Abstract
Cdc7-Dbf4 is a conserved, two-subunit kinase required for initiating eukaryotic DNA replication. Recent studies have shown that Cdc7-Dbf4 also regulates the mitotic exit network (MEN) and monopolar homolog orientation in meiosis I (Matos, J., Lipp, J. J., Bogdanova, A., Guillot, S., Okaz, E., Junqueira, M., Shevchenko, A., and Zachariae, W. (2008) Cell 135, 662–678 and Miller, C. T., Gabrielse, C., Chen, Y. C., and Weinreich, M. (2009) PLoS Genet. 5, e1000498). Both activities likely involve a Cdc7-Dbf4 interaction with Cdc5, the single Polo-like kinase in budding yeast. We previously showed that Dbf4 binds the Cdc5 polo-box domain (PBD) via an ∼40-residue N-terminal sequence, which lacks a PBD consensus binding site (S(pS/pT)(P/X)), and that Dbf4 inhibits Cdc5 function during mitosis. Here we identify a non-consensus PBD binding site within Dbf4 and demonstrate that the PBD-Dbf4 interaction occurs via a distinct PBD surface from that used to bind phosphoproteins. Genetic and biochemical analysis of multiple dbf4 mutants indicate that Dbf4 inhibits Cdc5 function through direct binding. Surprisingly, mutation of invariant Cdc5 residues required for binding phosphorylated substrates has little effect on yeast viability or growth rate. Instead, cdc5 mutants defective for binding phosphoproteins exhibit enhanced resistance to microtubule disruption and an increased rate of spindle elongation. This study, therefore, details the molecular nature of a new type of PBD binding and reveals that Cdc5 targeting to phosphorylated substrates likely regulates spindle dynamics.
Keywords: Cell Cycle, Cell Division, DNA Replication, Genetics, Yeast Genetics
Introduction
Cell cycle progression requires the highly accurate replication and segregation of chromosomes. Although these two events occur at different times, several cell cycle kinases regulate both DNA synthesis and chromosome segregation (3). In budding yeast the Cdc7-Dbf4 kinase (also called Dbf4-dependent kinase or DDK)2 plays such a dual role in the cell cycle. The Dbf4 regulatory subunit binds to and activates Cdc7 kinase to initiate DNA replication (4, 5). DDK also promotes other aspects of chromosome biology including cohesin loading during early S-phase in Xenopus laevis (6), centromeric cohesion in Schizosaccharomyces pombe (6), and meiotic recombination (7, 8) and the Ndt80 (early meiotic) transcriptional program in Saccharomyces cerevisiae (9). Budding yeast DDK also promotes monopolar orientation of homologs in meiosis I and inhibits chromosome segregation in the mitotic cycle (1, 2, 10, 11). Both activities are likely mediated through an interaction with Cdc5, the single Polo-like kinase in S. cerevisiae. Polo-like kinases (Plks) regulate mitotic events and are also involved in the response to DNA damage and checkpoint adaptation (12–14). Genetic and physical interactions between Dbf4 and Cdc5 were described many years ago (15, 16) raising the possibility that DDK acted beyond S phase. The DDK-Cdc5 interaction raises interesting questions regarding how these distinct kinases interact and coordinate accurate cell cycle progression.
The Polo gene was named for a Drosophila melanogaster mutant that exhibited abnormal spindle pole behavior (17), implying that Polo had a critical role in mitotic organization. Polo kinases are now known to comprise a large protein family that regulates centrosome maturation and duplication, mitotic entry, chromosome segregation, spindle dynamics, and mitotic exit (18). Budding yeast, fission yeast, and Drosophila each have a single Polo orthologs, but there are four Polo-like kinases (Plk1–4) in mammalian cells (18). Consistent with their diverse functions, individual Plks show different and sometimes dynamic subcellular localization (19). Polo kinases share a two-domain structure consisting of an N-terminal kinase domain and a C-terminal substrate binding domain. A unique C-terminal polo-box domain (PBD) comprised of one or two polo-box motifs was found in all Polo family members by multiple sequence alignment (20) and is required for Plk subcellular localization and substrate targeting (13, 21, 22). The PBD is one of many domains that binds phosphorylated substrates (23). The interaction between an optimal phosphothreonine peptide and the PBD of Plk1 has been defined by structural and mutational studies (24, 25). The polo-box domains of Plk1–3 orthologs are constituted from two highly conserved polo-box sequences, called PB1 and PB2, together with a polo cap region that stabilizes the folded domain. More than 600 Plk substrates were suggested in a proteomic study using the phosphorylation recognition feature of the PBD (26), suggesting that Plks regulate many substrates. Because Plk1 overexpression occurs in human tumors, Polo kinases are attractive targets for cancer therapy (27). In fact, different molecular approaches are being developed to inhibit both Plk1 kinase activity and its noncatalytic substrate binding domain (27–30).
