled by anti-lysosomal membrane associated protein 2 antibody (Fig. 4C). These data, therefore, support the conclusion that cystatin B was located at least partly in lysosomal compartments as well as in cytoplasm, where entry into the autophagy system may be expected during normal autophagic turnover of cytoplasm.
Figure 4.
Subcellular localization of cystatin B. Cultured mouse primary neurons (A and B) were transfected for 1 day with red fluorescent protein (RFP)-tagged mouse cystatin B (CstB) DNA plasmid (OriGene, custom-made ExactORF cDNA clone) according to the procedure provided by the manufacturer, and were then incubated with LysoSensor (Invitrogen) for 2 h to label lysosomal structures, left unfixed but were mounted with anti-fade mounting medium and examined under a confocal microscope (A), or fixed for double labelling using anti-cathepsin D antibody RU2 (B). (C) Brain sections from a wild-type mouse were double-immunolabelled with a homemade affinity purified anti-rat cystatin B antibody and an anti-lysosomal membrane associated protein 2 (LAMP-2) antibody. Arrowheads depict cystatin B-positive granular compartments co-labelled with LysoSensor (A). Scale bar: 10 µm (A–C).
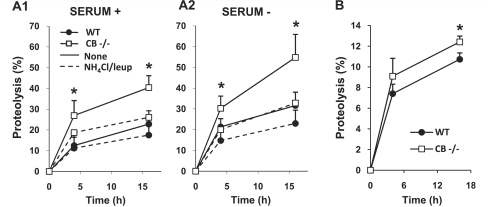
Reduced cystatin B expression has been previously shown to correlate with increased cathepsin activities (Rinne et al., 2002). We confirmed and extended this finding by revealing that enzymatic activities of cathepsin B, cathepsin L and cathepsin D were higher in 6-month-old CBKO brains (n = 14) than in wild-type brains (n = 16), which were conducted separately from, but similar to, those shown in Fig. 7 and therefore not shown. The higher cathepsin D activity found in CBKO brain is interesting given that cathepsin D is an aspartyl protease. Moreover, in metabolic analyses of protein turnover (Cuervo et al., 2004) in immortalized fibroblasts from wild-type and CBKO mice, degradation of long-lived proteins, the principal substrates for lysosomal proteases, was significantly higher in cells from CBKO mice than in wild-type mice under both basal (Fig. 5A1) and inducible (Fig. 5A2) conditions. These increases in protein degradation were mainly of lysosomal origin because differences between CBKO and wild-type cells were almost completely eliminated when lysosomal proteolysis was blocked with the serine protease inhibitor leupeptin and ammonium chloride, which neutralizes the acidic pH of lysosomes (Fig. 5A1 and A2). Similarly, in primary neuronal cultures maintained in neurobasal medium supplemented with B27, degradation of long-lived proteins was significantly higher in neurons from CBKO mice than in wild-type mice (Fig. 5B).
Figure 7.
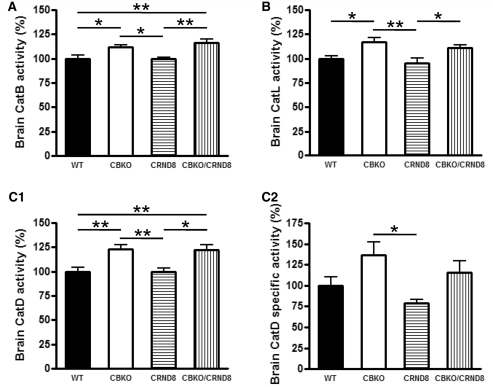
Cystatin B deletion in TgCRND8 elevating cathepsin activities. Results of enzymatic activity assays for cathepsin B (CatB; A), cathepsin L (CatL; B) and cathepsin D (CatD; C1) activities in brain homogenates of wild-type, CBKO, TgCRND8 and CBKO/TgCRND8 (all 6-month-old, n = 9–12 for each genotype). (C2) The cathepsin D activity shown in (C1) was recalculated against the tubulin-normalized cathepsin D values obtained from immunoblots probed with anti-cathepsin D (RU2) and anti-tubulin to yield the cathepsin D-specific activity. Values are the mean ± SEM for each group. Mean differences between genotypes were analysed by one way ANOVA followed by post hoc Bonferroni’s multiple comparison tests. *P < 0.05, **P < 0.01.
Figure 5.
Cystatin B deletion elevating lysosomal protein degradation. (A) Rates of proteolysis of long-lived proteins in fibroblasts from wild-type (WT) and CBKO (CB−/−) mice maintained in the presence (A1) or absence (A2) of serum. Where indicated a combination of ammonium chloride and leupeptin (leup) was added to inhibit proteolysis in the lysosomal compartment. Values are mean + SEM of three different experiments with triplicate samples. Mean differences between the untreated wild-type and CBKO (i.e. without lysosomal inhibitors) at each time point were analysed by two-tailed Student’s t-test. *P < 0.05. (B) Rates of proteolysis of long-lived proteins in primary neurons from wild-type and CBKO mice maintained in neurobasal medium supplemented with B27. Mean differences between wild-type and CBKO at each time point were analysed by two-tailed Student’s t-test. *P < 0.05.
Deletion of cystatin B reverses lysosomal system pathology and intracellular amyloid-β peptide accumulation in TgCRND8 mice
To examine effects of cystatin B deletion on lysosomal pathology in the TgCRND8 brain, we immunolabelled brain sections from groups of wild-type, CBKO, TgCRND8 and CBKO/TgCRND8 mice at 6 months of age with various cathepsin antibodies. The abundant numbers of giant autolysosomes seen in TgCRND8 (Fig. 6A3) were strikingly reduced in CBKO/TgCRND8 mice, while normal-sized lysosomes increased in abundance (Fig. 6A4) yielding an intracellular lysosomal pattern similar to that of wild-type mice (Fig. 6A1) or CBKO mice (Fig. 6A2). Antibodies to cathepsin B (Fig. 6B) and cathepsin L (Fig. 6C) yielded similar results. In addition to reversing intracellular autophagic-lysosomal pathology, cystatin B deletion nearly completely eliminated intracellular 4G8-immunoreactivity in the hippocampus of CBKO/TgCRND8 mice in immunocytochemical analyses (Fig. 6D).
Cystatin B deletion in TgCRND8 enhances brain lysosomal activities and promotes clearance of proteins within lysosomal compartments
Enzymatic activities of cathepsin B (Fig. 7A), cathepsin L (Fig. 7B) and cathepsin D (Fig. 7C1) in brain homogenates from 6-month-old mice were higher in CBKO brains than in wild-type and TgCRND8 brains. Cystatin B deletion also elevated the activities of these cathepsins in CBKO/TgCRND8 brains relative to the corresponding protease activities in TgCRND8 mice (Fig. 7A-C1). The specific enzymatic activity of cathepsin D was also determined as cathepsin D enzymatic activity per unit cathepsin D protein, as detected by quantitative immunoblot analysis. The results (Fig. 7C2) showed significantly higher specific activity of cathepsin D in CBKO compared to TgCRND8 (P < 0.05, one way ANOVA followed by post hoc Bonferroni’s multiple comparison tests). Comparisons made between two groups by unpaired two-tailed Student’s t-test also revealed differences between CBKO and TgCRND8 (P = 0.004), or CBKO/TgCRND and TgCRND8 (P = 0.03), and a trend of differences between wild-type and CBKO (P = 0.07) or TgCRND8 (P = 0.08). Due to the unavailability of reliable cathepsin B or cathepsin L antibodies for detecting mouse cathepsin B or cathepsin L by immunoblotting, we could not obtain the specific enzymatic activity for cathepsin B or cathepsin L. However, based on the similar results shown in Fig. 7A and B compared to Fig. 7C1, it is predicted that cathepsin B and/or cathepsin L may display trends in specific activity similar to that of cathepsin D for each genotype.
To assess if there were beneficial effects of these cystatin B deletion-induced changes in cathepsin activities on protein degradation/clearance in TgCRND8, we examined key proteins in isolated autophagic vacuole and lysosomes by western blotting. First, intracellular Aβ abundance was markedly reduced in the autophagic vacuole and lysosome fractions isolated from CBKO/TgCRND8 brains compared with those from TgCRND8 mice (Fig. 8A), consistent with the extent of intracellular Aβ clearance seen by immunocytochemistry (Fig. 6D). Similarly, levels of high molecular weight ubiquitinated proteins (sizes > 150 kDa) were decreased in the autophagic vacuole and lysosome fractions from CBKO/TgCRND8 brains (Fig. 8B, panel 1 versus 3). In addition, LC3-II immunoreactivity in the autophagic vacuole and lysosomal compartments from CBKO/TgCRND8 brains was also markedly reduced compared with amounts in corresponding TgCRND8 fractions (Fig. 8C, panel 1 versus 3).
Due to the difficulty in finding a common loading control for subcellular fractions, ponceau S stained blots were used as loading controls (Fig. 3A and Fig. 8A–C). To further validate whether or not same/similar amounts of proteins were loaded for samples from the different genotypes (e.g. CBKO/TgCRND8 versus TgCRND8 in Fig. 8A–C), AV20 and Lyso fractions from the four genotypes of mice were loaded onto one gel and the blots were probed with an anti-cathepsin D or anti–LC3 antibody (Fig. 8D). While the corresponding ponceau S-stained blots (Fig. 8D) displayed similar staining intensities and patterns of bands among the four genotypes—indicating comparable loading, the anti-cathepsin D antibody revealed that in both the AV20 and Lyso fractions the levels of the 50 kDa cathepsin D proform and the 32 kDa cleaved form were higher in samples of CBKO (compared to wild-type) and CBKO/TgCRND8 (compared to TgCRND8), the latter is in contrast to the observed low levels of Aβ (Fig. 8A), ubiquitinated proteins (Fig. 8B) and LC3-II (Fig. 8C) in CBKO/TgCRND8 (compared to TgCRND8). Differences in the levels of LC3, especially LC3-II, were also observed among the four genotypes (Fig. 8D). Comparing the cathepsin D signals with the LC3-II signals (Fig. 8D) revealed that higher levels of cathepsin D in CBKO and CBKO/TgCRND8 are associated with lower levels of LC3-II, consistent with efficient degradation of LC3-II in these mice. In contrast, in the presence of apparently higher cathepsin D protein levels in TgCRND8 than in wild-type, the levels of LC3-II are still higher in TgCRND8 than in wild-type (similar to the results in Fig. 3A), suggesting inefficient cathepsin D proteolytic activity toward LC3-II.
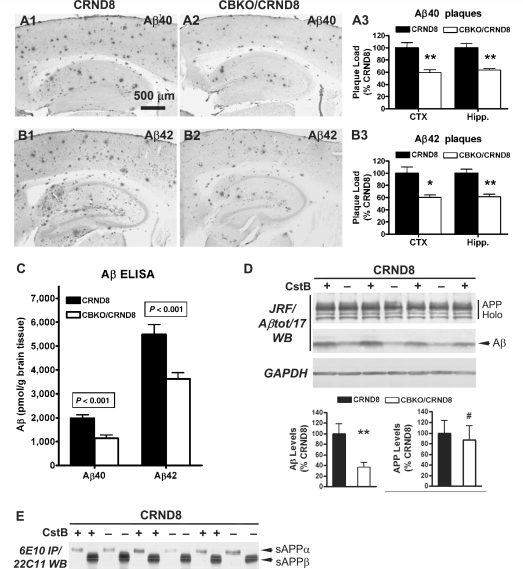
Cystatin B deletion decreases amyloid plaque load and total amyloid-β peptide levels in TgCRND8 brains
Antibodies to Aβ40 (Fig. 9A) or Aβ42 (Fig. 9B) revealed fewer amyloid plaques in 6-month-old CBKO/TgCRND8 mice (Fig. 9A2and B2) compared with age-matched TgCRND8 mice (Fig. 9A1 and B1). Amyloid load in cortical or hippocampal regions in CBKO/TgCRND8 mice (n = 12; 3 males + 9 females) was significantly lower compared to that in TgCRND8 mice (n = 10; 9 males + 1 female) based on quantifications performed with antibodies to Aβ40 (Fig. 9A3, P < 0.001 for both cortex and hippocampus) or Aβ42 (Fig. 9B3, P < 0.005 or <0.001 for either cortex or hippocampus, respectively).
Figure 9.
Reduction of amyloid load and Aβ levels in the brains of CBKO/TgCRND8. Immunocytochemistry using antibodies to Aβ40 (A) or Aβ42 (B) revealed reduction of amyloid deposition in 6-month-old CBKO/TgCRND8 compared to age-matched TgCRND8. (A3 and B3) Results of quantification of amyloid load detected with antibodies to Aβ40 (A3) or Aβ42 (B3) in the cerebral cortex (CTX) and the hippocampus (Hipp) from TgCRND8 (n = 10; 9 males + 1 female) and CBKO/TgCRND8 (n = 12; 3 males + 9 females), expressed as the percent difference from TgCRND8. Values are the mean ± SEM for each group. Significant differences were analysed by two-tailed Student’s t-test. *P < 0.005, **P < 0.001. (C) Levels of Aβ40 and Aβ42 in the brains of 6-month-old TgCRND8 (n = 11; 9 males + 2 females) and CBKO/TgCRND8 (n = 13; 3 males + 10 females) were measured in formic acid-extracted, sucrose homogenates using an enzyme-linked immunosorbent assay method. Values are the mean ± SEM for each group. Significant differences were analysed by two-tailed Student’s t-test. (D) Western blotting (WB) using the antibody JRF/Aβtot/17 (top) on brain homogenates from 6-month-old TgCRND8 and CBKO/TgCRND8. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) blot serves as a loading control. Graphs are results of densitometric analyses of Aβ (bottom left) or APP holoprotein (bottom right) signals normalized by the glyceraldehyde 3-phosphate dehydrogenase signals (**P < 0.01, #P = 0.72, n = 8 per genotype, two-tailed Student’s t-test). (E) Immunoprecipitation (IP) of brain homogenates with 6E10 followed by western blotting (WB) with 22C11. APP Holo = APP holoprotein and holoprotein derivatives; CRND8 = TgCRND8; CstB = cystatin B. Scale bars: 500 µm (A1, A2, B1 and B2).