The CDC5 gene was first described in a cell division cycle mutant screen by Hartwell et al. (31) through the isolation of a single cdc5-1 temperature-sensitive allele. Like the other Polo family members, Cdc5 has multiple roles in mitosis and cytokinesis (13). Human Plk1 can complement the growth defect of the yeast cdc5-1 mutant, which provided further evidence that Polo functional interactions were conserved during evolution (32, 33). Despite a broad spectrum of potential Cdc5 substrates, only a few PBD binding interactions have been characterized in detail (34–39). We recently performed a two-hybrid screen using the Dbf4 N terminus and defined a Dbf4 interaction with the Cdc5 PBD (2). We further found that Dbf4 residues 66–109 were necessary and sufficient for this interaction. However, this Dbf4 region did not contain a recognizable PBD consensus binding sequence, i.e. (S(pS/pT)(P/X) (p denotes phosphorylation, and X indicates any amino acid), and mediated an interaction with the PBD without a requirement for phosphorylation. Similarly, Glover and co-workers (40) reported that the PBD of Drosophila Polo mediates an interaction with Map205 (a microtubule-associated protein) that occurs in the absence of Map205 phosphorylation.
Here we systematically map Dbf4 residues required for binding the PBD using genetic and direct peptide binding assays. Although targeted deletion of Dbf4 residues 83–88 or 89–93 completely abrogates Dbf4-Cdc5 binding in vivo, only residues 83–88 are critical for a direct PBD interaction and comprise the core of a new type of PBD binding sequence. Furthermore, the PBD interacts with Dbf4 independently of residues that mediate its interaction with phosphorylated proteins using a distinct molecular surface. Surprisingly, highly conserved Cdc5 residues (Trp-517, His-641, Lys-643) in the PBD, required for binding proteins with an S(pS/pT)P/X consensus sequence, are not required for yeast viability or wild type growth rates. This strongly suggests that Cdc5 binding to phosphorylated (primed) substrates is not essential in yeast. Instead, the cdc5-HK and cdc5-WHK mutants exhibit enhanced resistance to spindle poisons and display altered spindle dynamics. These data define an alternative mode for PBD-protein interactions and raise the possibility that Cdc5 may bind some essential mitotic substrates through a Dbf4-like consensus sequence.
EXPERIMENTAL PROCEDURES
Construction of Yeast Strains, Plasmids, and Baculoviruses
Strains and plasmids used in this study are listed in supplemental Tables 1 and 2. PJ69–4a cells (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met::GAL7-lacZ) were used for two-hybrid experiments. All other strains were derivatives of W303 (MATa ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3). The CDC5 shuffle stain M1672 (cdc5Δ::kanMX6/pMW536 [CDC5 URA3 ARS-CEN]) was constructed using the same procedure for the DBF4 shuffle strain, M895, as previously described (41). To integrate dbf4 mutants, HindIII-XbaI fragments containing full-length dbf4-Δ82–88 or dbf4-R83E were co-transformed into M895 (dbf4Δ::kanMX6/pMW490 [DBF4 URA3 ARS-CEN]) together with pRS415. Leu-positive transformants were replica-plated on 5-fluoro-orotic acid. Multiple 5-fluoro-orotic acid-resistant colonies were recovered to YPD ((1% yeast extract, 2% Bacto-peptone and 2% glucose)) plates and then tested on YPD plates containing 0.2 mg/ml Geneticin to score loss of the kanMX6 marker. The resulting Geneticin-sensitive candidates were confirmed as correct recombinants following PCR amplification of the DBF4 locus and then backcrossed to W303. The epitope-tagged Cdc5 strains were made by the method of Longtine et al. (42).
Deletions and point mutations within DBF4 and CDC5 were generated by site-directed mutagenesis using the QuikChange system (Stratagene). PCR-amplified NcoI-PstI fragments containing the full-length DBF4 coding sequence or various dbf4 mutants were cloned into the same sites of pGBKT7 (ClontechClontech) to give the GalDBD-Dbf4 fusions. CDC5 (−332 to +2360) was PCR-amplified from genomic DNA and cloned into the HindIII-XbaI sites of pRS415 and pRS416 to give pMW535 and pMW536, respectively. Spc72 residues 1–400 were PCR-amplified from genomic DNA and cloned into the NdeI-BamHI sites of pGBKT7 to give pYJ356. For high-copy number plasmids, HindIII-NotI fragments containing entire WT DBF4 or various dbf4 mutants were cloned into the same sites of pRS425. Cdc5 residues 357–705 were cloned on a BamHI-XhoI into pET24a-GST (a gift of Eric Xu, Van Andel Research Institute) for expression of His6-GST-PBD.
Construction of baculovirus plasmids encoding WT Dbf4, Dbf4-NΔ109, HA-Cdc7, and 3Myc-Cdc5 was previously described (41). An NcoI-NotI fragment containing dbf4-Δ82–88 was cloned in the baculovirus transfer vector, pAcSG2. High-titer baculoviruses were generated by transfection of Sf9 cells using the BaculoGold kit (BD Biosciences) followed by plaque purification and virus amplification.
Growth Conditions, Cell Cycle Synchronization, and Immunofluorescence
Cells were cultured in YPD. Synthetic Complete medium (SCM) (43) was supplemented with 5% glucose or 2.5% galactose. Benomyl (Sigma) was added directly to plates immediately before pouring (final 0.2% DMSO (v/v)). Synchronous G1 or G2/M cultures were obtained after the addition of 5 μg/ml α-factor or 15 μg/ml nocodazole, respectively, for 3 h. DNA content was analyzed by flow cytometry as previously described (44). Tubulin and DAPI staining was previously described (45). Spindle length was measured by 60× objective using a Nikon Eclipse TE300 fluorescence microscope and OpenLab Version 3.1.7 image analysis software.
Two-hybrid Analysis
Various DBF4 bait constructs containing Gal4 DNA binding domain were transformed with pGAD-Cdc5.3 (Gal activation domain fusion to Cdc5357–705) in PJ69–4a and selected on SCM plates lacking tryptophan and leucine. These were spotted at 10-fold serial dilutions on the same plates and also on plates also lacking histidine but containing 2 mm 3-aminotriazole at 30 °C and cultured for 2–3 days.
Yeast Whole-cell Extracts, Immunoprecipitations from Sf9 Cells, and Western Blotting
Yeast protein extracts were prepared for Western blotting by trichloroacetic acid extraction (46). Blots were probed in phosphate-buffered saline containing 0.1% Tween containing 1% dried milk. Dbf4 bait constructs containing a Myc tag were detected using anti-Myc monoclonal antibody (9E10, 1:2000) followed by anti-mouse-HRP secondary antibody. Sf9 cells were co-infected with HA-Cdc7, 3Myc-Cdc5, and Dbf4 mutants as previously described (41). Whole cell extracts and immunoprecipitations were probed with polyclonal antibodies against Cdc7 (1:4000) and Dbf4 (1:1000). 3Myc-Cdc5 was detected with 9E10.
Protein Purification and Peptide Binding Assays
His6-GST-Cdc5 (PBD) was induced in BL21 cells for 3 h at 30 °C using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were sonicated in PBS containing 1% Triton X-100, and GST proteins were purified from soluble extracts by binding to glutathione-agarose (Amersham Biosciences) and eluted in the buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, and 10% glycerol) containing 5 mm glutathione and dialyzed against 50 mm MOPS (pH 7.4), 100 mm NaCl, and 10% glycerol.
Dbf4 peptide-PBD binding was quantitated using the AlphaScreen luminescence proximity assay (PerkinElmer Life Sciences) using a histidine detection kit as described (47). Binding mixtures containing 50 nm N-terminal-biotinylated Dbf4 peptide (Biotin-EKKRARIERARSIEGAVQVSKGTG), 50 nm His6-GST-PBD, 15 μg/ml streptavidin-coated donor beads, and nickel-chelate-coated acceptor beads were incubated in buffer containing 50 mm MOPS (pH 7.4), 100 mm NaCl, 0.1 mg/ml BSA for 1 h. Luminescence was recorded in a 384-well plate using an Envision 2104 plate reader (PerkinElmer Life Sciences). For competition assays, titrated unlabeled peptides were added and incubated at room temperature for 1 h before measurement. Nonlinear regression as implemented in Prism 5.0 (GraphPad Software, San Diego) was used to fit the data to a variable slope dose-response inhibition equation to determine IC50 values. All peptides used in this study are listed in supplemental Table 3.
RESULTS
Dbf4 Residues 82–96 Are Required to Interact with the Cdc5 PBD
We previously recovered multiple clones of the Cdc5 PBD in a two-hybrid screen using the Dbf4 N terminus as bait and found that residues 66–109 are necessary and sufficient for a direct interaction with Cdc5 PBD (2). Because Dbf4 N-terminal residues 1–109 are dispensable for DNA replication (41), the Dbf4 N terminus interacts with Cdc5 to perform nonessential functions in budding yeast. To define the exact molecular basis of the Dbf4-Cdc5 interaction, we constructed a series of N-terminal Dbf4 deletion mutants and tested their ability to interact with the Cdc5 PBD using a two-hybrid assay. Deletions to residue 82 did not significantly affect the PBD two-hybrid interaction; however, N-terminal deletions extending beyond residue 82 lost the ability to interact with the PBD (Fig. 1, A and B). We then truncated the C terminus and found that Dbf4 residues 66–96 were sufficient for PBD binding. Deletion of residues 82–88 (as shown previously (2)), 89–93, or 82–96 eliminated PBD binding (Fig. 1, A and B). This data indicated that sequences between residues 82 and 96 were essential for the Dbf4-Cdc5 interaction but did not define which residues directly contact the PBD.
FIGURE 1.