Aβ40 and Aβ42 level measurements by enzyme-linked immunosorbent assay (Fig. 9C) confirmed plaque quantifications by showing significantly lower total Aβ40 (43%, P < 0.001) and Aβ42 (34%, P < 0.001) in brain homogenates of 6-month-old CBKO/TgCRND8 mice (n = 13; 3 males + 10 females) than in TgCRND8 mice (n = 11; 9 males + 2 females). Consistent with these enzyme-linked immunosorbent assay data, immunoblot analysis of brain homogenates using JRF/Aβtot/17 (Fig. 9D, top) revealed substantially reduced signals from the Aβ immunoreactive bands in 6-month-old CBKO/TgCRND8 mice than in TgCRND8 mice. Quantification of the Aβ signals after being normalized by the glyceraldehyde 3-phosphate dehydrogenase signals showed significant differences in the Aβ levels between CBKO/TgCRND8 and TgCRND8 (Fig. 9D, bottom left; **P < 0.01, n = 8 per genotype). In contrast, TgCRND8 and CBKO/TgCRND8 displayed comparable levels of APP holoprotein (Fig. 9D, top and bottom right; #P = 0.72, n = 8 per genotype) and sAPPα or sAPPβ as measured by immunoprecipitation–western blot analysis (Fig. 9E). Immunoblot analysis of brain homogenates using 6E10 also revealed differences in the levels of Aβ, but not APP holoprotein, between TgCRND8 and CBKO/TgCRND8 (not shown), similar to the above results detected by JRF/Aβtot/17.
Cystatin B deletion restores learning and memory functions in TgCRND8 mice
Contextual fear conditioning
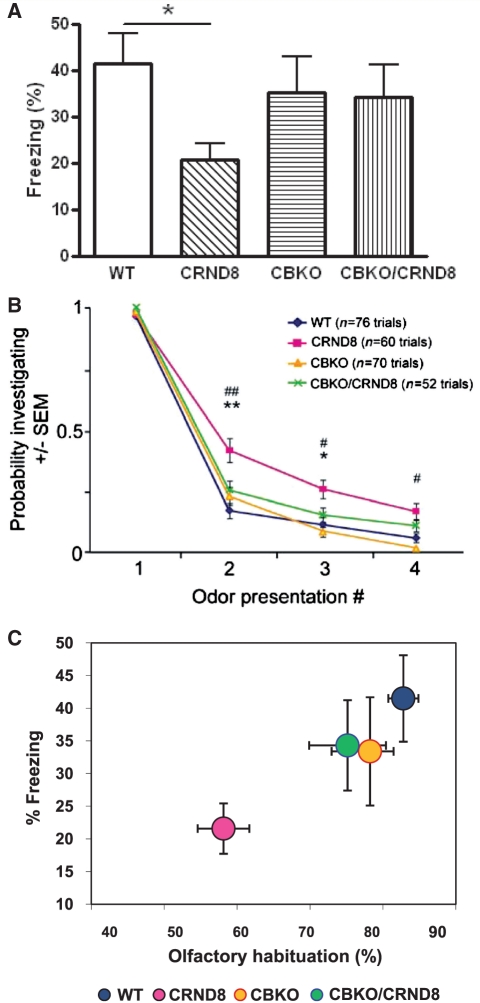
To test the effects of CBKO on memory, we analysed mice with a hippocampus-dependent contextual fear conditioning paradigm (Fig. 10A), which has been widely used for evaluating learning and memory deficits in APP transgenic mice (Kobayashi and Chen, 2005). In this test, mice learn to associate a distinct context (conditioned stimulus or chamber) with an aversive unconditioned stimulus (or foot-shock) that takes place in the context. Wild-type mice exhibited a robust conditioned fear response as measured by freezing (the absence of all movement except for that needed for breathing) when placed back into the conditioning chamber 24 h after training. TgCRND8 mice showed a significantly reduced duration of contextual freezing in comparison to wild-type littermate controls (55%, P < 0.05). In contrast, CBKO and CBKO/TgCRND8 mice showed equivalent durations of freezing that were not significantly different from those of wild-type controls. A fairly strong trend toward increases in freezing levels of CBKO/TgCRND8 mice compared to those of TgCRND8 mice (P = 0.08, two-tailed Student’s t-test) indicated the abilities of CBKO mice to improve contextual memory deficits found in TgCRND8 mice.
Figure 10.
Improved behavioural performance in the CBKO/TgCRND8. (A) Effects of CBKO on impaired contextual fear memory in TgCRND8 mice. The 6-month-old wild-type (WT) (n = 10), TgCRND8 (n = 9), CBKO (n = 10) and CBKO/TgCRND8 (n = 7) were trained with two conditioned stimulus-unconditioned stimulus pairings for contextual fear conditioning. Only TgCRND8 mice show significantly lower levels of contextual freezing than wild-type control mice when tested 24 h after training (*P < 0.05; two-tailed Student’s t-test). There is a trend toward increases in freezing of CBKO/TgCRND8 mice as compared to that of TgCRND8 mice (P = 0.08). (B) Reversal of APP olfactory behaviour phenotype in CBKO/TgCRND8 mice. Results from an odour habituation test from 6-month-old wild-type (n = 10), TgCRND8 (n = 8), CBKO (n = 9) and CBKO/TgCRND8 mice (n = 7). Odour investigation duration was normalized to a maximum value of ‘1’ per odour for each mouse. TgCRND8 mice (pink) displayed an increased latency to habituate to novel odours in comparison to wild-type (blue) and CBKO mice (orange), which was reversed when TgCRND8 was crossed with CBKO (CBKO/TgCRND8, green). *P < 0.05, **P < 0.01, CBKO/TgCRND8 versus TgCRND8; #P < 0.01, ##P < 0.0001, wild-type versus TgCRND8; ANOVA and Fisher’s protected least significant difference. (C) Comparison of the results from the two behavioural assays. To test a correlation between contextual and olfactory memory performances, percent freezing levels in the contextual fear conditioning are plotted against percent reductions of habituation from Trial 1 to Trial 2 in the odour habituation test. CRND8 = TgCRND8.
Odour habituation test
Recent work has shown that novel odour investigation behaviour positively correlates with Aβ burden in the brain (Wesson et al., 2010). We, therefore, measured novel odour investigation times in an odour habituation task (Wesson et al., 2010) with a particular focus on whether olfactory behaviour mirrored the pathological reversal in the CBKO/TgCRND8 mice.