Mapping the interaction between Dbf4 and the Cdc5 PBD. A, N-terminal Dbf4 deletion mutants were tested for a two-hybrid interaction with the PBD. 10-Fold serial dilutions of saturated cultures were spotted onto SCM−Trp−Leu plates to visualize total cells and SCM−Trp−Leu−His + 2 mm 3-aminotriazole (3-AT) plates to score the two-hybrid interaction. aa, amino acids. B, a schematic of the features in Dbf4 N terminus is shown including two potential destruction boxes (D-boxes), a conserved BRCT-like domain, and motifs N, M, and C along with a summary of the Dbf4-PBD two-hybrid data. C, two-hybrid results for various point mutants spanning Dbf4 residues 82–96 are summarized. Arg-83, Ile-85, Gly-87, and Ala-88 are critical for PBD binding. D, HA-Cdc7-Dbf4 complexes were immunoprecipitated (IP) from baculovirus-infected Sf9 cells and examined for co-immunoprecipitation of 3Myc-Cdc5. Cdc5 was co- immunoprecipitated by wild type Dbf4 but not by Dbf4-Δ82–88 and Dbf4-NΔ109 mutant proteins. WCE, whole cell lysates; WB, Western blot.
A Novel Binding Motif for the Cdc5 PBD
Cdc5 is most closely related to the Plk1 family, and its C-terminal PBD shares about 36% identity with the PBD of human Plk1 (13). Using an oriented peptide-library screen, the PBD of Cdc5 and Plk1 were found to preferentially bind S(pS/pT)(P/X) peptides (25). For both Plk1 and Cdc5, the serine preceding the phosphorylated residue is absolutely required for PBD binding in vitro (25, 48). The current model for Polo targeting suggests that a priming kinase, such as a cyclin-dependent kinase or mitogen-activated protein kinase, phosphorylates selected Ser/Thr residues on Polo substrates to create a high affinity PBD recognition motif (19, 25). Plk1 also uses self-priming to create its own high affinity binding site on PBIPB1 (49). However, several Cdc5 or Plk substrates apparently do not require the priming kinases for PBD binding (40, 50, 51), and Dbf4 residues 82–96 do not contain a match to the PBD consensus binding site.
To determine individual Dbf4 residues required for PBD binding, we constructed a series of point mutants spanning residues 82–96 and quantitated the Dbf4 two-hybrid interaction with the PBD. Mutations of residues Arg-83, Ile-85, Gly-87, and Ala-88 completely abrogated the interaction with the PBD similar to deletion of residues 82–88 (Fig. 1C; see supplemental Fig. S1 for two-hybrid spotting data), although the mutant proteins were expressed similarly to the wild type (supplemental Fig. S2 and data not shown). In contrast, the A82V, S84A, S84E, E86A, and E86K mutations had little effect on the Dbf4-PBD interaction (Fig. 1C). Although the V89A/Q90A/V91A mutation had a modest effect on PBD binding, the S92A/K93A and G94A/T95A/G96A mutations had no effect (Fig. 1C). The V89A, Q90A, and V91A single point mutations also had no effect on the Dbf4-PBD interaction (supplemental Fig. S1). Together, these observations suggest that Dbf4 residues 83–88 directly bind the Cdc5 PBD in a phosphorylation-independent manner. Because deletion of residues 89–93 eliminated the PBD interaction but mutation of individual amino acids within this sequence had no effect on binding, it is likely that residues 89–93 do not directly contact the PBD or they make nonessential contacts. Based on this point mutant analysis we suggest that Dbf4 residues 83RSIEGA88 comprise the core of a novel PBD binding motif.
Last, we tested whether residues 82 and following were required for PBD binding in the context of full-length Dbf4. Although full-length Dbf4 interacted with the PBD, dbf4 mutants deleting past residue 82 failed to interact (supplemental Fig. S3), indicating that these residues were critical for the interaction in full-length Dbf4. To examine Dbf4-Cdc5 in the context of functional Cdc7-Dbf4 kinase (DDK), we tested the ability of wild type Dbf4 and Dbf4-Δ82–88 proteins to co-immunoprecipitate Cdc5 using a baculovirus expression system. HA-Cdc7, Dbf4, and Myc-Cdc5 proteins were co-expressed in Sf9 cells, and the HA-Cdc7-Dbf4 complex was immunoprecipitated using an anti-HA monoclonal antibody. Cdc5 was co-immunoprecipitated by DDK complexes containing wild type Dbf4 but not by DDK complexes containing the Dbf4-Δ82–88 and Dbf4-NΔ109 mutant proteins (Fig. 1D). These data indicate the DDK-Cdc5 interaction requires Dbf4 residues 82–88.