Mice were presented in their home cages with cotton-applicator sticks laced with odour. Each odour was delivered across four successive trials and the duration of time spent investigating the odour recorded across trials was used to quantify olfactory habituation. Similar to previous reports in another APP transgenic mouse line (i.e. Tg2576) (Wesson et al., 2010), we found that TgCRND8 mice had an increased latency to habituate to novel odours in comparison to age-matched controls (Fig. 10B). There was a significant overall group effect, reported as [F(groups, degrees of freedom) = score, P-value], [F(3, 237) = 14.59, P ≤ 0.0001], a repeated measures effect [F(3, 237) = 1000.33, P < 0.0001], as well as a significant interaction [F(9,237) = 3.45, P = 0.0004]. The significant effect of repeated measures indicated that all groups were eventually able to habituate to odours by the fourth trial, however, with slight differences (Fig. 10B). Post hoc analyses of the group effect showed that wild-type, CBKO and CBKO/TgCRND8 mice habituated more rapidly to novel odours in comparison to TgCRND8 mice, achieving statistical significance by the second trial: wild-type [F(1, 120) = 19.45, P = <0.0001]; CBKO [F(1, 121) = 10.23, P = 0.0018]; CBKO/TgCRND8 [F(1, 104) = 7.05, P = 0.009]. Importantly, CBKO and CBKO/TgCRND8 mice did not significantly differ from wild-type mice across either Trials 2 or 3 (P > 0.05). These data demonstrate a reversal of the olfactory behaviour phenotype observed in TgCRND8 by virtue of crossing TgCRND8 with CBKO mice.
To examine the relationship between the memory phenotypes in both behavioural assays, we plotted freezing levels in the contextual fear conditioning (a hippocampal memory measure) versus reductions of probability investigating during Trial 2 compared to Trial 1 in the odour habituation test (an olfactory short-term working memory measure) (Fig. 10C). Both tasks clearly revealed poor performances for TgCRND8 mice, while the other three groups performed well and did not differ from each other. Therefore, our results consistently demonstrate that cystatin B deletion improves memory in TgCRND8 transgenic mice in two different learning paradigms.
Discussion
We have shown that aggressive amyloidosis and neuritic plaque development in TgCRND8 mice (Chishti et al., 2001) is accompanied by extensive autophagic-lysosomal pathology similar to that seen in Alzheimer’s disease brain (Cataldo et al., 1996; Nixon et al., 2005), reflecting defective proteolytic clearance of autophagic substrate. Moreover, we establish proof of concept that partially restoring lysosomal proteolytic function in TgCRND8 mice significantly ameliorates lysosomal system pathology, intraneuronal Aβ accumulation, amyloid plaque formation, and memory and learning deficits in the TgCRND8 model. These effects underscore the pathogenic significance of lysosomal system dysfunction in Alzheimer’s disease and they demonstrate the value of reversing this dysfunction as a potential therapy for Alzheimer’s disease.
Autophagic-lysosomal pathology in the TgCRND8 mice, carrying APP with two familial Alzheimer's disease mutations, is nearly as florid as that seen in the presenilin/APP mouse model of Alzheimer’s disease (Nixon et al., 2005; Yu et al., 2005), indicating that APP mutations alone can promote autophagic-lysosomal pathology in the absence of mutant presenilin, a genetic factor that greatly accelerates this pathology (Cataldo et al., 2004b; Nixon, 2007). Autophagic vacuole-filled dystrophic neurites are more prominent and widespread in Alzheimer’s disease than in other adult onset neurodegenerative diseases, and achieve the severity of lysosomal system pathology seen in lysosomal storage disorders caused by mutations of cathepsins or other mechanisms that reduce lysosomal proteolytic activity (Nixon et al., 2008). The marked autophagic-lysosomal system pathology in TgCRND8 mice included development of strikingly enlarged lysosome-related compartments in cortical and hippocampal neurons, which have been seen in several other models of β-amyloidosis with prominent intracellular Aβ accumulation (Langui et al., 2004; Belinson et al., 2008). We found that these compartments were positive not only for typical components of lysosomes, but also for markers of autophagosomes/autolysosomes (i.e. LC3) and late endosomes (i.e. lysobisphosphatidic acid) indicating an impairment in the clearance of autophagic substrates as well as endocytosed cargoes that reach lysosomes when late endosomes/multivesicular bodies fuse with autophagosomes (Filimonenko et al., 2007; Fader and Colombo, 2009).
Giant neuronal autolysosomes reflect reduced lysosomal proteolysis
Reduced lysosomal degradation of autophagic substrates in neurons of TgCRND8 mice is indicated by the delayed turnover of LC3-II—a marker of autophagosomes that is normally rapidly degraded after fusion with lysosomes (Mizushima and Yoshimori, 2007). LC3-II accumulated in autophagic vacuoles and lysosomes, and was at highest levels in the higher density autophagic vacuole fraction (i.e. AV20) containing the highest proportions of post-fusion autophagic vacuoles, indicating impaired degradation within autolysosomes. Elevated levels of ubiquitinated proteins in isolated autophagic vacuoles or lysosomes from TgCRND8 mice are further evidence for impaired autophagy in light of previous evidence that ubiquitinated proteins can be autophagy substrates and accumulate in neurons of autophagy-deficient mice (Klionsky, 2006; Komatsu et al., 2006). Also supporting our conclusion, the morphology of giant autolysosomes resembles that of ceroid-containing autophagic vacuoles that develop when lysosomal degradation is inhibited by deleting one or more cathepsins (Felbor et al., 2002; Koike et al., 2005) or by using cysteine protease inhibitors or general lysosomal enzyme inhibitors (Ivy et al., 1984; Boland et al., 2008).
Clearance of amyloid-β peptide within autolysosomes/lysosomes and its impairment in TgCRND8 mice
Evidence is emerging that accumulation of intraneuronal Aβ in Alzheimer’s disease is an early event that precedes extracellular Aβ deposition (Takahashi et al., 2002; Cataldo et al., 2004a; Billings et al., 2005) and is strongly implicated in the mechanism of neurodegeneration in these models (Christensen et al., 2008). Aβ immunoreactivity has been previously detected by immunocytochemistry in early endosomes, late endosome/multivesicular bodies and autophagic vacuoles in the brains of Alzheimer’s disease and Alzheimer’s disease mouse models (Takahashi et al., 2002; Cataldo et al., 2004a; Yu et al., 2005; Belinson et al., 2008). We detected Aβ accumulation in intracellular cathepsin D-positive compartments in TgCRND8 immunocytochemically, and for the first time, directly demonstrated Aβ accumulation within isolated autophagic vacuoles and lysosomes, showing also that Aβ was the predominant APP-related metabolite in these compartments.