A 14-mer Dbf4 Peptide Containing Residues 83–88 Is Sufficient for the PBD Interaction
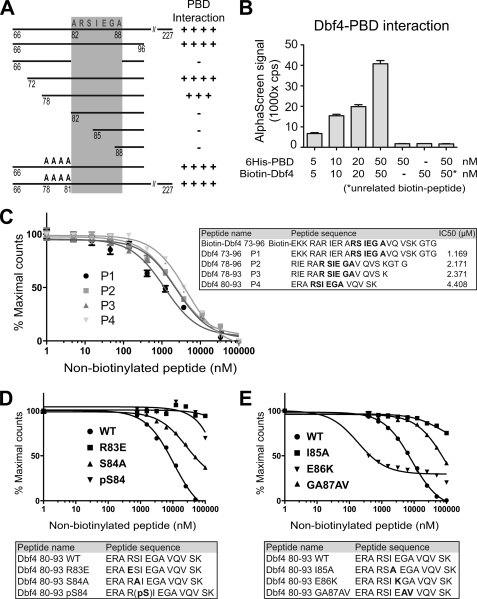
We next defined the minimal Dbf4-interacting peptide using the two-hybrid assay. A short Dbf4 peptide containing only residues 78–96 was sufficient for PBD binding (Fig. 2A). Although residues 82–96 did not bind the PBD, this was likely due to assay constraints and not due to loss of critical residues from 78–81, i.e. a quadruple alanine mutant of residues 78–81 (RIER to AAAA) bound the PBD as well as wild type Dbf4 66–96 (Fig. 2A). This demonstrates that a 19-amino acid peptide (78–96) containing the Dbf4 sequence 83RSIEGA88 is sufficient for PBD binding.
FIGURE 2.
A novel, non-consensus polo-box binding sequence in Dbf4. A, Dbf4 N-terminal deletion mutants were constructed to determine the minimal region (residues 78–96) required for a two-hybrid interaction with the PBD. B, a biotinylated Dbf4 peptide (residues 73–96, panel C) was tested for interaction with purified His6-PBD using the AlphaScreen assay. Data represent the average of three independent experiments ± S.D. C, the Dbf4(73–96)-PBD interaction was competed with the indicated non-biotinylated, wild type Dbf4 peptides (P1–P4), or mutant Dbf4 peptides as indicated in (D and E).
To verify that Dbf4 residues 82–88 comprise a unique PBD binding motif, we first tested the ability of Dbf4 peptides to directly interact with the purified Cdc5 PBD using the AlphaScreen assay (52). In this assay, a biotinylated Dbf4 peptide and purified His6-PBD are bound to streptavidin (donor) and Ni2+ (acceptor) beads, respectively. Excitation with 680-nm light causes donor beads to emit singlet oxygen, which activates fluorophores in proximally bound acceptor beads to emit light at 520–620 nm. A biotinylated Dbf4 peptide (73–96), but not an unrelated peptide, interacted with purified Cdc5 polo-box domain (residues 357–705) in a dose-dependent manner (Fig. 2B).
To confirm the specificity of the binding assays, non-biotinylated Dbf4 peptides of differing lengths but containing the Dbf4-RSIEGA sequence were tested for their ability to compete the biotinylated-peptide PBD interaction. Four different peptides ranging from 24 to 14 residues showed a similar ability to compete the Dbf4-PBD interaction (Fig. 2C), strongly suggesting that the RSIEGA residues directly bind the PBD. The affinity of Dbf4 peptide binding to the PBD in this assay was 1–5 μm as determined by competition with an unlabeled Dbf4 peptide (Fig. 2C).
Dbf4 Uses Four Key Residues to Bind the PBD, and Binding Is Inhibited by Phosphorylation
We used peptide competition assays to determine how mutations in the Dbf4-RSIEGA sequence affected PBD binding. Dbf4-R83E, -I85A, and -GA87AV peptides (containing mutations that disrupted the Dbf4-PBD two-hybrid interaction) lost the ability to compete with the Dbf4-PBD interaction even when the peptide concentration was increased to 10 μm (Fig. 2, D and E). However, the Dbf4-S84A and -E86K peptides, which still interacted with Polo in the two-hybrid assay, competed the Dbf4-PBD interaction in vitro (Fig. 2, D and E). These data are in complete agreement with the interaction map produced by two-hybrid data.
Interestingly, the Dbf4-E86K peptide bound to the PBD with higher affinity than the wild type peptide (10–100 nm, Fig. 2E), and importantly, the Ser-84-phosphorylated peptide lost the binding ability to PBD (Fig. 2D). Therefore, although the S84A and S84E (phosphomimetic) mutants did not noticeably affect the Dbf4-PBD interaction in the two-hybrid assay (Fig. 1C), a Ser(P)-84 residue blocked the interaction in vitro. These data indicate that Ser-84 phosphorylation is not simply dispensable for the Dbf4-PBD interaction, but it blocks the interaction in vitro, suggesting that an entirely different type of PBD-protein interaction occurs.