Our data support previous evidence that APP is delivered to autophagosomes (Pickford et al., 2008) and that Aβ is both generated and degraded during autophagy (Yu et al., 2005; Nixon, 2007). Both endocytic and autophagic pathways are sites for APP processing and Aβ production, with dysfunction of the autophagy pathway now being increasingly regarded as an important contributor to Aβ accumulation in pathological conditions (Nixon, 2007). Aβ and other APP metabolites generated in these compartments are presumably delivered at least in part to lysosomes for degradation. Lysosomes are also the destination for internalized substrates, including Aβ that may be secreted from glia and neurons (Burdick et al., 1997; Chung et al., 1999). Several cathepsins, including cathepsin B, cathepsin D and cathepsin E, can cleave in vitro Aβ (Mackay et al., 1997; Mueller-Steiner et al., 2006). Moreover, increasing cathepsin B expression lowers brain Aβ levels (Mueller-Steiner et al., 2006), while Aβ levels increase after cathepsin B deletion (Mueller-Steiner et al., 2006) or after perturbing lysosomal degradative function with specific or general protease inhibitors in brains of mice in vivo (Mielke et al., 1997). Aβ from all of the aforementioned potential sources would, therefore, be expected to accumulate within lysosomal compartments if there were deficits in lysosomal degradative function.
Reversal of Alzheimer’s disease-related pathologies in TgCRND8 mice by cystatin B deletion
In this study, we have taken advantage of the CBKO mouse, despite its limitations as being also a developmental disease model, to determine whether or not increasing lysosomal proteolysis can rescue the pathophysiology seen in TgCRND8 mice. Here we show that cystatin B deletion in TgCRND8 mice largely reversed intracellular autophagic-lysosomal pathology and accumulation of incompletely digested substrates within autophagic-lysosomal compartments, including the autophagosome marker LC3-II, ubiquitinated proteins that accumulate when autophagy is blocked (Komatsu et al., 2006), as well as Aβ that may be generated in, or delivered to, lysosomes. It is interesting that not only the intraneuronal Aβ associated with giant autolysosomes decreased in the CBKO/TgCRND8 brain but extracellular amyloid load was also reduced, supporting a link between intracellular Aβ accumulation and extracellular amyloid deposition. In this regard, it has been shown that the intralumenal vesicles (exosomes) in multivesicular bodies contain Aβ that can be released into the extracellular space via exocytosis (Rajendran et al., 2006). It is possible that autolysosomes can discharge their contents into the extracellular space directly or indirectly by exchanging contents with other compartments in the endocytic and autophagic pathways (Nixon, 2007; Nixon et al., 2008). Reduced intracellular and extracellular Aβ load in CBKO/TgCRND8 brains was not related to a change in APP processing because levels of APP holoprotein, sAPPα, sAPPβ and APP CTF were similar in the brains of TgCRND8 and CBKO/TgCRND8 mice.
Although autophagy modulation and not cystatin B is the main focus of our study, our observations have provided additional evidence for the understanding of the function of cystatin B and its role in regulating lysosomal proteolysis in vivo. Unlike other members of the cystatin superfamily, such as the secreted cystatin C belonging to the cystatin family (Type 2) of the cystatin superfamily (Turk et al., 2008), we found using highly characterized affinity purified antibodies, that cystatin B is localized mainly to lysosomes and that its deletion stimulates lysosomal turnover of proteins. Cysteine cathepsin activities were higher in the brains of CBKO mice than wild-type and TgCRND8 mice, and similarly, were increased in CBKO/TgCRND8 mice compared to TgCRND8. These findings are consistent with those in cells from patients with myoclonus epilepsy of type 1 (Rinne et al., 2002), in which decreased levels of cystatin B messenger RNA and reduced cystatin B activity correlated with increased activities of cathepsin B, cathepsin L and cathepsin S. Interestingly, deletion of cystatin B in our study also increases the activity of cathepsin D, an aspartyl protease, which may suggest that enhanced activities of cysteine proteases such as cathepsin B and cathepsin L as a result of cystatin B deletion may promote the processing of cathepsin D from its proform to its cleaved active forms—a process for the activation of proteases that generally requires the involvement of processing enzymes(s) (Erickson, 1989). Our observation that not only the 32 kDa cleaved form, but also the 50 kDa proform, of cathepsin D are increased in AV20 and Lyso fractions from CBKO and CBKO/TgCRND8 brains (Fig. 8D), however, may imply the involvement of additional mechanisms underlying the effects of cystatin B deletion on cathepsin D, which remain to be explored in future studies. Enhanced lysosomal clearance of autophagic substrates in CBKO/TgCRND8 mice compared with TgCRND8 mice also accords with a previous report that the proteolysis of myristoylated alanine-rich C kinase substrate (MARCKS), a known substrate of cysteine cathepsins, is increased in the brain and macrophages from CBKO mice and is blocked by a cysteine protease inhibitor E64-d (Kopitar-Jerala and Turk, 2007).
The observations of reduced brain Aβ levels and plaque load in CBKO/TgCRND8 are similar to the findings from the study of ablation of cystatin C in the hAPP-J20 Alzheimer’s disease mouse model (Sun et al., 2008). However, the underlying mechanisms involved may or may not be the same given the differences in a number of aspects between these two cystatins, which belong to two families of the cystatin superfamily. These two cystatins differ in the number of amino acids, chemical bonds, secretory targeting signal, concentrations in body fluids, inhibitory properties, subcellular distribution, path of delivery into lysosomes, functions in autophagy and types of diseases in which they are involved/implicated (Machleidt et al., 1991; Abrahamson, 1994; Turk et al., 1997, 2008; Tizon and Levy, 2006). As one of the examples most relevant to our study, cystatin C has functions in inducing autophagy via the mTOR signalling pathway (Tizon et al., 2010), while cystatin B does not (unpublished data). We have demonstrated a general defect in autophagy as a basis for the pathology in the TgCRND8 mouse and reversal of the deficit in autophagy as the basis for the therapeutic effects of cystatin B deletion. We obtained other lines of evidence showing that cystatin B deletion improves clearance of abnormally accumulated proteins including Aβ within autophagic-lysosomal compartments, all supporting an autophagic mechanism for explaining the beneficial effects of cystatin B deletion. The main goal of our study was to investigate the impairments in autophagy in an aggressive model of Alzheimer’s disease pathology—TgCRND8 and possible therapeutic effects of lysosomal protease enhancement rather than to compare the mechanisms of action of the two cystatins. Nevertheless, it is necessary and interesting to clarify this latter issue in future studies to advance our knowledge about the roles of these endogenous protease inhibitors in normal neuronal functioning and in neurodegenerative diseases.
Rescued memory performance in TgCRND8 mice by cystatin B deletion
Memory performance in two distinct testing paradigms was almost completely rescued in TgCRND8 mice after cystatin B deletion. Memory deficits have been previously reported in TgCRND8 mice after the appearance of amyloid plaques (Janus et al., 2000; Hyde et al., 2005), although deficits do not necessarily correlate with levels of insoluble Aβ in these mice (Hanna et al., 2009) or other mouse models of amyloidosis (Holcomb et al., 1999; Lesne et al., 2006). Whereas cystatin B deletion halved amyloid plaque burden, it rescued lysosomal pathology and reversed intraneuronal Aβ accumulation more completely. In this regard, increasing evidence shows correlations between memory deficits and intracellular Aβ levels in mouse models in which intraneuronal Aβ accumulates at an early age, including the APP/PS1KI mice (Bayer and Wirths, 2008), the arcAβ mice (Knobloch et al., 2007) and the 3× Tg-Alzheimer’s disease mice (Billings et al., 2005). Although it is difficult to unequivocally determine whether intracellular, extracellular Aβ species, or both are responsible for causing memory deficits, our findings support the idea that the extensive clearance of intraneuronal Aβ accumulation by restoring autolysosomal proteolysis can lead to improved memory functions.