Mutants Altering Critical Residues in the Dbf4 PBD Binding Motif Suppress the cdc5-1 Temperature Sensitivity
Deletion of the Dbf4 N-terminal 109 amino acids suppresses the temperature-sensitive (ts) lethal phenotype of the cdc5-1 mutant (Ref. 2 and Fig. 3A). Cdc5-1 protein contains a P511L missense mutation immediately preceding the PB1 motif and retains significant kinase activity at the non-permissive temperature (53) but is unable to promote mitotic exit (54). The suppression data suggest that Dbf4 (DDK) might inhibit Cdc5-1 binding to critical targets required for mitotic exit. Using integrated alleles, we found dbf4-Δ82–88 and dbf4-R83E that are defective for the Polo interaction suppressed the cdc5-1 ts. The cdc5-1 mutant grew poorly at 30 °C, but the double mutants grew well at 30 °C and also suppressed the cdc5-1 ts at 32 °C (Fig. 3A). Although the dbf4-Δ82–88 and dbf4-R83E mutants suppressed the cdc5-1 ts at 32 °C, there was little suppression at 34 °C compared with the dbf4-NΔ109 mutant. This suggests that additional residues in the Dbf4 N-terminal 109 contribute to robust suppression of the cdc5-1 ts at 34 °C.
FIGURE 3.
Dbf4-RSIEGA mutants suppress the cdc5-1 temperature sensitivity. A, the indicated strains W303-1A, dbf4-Δ82–88 (M2805), cdc5-1 (M1614), cdc5-1 dbf4-Δ82–88 (M3112 and M3114), cdc5-1 dbf4-R83E (M3116, M3117), and cdc5-1 dbf4-NΔ109 (M2655, M2656) were spotted onto YPD plates and scored for growth at the indicated temperatures. B, various dbf4 deletions on an ARS CEN plasmid (pMW489) were introduced into M2600 (cdc5-1 dbf4Δ::kanMX6) and scored for growth by spotting serial dilutions on YPD media at the indicated temperatures. C, shown is a summary of dbf4 mutations, their effect on the Dbf4-PBD interaction, and suppression of the cdc5-1 ts. dbf4 mutants were scored for growth in the M2600 (cdc5-1 dbf4Δ::kanMX6) background by spotting serial dilutions on YPD media at increasing temperatures (supplemental Fig. S4). D, high copy plasmids expressing wild type DBF4, and the indicated mutants were transformed into M1614 (cdc5-1). Cultures were spotted onto SCM−Leu plates at 25 °C, indicating that high copy dbf4-NΔ65 lethality is alleviated by deleting residues 82–88. E, expression of the Dbf4 N terminus from the GAL1,10 promoter is lethal to cdc5-1 cells only if Dbf4 retains the ability to interact with Cdc5 as occurs in the WT, S84A, S84E, E86A, and E86K mutants.
To more closely examine the correlation between loss of the Polo interaction and the ability to suppress the cdc5-1 ts, we created a DBF4 plasmid shuffle system in the cdc5-1 background and tested the ability of various dbf4 mutants (expressed from the endogenous DBF4 promoter) to suppress the cdc5-1 ts. We found that the dbf4-Δ66–109, dbf4-Δ76–109, and dbf4-Δ82–109 alleles suppressed the cdc5-1 ts similar to the dbf4-NΔ109 allele. However, dbf4-Δ94–109 or dbf4-Δ100–109 that retain residues 83–88 did not (Fig. 3B). These latter two mutants retain the ability to bind Cdc5 in the two-hybrid assay (data not shown). In the cdc5-1 plasmid shuffle strain, the dbf4-NΔ109, dbf4-Δ66–109, dbf4-Δ76-109, and dbf4-Δ82-109 mutants exhibit very similar growth properties at 32 °C (Fig. 3B) but did not grow at 34 °C (not shown). So in this system, the larger N-terminal deletion is phenocopied by smaller deletions removing the Cdc5 binding site. We then examined dbf4-R83A, -R83E, -I85A, -G87A, and -A88V alleles, which alter residues critical for PBD binding in the two-hybrid and AlphaScreen assays. As expected, these mutants suppressed the cdc5-1 ts at 30 °C and 32 °C like the dbf4-Δ82–88 cdc5-1 mutant (summarized in Fig. 3C). In contrast, we observed no ts suppression by the dbf4-S84A and dbf4-E86K alleles, which still interacted with the PBD. We observed a strict correlation among mutants in residues 82–88 between loss of Polo binding and suppression of the cdc5-1 ts (Fig. 3C). This indicates that loss of the Dbf4 physical interaction with Cdc5 suppresses the cdc5-1 growth defect at restrictive temperatures.
The dbf4-S84A and dbf4-S84E mutants that still interact with Cdc5 did not suppress the cdc5-1 ts. Accordingly, the dbf4-E86K mutant exacerbated the cdc5-1 growth defect (supplemental Fig. S4B), consistent with the fact that the Dbf4-E86K peptide bound with higher affinity to the PBD than the wild type. These observations underscore that although alterations of Dbf4 residues Ser-84 and Glu-86 within the PBD binding motif can influence the PBD interaction, the Ser-84 and Glu-86 residues do not make essential contacts required for the PBD interaction.