Targeting the autophagic-lysosomal pathway at degradative stages
Autophagy has been implicated in various neurodegenerative diseases, including Alzheimer’s disease (Nixon, 2006; Mizushima et al., 2008; Nixon et al., 2008; Shacka et al., 2008); however, the site of impairment within the pathway varies in the different disorders, which underscores the importance of correct targeting of a putative therapeutic modulation of autophagy. The observed therapeutic effects of improving lysosomal efficiency by cystatin B deletion are consistent with evidence for impaired autophagic substrate proteolysis as a basis for dystrophic neurite pathology in Alzheimer’s disease and Alzheimer’s disease mouse models (Nixon, 2007; Boland et al., 2008). Our results established proof of concept for lysosomal proteolysis restoration therapy in Alzheimer’s disease. It remains to be seen whether inducing autophagy in the absence of a correction of this proteolytic impairment would be therapeutic as seen in Huntington’s disease and several other proteopathies (Sarkar and Rubinsztein, 2008).
We used cystatin B gene deletion simply as a tool to selectively restore a more normal level of lysosomal proteolysis in TgCRND8 mice. However, it should be noted that it has been recently reported that cystatin B deficiency sensitizes cerebellar granule neurons to oxidative stress-induced cell death (Lehtinen et al., 2009), while other reports showed that exogenously adding cystatin B into cell culture is associated with cellular toxicity (Anderluh et al., 2005; Ceru et al., 2008). Further, it has also been reported that cystatin B is aggregate prone and overexpression of cystatin B in neuroblastoma cells generates cytoplasmic aggregates (Cipollini et al., 2008), and in cultured cells, cystatin B colocalizes with Aβ intracellular inclusions (Skerget et al., 2010). More importantly, in humans, loss-of-function mutations in the gene encoding cystatin B cause a developmental disorder, myoclonus epilepsy of type 1 (Joensuu et al., 2008), and cystatin B deletion in mice produces similar developmental degenerative changes in the cerebellum and brain atrophy and exhibits myoclonic seizure and ataxia (Pennacchio et al., 1998; Shannon et al., 2002; Houseweart et al., 2003). Previous studies have also reported gliosis in the cerebral hemisphere of CBKO mice (Lieuallen et al., 2001; Shannon et al., 2002) and a neuronal phenotype in which neuronal atrophy occurs in the superficial neurons of the prosubiculum, and apoptotic bodies could also be found ‘after extensive, careful searching’ (Shannon et al., 2002).
Consistent with these reports, we found neuronal cell death in the cerebellar granular cell layer in the CBKO and, similarly, also in the CBKO/TgCRND8 mice (not shown). However, examination of neuronal nuclei staining in the hippocampus of 4- and 6-month-old mice, an area of interest in our study, revealed similar staining patterns and cell densities among all four genotypes of mice, without appreciable signs of cell loss in CBKO or CBKO/TgCRND8 (Supplementary Fig. 4), which is consistent with our electron microscopy observations (Supplementary Fig. 1), and with the finding that CBKO mice have no measurable deficits in contextual fear conditioning (Fig. 10)—a hippocampus-dependent paradigm. As expected, CBKO mice showed more glial fibrillary acidic protein-staining in the cerebral cortex compared to wild-type mice. However, similar glial fibrillary acidic protein staining patterns were observed in the hippocampus of both CBKO and wild-type mice. Among the four genotypes, the strongest staining was observed in both the cerebral cortex and the hippocampus of TgCRND8 and it should be noted that this enhanced gliosis was reversed in CBKO/TgCRND8 (Supplementary Fig. 5).
Remarkably, despite the reported negative effects associated with cystatin B deletion, the data from the present study demonstrate that cystatin B deletion in TgCRND8 nevertheless improved lysosome/autophagy function and Aβ clearance, and rescued behavioural effects. A more practical potential approach to Alzheimer’s disease therapy, however, would need to involve the subacute modulation of cystatin B in mature brain or an equivalent approach to enhancing proteolytic activities in lysosomes. Although over-activating lysosomal proteolysis has potential limitations, evidence linking the lysosomal system to cell death relates to lysosomal membrane destabilization rather than to cathepsin overactivation (Kroemer and Jaattela, 2005; Nixon, 2006; Turk and Turk, 2009). In our study, even modest enhancement of lysosomal proteolytic activity was associated with significant ameliorative effects on multiple facets, supporting the conclusion that targeting late stages of the autophagic-lysosomal pathway to improve lysosomal degradation of intracellular Aβ and other potentially toxic autophagic substrates is a potential therapeutic approach for Alzheimer’s disease. This study provides proof of principal evidence supporting further investigations of pharmacological agents that can also normalize lysosomal proteolysis in the setting of Alzheimer’s disease and may have therapeutic value in Alzheimer’s disease and other neurodegenerative diseases.
Funding
National Institute on Aging (P01 AG017617 to R.A.N); and the Alzheimer’s Association (IIRG-08-90771 to D.-S. Y.).
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
We are grateful to Nicole Piorkowski for assistance in article preparation. We are also grateful to Drs Gurjinder Kaur, Yutaka Sato, M. Azhar Chishti and Megan K. Houseweart for assistance and reagents for studies, and Kerry Mullaney, Arthur Saltzman for technical assistance. We thank Drs Richard M. Myers for providing a breeding colony of CBKO mice and Dr Marialuisa Melli for providing the rat cystatin B cDNA.
Glossary
Abbreviations
- Aβ
amyloid-β peptide
- APP
amyloid precursor protein
- CBKO
cystatin B knockout
- LC3
microtubule-associated protein 1 light chain 3
References
- Abrahamson M. Cystatins. Methods Enzymol. 1994;244:685–700. doi: 10.1016/0076-6879(94)44051-4. [DOI] [PubMed] [Google Scholar]
- Alakurtti K, Weber E, Rinne R, Theil G, de Haan GJ, Lindhout D, et al. Loss of lysosomal association of cystatin B proteins representing progressive myoclonus epilepsy, EPM1, mutations. Eur J Hum Genet. 2005;13:208–15. doi: 10.1038/sj.ejhg.5201300. [DOI] [PubMed] [Google Scholar]
- Anderluh G, Gutierrez-Aguirre I, Rabzelj S, Ceru S, Kopitar-Jerala N, Macek P, et al. Interaction of human stefin B in the prefibrillar oligomeric form with membranes. Correlation with cellular toxicity. FEBS J. 2005;272:3042–51. doi: 10.1111/j.1742-4658.2005.04717.x. [DOI] [PubMed] [Google Scholar]
- Auteri J, Okada A, Bochaki V, Dice J. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J Cell Physiol. 1983;115:167–74. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- Bayer TA, Wirths O. Review on the APP/PS1KI mouse model: intraneuronal Abeta accumulation triggers axonopathy, neuron loss and working memory impairment. Genes Brain Behav. 2008;7(Suppl. 1):6–11. doi: 10.1111/j.1601-183X.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- Belinson H, Lev D, Masliah E, Michaelson DM. Activation of the amyloid cascade in apolipoprotein E4 transgenic mice induces lysosomal activation and neurodegeneration resulting in marked cognitive deficits. J Neurosci. 2008;28:4690–701. doi: 10.1523/JNEUROSCI.5633-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings LM, Oddo S, Green KN, McGaugh JL, Laferla FM. Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron. 2005;45:675–88. doi: 10.1016/j.neuron.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s Disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick D, Kosmoski J, Knauer MF, Glabe CG. Preferential adsorption, internalization and resistance to degradation of the major isoform of the Alzheimer’s amyloid peptide, A beta 1-42, in differentiated PC12 cells. Brain Res. 1997;746:275–84. doi: 10.1016/s0006-8993(96)01262-0. [DOI] [PubMed] [Google Scholar]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–93. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Properties of the endosomal-lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer’s disease. J Neurosci. 1996;16:186–99. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, et al. Down syndrome fibroblast model of Alzheimer-related endosome pathology. Accelerated endocytosis promotes late endocytic defects. Am J Pathol. 2008;173:370–84. doi: 10.2353/ajpath.2008.071053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Terio NB, Peterhoff CM, Durham R, Mercken M, et al. Abeta localization in abnormal endosomes: association with earliest Abeta elevations in AD and Down syndrome. Neurobiol Aging. 2004a;25:1263–72. doi: 10.1016/j.neurobiolaging.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Schmidt SD, Terio NB, Duff K, Beard M, et al. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol. 2004b;63:821–30. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]
- Ceru S, Kokalj SJ, Rabzelj S, Skarabot M, Gutierrez-Aguirre I, Kopitar-Jerala N, et al. Size and morphology of toxic oligomers of amyloidogenic proteins: a case study of human stefin B. Amyloid. 2008;15:147–59. doi: 10.1080/13506120802193555. [DOI] [PubMed] [Google Scholar]
- Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J, et al. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem. 2001;276:21562–70. doi: 10.1074/jbc.M100710200. [DOI] [PubMed] [Google Scholar]
- Christensen DZ, Kraus SL, Flohr A, Cotel MC, Wirths O, Bayer TA. Transient intraneuronal A beta rather than extracellular plaque pathology correlates with neuron loss in the frontal cortex of APP/PS1KI mice. Acta Neuropathol. 2008;116:647–55. doi: 10.1007/s00401-008-0451-6. [DOI] [PubMed] [Google Scholar]
- Chung H, Brazil MI, Soe TT, Maxfield FR. Uptake, degradation, and release of fibrillar and soluble forms of Alzheimer's amyloid beta-peptide by microglial cells. J Biol Chem. 1999;274:32301–8. doi: 10.1074/jbc.274.45.32301. [DOI] [PubMed] [Google Scholar]
- Cipollini E, Riccio M, Di Giaimo R, Dal Piaz F, Pulice G, Cathepsinania S, et al. Cystatin B and its EPM1 mutants are polymeric and aggregate prone in vivo. Biochim Biophys Acta. 2008;1783:312–22. doi: 10.1016/j.bbamcr.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21:142–50. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- DeDuve C, Wattiaux R. Functions of lysosomes. Annual Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Erickson AH. Biosynthesis of lysosomal endopeptidases. J Cell Biochem. 1989;40:31–41. doi: 10.1002/jcb.240400104. [DOI] [PubMed] [Google Scholar]
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–8. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, et al. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA. 2002;99:7883–8. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EMC, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna A, Horne P, Yager D, Eckman C, Eckman E, Janus C. Amyloid beta and impairment in multiple memory systems in older transgenic APP TgCRND8 mice. Genes Brain Behav. 2009;8:676–84. doi: 10.1111/j.1601-183X.2009.00510.x. [DOI] [PubMed] [Google Scholar]
- Holcomb LA, Gordon MN, Jantzen P, Hsiao K, Duff K, Morgan D. Behavioral changes in transgenic mice expressing both amyloid precursor protein and presenilin-1 mutations: lack of association with amyloid deposits. Behav Genet. 1999;29:177–85. doi: 10.1023/a:1021691918517. [DOI] [PubMed] [Google Scholar]
- Houseweart MK, Pennacchio LA, Vilaythong A, Peters C, Noebels JL, Myers RM. Cathepsin B but not cathepsins L or S contributes to the pathogenesis of Unverricht-Lundborg progressive myoclonus epilepsy (EPM1) J Neurobiol. 2003;56:315–27. doi: 10.1002/neu.10253. [DOI] [PubMed] [Google Scholar]
- Hyde LA, Kazdoba TM, Grilli M, Lozza G, Brusa R, Zhang Q, et al. Age-progressing cognitive impairments and neuropathology in transgenic CRND8 mice. Behav Brain Res. 2005;160:344–55. doi: 10.1016/j.bbr.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Ivy GO, Schottler F, Wenzel J, Baudry M, Lynch G. Inhibitors of lysosomal enzymes: accumulation of lipofuscin-like dense bodies in the brain. Science. 1984;226:985–7. doi: 10.1126/science.6505679. [DOI] [PubMed] [Google Scholar]
- Janus C, Pearson J, McLaurin J, Mathews PM, Jiang Y, Schmidt SD, et al. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer’s disease. Nature. 2000;408:979–82. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A, et al. Alzheimer’s-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci USA. 2010;107:1630–5. doi: 10.1073/pnas.0908953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu T, Lehesjoki AE, Kopra O. Molecular background of EPM1-Unverricht-Lundborg disease. Epilepsia. 2008;49:557–63. doi: 10.1111/j.1528-1167.2007.01422.x. [DOI] [PubMed] [Google Scholar]
- Kimura R, Ohno M. Impairments in remote memory stabilization precede hippocampal synaptic and cognitive failures in 5XFAD Alzheimer mouse model. Neurobiol Dis. 2009;33:229–35. doi: 10.1016/j.nbd.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Neurodegeneration: good riddance to bad rubbish. Nature. 2006;441:819–20. doi: 10.1038/441819a. [DOI] [PubMed] [Google Scholar]