Dbf4 Inhibits Cdc5 by Directly Binding the PBD
We previously found that a chromosomal dbf4-NΔ65 mutant that interacts with Cdc5 but is stabilized by deleting several d-boxes was synthetic sick or lethal in combination with cdc5-1 (2). This supports the model that Dbf4 inhibits the essential function of Cdc5. This hypothesis is supported by a recent report that a dbf4-NΔ65 mutant can inhibit ribosomal DNA segregation under certain circumstances (10), as rDNA segregation during anaphase is regulated by Cdc5 activation of the FEAR (Cdc14 early anaphase release) pathway (55). We investigated whether Dbf4 residues 82–88 are required to inhibit Cdc5 activity by overexpressing Dbf4 from high-copy plasmids. Increased expression of wild type DBF4 was deleterious to cdc5-1, but overexpression of dbf4-NΔ65 was lethal to cdc5-1 (Fig. 3D). Deletion of residues 82–88 rescued the synthetic lethality between dbf4-NΔ65 and cdc5-1, strongly suggesting that Dbf4 inhibits Cdc5 function through a direct interaction. Similarly, overproduction of the isolated Dbf4 N terminus (residues 1–225) from the GAL1, 10 promoter was lethal to cdc5-1 cells (Fig. 3E) (2). In contrast, overproduction of the Dbf4-Δ82–88 and Dbf4-R83E peptides that fail to interact with Cdc5 was not lethal. Taken together with the cdc5-1 ts suppression results, these data indicate that the Dbf4-RSIEGA sequence is required to inhibit Cdc5 activity by direct binding to the PBD.
The PBD Interacts with Dbf4 Using a Surface Distinct from Its Phosphopeptide Binding Surface
Among the Cdc5 PBD substrates that have been described in detail, the spindle pole body protein Spc72 was found to bind the PBD through its SpSP motif (Snead et al. (34)). We confirmed that purified His6-PBD bound the Spc72 phosphopeptide in vitro (Fig. 4A) with an IC50 of ∼2 μm (data not shown) that is very similar to the Dbf4-PBD interaction. To test whether Dbf4 and Spc72 peptides bound to distinct surfaces of the PBD, we performed competition assays using a non-biotinylated Spc72 phosphopeptide to compete the Dbf4-PBD interaction. Although a wild type Dbf4 peptide spanning residues 80–93 effectively competed the Dbf4-PBD interaction, the phosphorylated Spc72 peptide (containing S-pS-P) did not (Fig. 4B). This non-competitive result strongly suggests that two specific binding sites exist on the PBD, one for Dbf4 and another for phosphorylated substrates.
FIGURE 4.
Dbf4 binds a surface on the PBD distinct from its phospho-protein binding site. A, a biotinylated Spc72 phospho-peptide (residues 223–242) bound the PBD in the AlphaScreen assay. B, the same (non-biotinylated) Spc72 phosphopeptide did not compete the Dbf4-PBD interaction. C, purified wild type PBD and PBD-HK proteins interact with Dbf4 in the AlphaScreen assay, but the PBD-HK mutant protein fails to interact with Spc72 phosphopeptide. D, two-hybrid Spc721–400 and Dbf466–227 interactions with the PBD were tested on the indicated plates. As in C, mutation of the PBD pincer residues H641A and K643M or W517F, H641A, and K643M had no affect on the Dbf4-PBD interaction but eliminated the Spc72-PBD interaction. 3-AT, 3-aminotriazole.
The co-crystal structure of the Plk1 PBD with a phosphothreonine peptide revealed that the “pincer” residues His-538 and Lys-540 (which are invariant among human and mouse Plk1, Polo, Plo1, and Cdc5) directly interact with the phosphate group on threonine (24, 25). However, the purified PBD-H641A, K643M protein, containing the analogous mutations to Plk1-H538A, K540M, interacted with Dbf4 like the wild type in vitro (Fig. 4C). In contrast, although the wild type PBD interacted with Spc72, the PBD-HK protein did not. Similarly, the PBD-HK mutant interacted with Dbf4 but not with Spc72 in yeast cells (Fig. 4D). The additional mutation of a conserved hydrophobic residue W517F, analogous to Trp-414 in Plk1 that interacts with the preceding serine (S*-pT-P) of the phosphopeptide, also did not disrupt the two-hybrid interaction with Dbf4 (Fig. 4D). Using random mutagenesis, we isolated additional PBD mutations that abrogate the PBD-Spc72 interaction but retain the PBD-Dbf4 interaction (supplemental Fig. S6). The effects of these new PBD mutants on phospho-substrate binding are consistent with structural studies of Plk1-phosphopeptide molecular interactions (24, 25). Together, these data indicate that the Cdc5 PBD contains a second binding surface that recognizes non-phosphorylated sequences, like the Dbf4 residues 83RSIEGA88.
cdc5-HK Pincer Mutant Has Normal Growth Rate but Shows Increased Resistance to Microtubule Disruption
Very surprisingly, mutation of the pincer residues (H641A and K643M), which eliminated interaction with the Spc72 phospho-peptide in vitro (Fig. 4C), was tolerated in yeast. In fact, the cdc5-HK allele completely complemented yeast viability and growth in a cdc5Δ plasmid shuffle strain (Fig. 5, A and B) and when integrated at the endogenous CDC5 locus (Fig. 5C). Similarly, the cdc5-W517F, H641A, K643M (cdc5-WHK) allele also fully complemented yeast viability and growth rate (Fig. 5, A and B) and exhibited no temperature sensitivity up to 37 °C (Fig. 5A). These data indicate that the invariant pincer residues are not required for Cdc5 to bind essential substrates in yeast.