- Knobloch M, Konietzko U, Krebs DC, Nitsch RM. Intracellular Abeta and cognitive deficits precede beta-amyloid deposition in transgenic arcAbeta mice. Neurobiol Aging. 2007;28:1297–306. doi: 10.1016/j.neurobiolaging.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Kobayashi DT, Chen KS. Behavioral phenotypes of amyloid-based genetically modified mouse models of Alzheimer’s disease. Genes Brain Behav. 2005;4:173–96. doi: 10.1111/j.1601-183X.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–28. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kopitar-Jerala N, Turk B. Cleavage of the myristoylated alanine-rich C kinase substrate (MARCKS) by cysteine cathepsins in cells and tissues of stefin B-deficient mice. Biol Chem. 2007;388:847–52. doi: 10.1515/BC.2007.092. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–97. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Langui D, Girardot N, El Hachimi KH, Allinquant B, Blanchard V, Pradier L, et al. Subcellular topography of neuronal Abeta peptide in APPxPS1 transgenic mice. Am J Pathol. 2004;165:1465–77. doi: 10.1016/s0002-9440(10)63405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Presenilin 1 (PS1) is required for v-ATPase targeting and autolysosome acidification: PS1 mutations in Alzheimer’s disease disrupt lysosomal proteolysis and autophagy. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Tegelberg S, Schipper H, Su H, Zukor H, Manninen O, et al. Cystatin B deficiency sensitizes neurons to oxidative stress in progressive myoclonus epilepsy, EPM1. J Neurosci. 2009;29:5910–5. doi: 10.1523/JNEUROSCI.0682-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieuallen K, Pennacchio LA, Park M, Myers RM, Lennon GG. Cystatin B-deficient mice have increased expression of apoptosis and glial activation genes. Hum Mol Genet. 2001;10:1867–71. doi: 10.1093/hmg/10.18.1867. [DOI] [PubMed] [Google Scholar]
- Machleidt W, Thiele U, Assfalg-Machleidt I, Forger D, Auerswald EA. Molecular mechanism of inhibition of cysteine proteinases by their protein inhibitors: kinetic studies with natural and recombinant variants of cystatins and stefins. Biomed Biochim Acta. 1991;50:613–20. [PubMed] [Google Scholar]
- Mackay EA, Ehrhard A, Moniatte M, Guenet C, Tardif C, Tarnus C, et al. A possible role for cathepsins D, E, and B in the processing of beta- amyloid precursor protein in Alzheimer’s disease. Eur J Biochem. 1997;244:414–25. doi: 10.1111/j.1432-1033.1997.00414.x. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–54. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews PM, Jiang Y, Schmidt SD, Grbovic OM, Mercken M, Nixon RA. Calpain activity regulates the cell surface distribution of amyloid precursor protein. Inhibition of calpains enhances endosomal generation of beta-cleaved C-terminal APP fragments. J Biol Chem. 2002;277:36415–24. doi: 10.1074/jbc.M205208200. [DOI] [PubMed] [Google Scholar]
- Mielke JG, Murphy MP, Maritz J, Bengualid KM, Ivy GO. Chloroquine administration in mice increases beta-amyloid immunoreactivity and attenuates kainate-induced blood-brain barrier dysfunction. Neurosci Lett. 1997;227:169–72. doi: 10.1016/s0304-3940(97)00340-6. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51:703–14. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–35. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–91. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Cataldo AM. Lysosomal system pathways: genes to neurodegeneration in Alzheimer’s disease. J Alzheimers Dis. 2006;9:277–89. doi: 10.3233/jad-2006-9s331. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–9. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Ohno M, Frankland PW, Chen AP, Costa RM, Silva AJ. Inducible, pharmacogenetic approaches to the study of learning and memory. Nat Neurosci. 2001;4:1238–43. doi: 10.1038/nn771. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bouley DM, Higgins KM, Scott MP, Noebels JL, Myers RM. Progressive ataxia, myoclonic epilepsy and cerebellar apoptosis in cystatin B-deficient mice. Nat Genet. 1998;20:251–8. doi: 10.1038/3059. [DOI] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Luzio JP. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic. 2001;2:612–21. doi: 10.1034/j.1600-0854.2001.20904.x. [DOI] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, et al. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103:11172–7. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio M, Di Giaimo R, Pianetti S, Palmieri PP, Melli M, Santi S. Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1) Exp Cell Res. 2001;262:84–94. doi: 10.1006/excr.2000.5085. [DOI] [PubMed] [Google Scholar]
- Rinne R, Saukko P, Jarvinen M, Lehesjoki AE. Reduced cystatin B activity correlates with enhanced cathepsin activity in progressive myoclonus epilepsy. Ann Med. 2002;34:380–5. doi: 10.1080/078538902320772124. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–86. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol Biosyst. 2008;4:895–901. doi: 10.1039/b804606a. [DOI] [PubMed] [Google Scholar]
- Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–36. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- Shannon P, Pennacchio LA, Houseweart MK, Minassian BA, Myers RM. Neuropathological changes in a mouse model of progressive myoclonus epilepsy: cystatin B deficiency and Unverricht-Lundborg disease. J Neuropathol Exp Neurol. 2002;61:1085–91. doi: 10.1093/jnen/61.12.1085. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerget K, Taler-Vercic A, Bavdek A, Hodnik V, Ceru S, Tusek-Znidaric M, et al. Interaction between oligomers of stefin B and amyloid-beta in vitro and in cells. J Biol Chem. 2010;285:3201–10. doi: 10.1074/jbc.M109.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Zhou Y, Halabisky B, Lo I, Cho SH, Mueller-Steiner S, et al. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron. 2008;60:247–57. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg H, Doving K, Novikov S, Ursin H. A method for studying responses and habituation to odors in rats. Behav Neural Biol. 1982;34:113–9. doi: 10.1016/s0163-1047(82)91501-1. [DOI] [PubMed] [Google Scholar]
- Takahashi RH, Milner TA, Li F, Nam EE, Edgar MA, Yamaguchi H, et al. Intraneuronal Alzheimer abeta42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol. 2002;161:1869–79. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizon B, Levy E. Protease inhibitors and their involvement in neurological disorders. In: Lajtha A, editor. Handbook of Neurochemistry and Molecular Neurobiology. New York: Springer Publishers; 2006. pp. 1–34. [Google Scholar]
- Tizon B, Sahoo S, Yu H, Gauthier S, Kumar AR, Mohan P, et al. Induction of autophagy by cystatin C: a mechanism that protects murine primary cortical neurons and neuronal cell lines. PLoS One. 2010;5:e9819. doi: 10.1371/journal.pone.0009819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, Turk V. Lysosomes as “suicide bags” in cell death: myth or reality? J Biol Chem. 2009;284:21783–7. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, Turk V, Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol Chem. 1997;378:141–50. [PubMed] [Google Scholar]
- Turk D, Turk B, Turk V. Papain-like lysosomal cysteine proteases and their inhibitors: drug discovery targets? Biochem Soc Symp. 2003:15–30. doi: 10.1042/bss0700015. [DOI] [PubMed] [Google Scholar]
- Turk V, Stoka V, Turk D. Cystatins: Biochemical and structural properties, and medical relevance. Front Biosci. 2008;13:5406–20. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- Wesson DW, Levy E, Nixon RA, Wilson DW. Olfactory perceptual correlates of β-amyloid plaque burden in Alzheimer’s disease mouse models. J Neurosci. 2010;30:505–14. doi: 10.1523/JNEUROSCI.4622-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DS, Lee JH, Nixon RA. Monitoring autophagy in Alzheimer’s disease and related neurodegenerative diseases. Methods Enzymol. 2009;453:111–44. doi: 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee J-H, et al. Macroautophagy–a novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.