FIGURE 5.
The Cdc5 pincer residues are not required for yeast viability. A, the cdc5-HK and cdc5-WHK mutants complemented a cdc5Δ by plasmid shuffle into M1672 (cdc5Δ::kanMX6/pMW536[CDC5 URA3]) evidenced by growth on 5-fluoro-orotic acid plates and at various temperatures on YPD plates after loss of pMW536. B, growth curves of M1672 strains containing only the indicated CDC5 alleles in YPD at 30 °C are shown. C, the M1672-transformed strains in panel A were spotted onto YPD plates with/without benomyl (top). WT (M138), cdc5-HK (M3502), cdc5-5 (M1680), and cdc15-4 (M1999) strains containing integrated alleles were similarly spotted onto YPD with/without benomyl (bottom). D, representative tetrads from diploid strains of genotype cdc5-HK/CDC5 dbf2–1/DBF2, cdc5-HK/CDC5 cdc15-4/CDC15 TAB6-1/CDC14, or cdc5-HK/CDC5 cdc14-1/CDC14 that were sporulated and dissected onto YPD plates at 25 °C. Recombinant genotypes are indicated.
Although there were little growth phenotypes, the cdc5-HK and cdc5-WHK mutants exhibited increased resistance over the wild type to microtubule disruption by benomyl (Fig. 5C). All three strains grew well on plates containing 15 μg/ml benomyl, but the mutant strains grew about 100-fold better in the presence of 30 and 37.5 μg/ml benomyl. This phenotype was also observed with the integrated cdc5-HK allele but not with the temperature-sensitive (hypomorphic) cdc5-5 or cdc15-4 mutants, which were more sensitive or as sensitive to benomyl compared with the wild type strain (Fig. 5C). These data raise the possibility that the PBD pincer residues target Cdc5 to a substrate (perhaps a microtubule-associated protein) that regulates spindle dynamics.
In M-phase, Cdc5 promotes loss of sister chromatid cohesion, regulates spindle dynamics, and is essential to promote mitotic exit (18). Therefore, we tested for cdc5-HK synthetic growth interactions with spindle checkpoint mutants and the cdc15, dbf2, and cdc14 ts MEN mutants. The cdc5-HK allele exhibited no synthetic growth interaction with the mad1Δ, mad2Δ, bub1Δ, or bub2Δ spindle checkpoint mutants (data not shown). Although we saw no synthetic growth interaction with cdc14-1, cdc5-HK was synthetically lethal with cdc15-4 (cdc15-2, not shown) and dbf2–1 alleles (Fig. 5D). These synthetic lethal interactions were alleviated by TAB6-1, a dominant cdc14 mutant that suppresses some MEN defects (56). Importantly, TAB6-1 did not significantly affect the growth of the cdc15-2 or cdc15-4 mutants at 25 °C (data not shown), arguing that TAB6-1 bypasses the synthetic lethality we observe by suppressing a cdc5-HK defect. These data, therefore, suggest that cdc5-HK is defective in promoting mitotic exit.
Because cdc5-HK exhibited increased resistance to benomyl, we also directly examined spindle length in asynchronous wild type and cdc5-HK cells. The cell cycle distribution of cdc5-HK cells revealed a larger percentage of cells in G2/M phase relative to the wild type, suggesting a mitotic delay in the mutant (Fig. 6A). We quantitated spindle length in large-budded, mitotic cells and observed that the fraction of cells with short spindles (<2 μm) was the same in both strains, indicating there was no defect in mitotic entry (Fig. 6B). In contrast, the average spindle length in the mutant was about 38% greater than the wild type (7 versus 5.1 μm) (Fig. 6B). This could indicate a defect in exiting mitosis (spindle disassembly) or a defect in properly restraining spindle elongation. To address whether spindle elongation occurred with faster kinetics in the mutant, we measured the rate of spindle growth after release from a G2/M block using nocodazole. Wild type and cdc5-HK cells were arrested for 3 h using 15 μg/ml nocodazole and then released into the cell cycle in the absence of nocodazole at 30 °C. Flow cytometry profiles are shown in Fig. 6C. The spindles in both strains were depolymerized at 0 min; however, spindle length increased more rapidly for the cdc5-HK mutant, varying from 300% over wild type lengths at early time points to 40% greater length at 60 min (Fig. 6, D and E). These data indicate that mutation of the pincer residues also causes aberrant spindle elongation.
FIGURE 6.
Mutation of the Cdc5 pincer residues causes a G2/M delay and alters spindle dynamics. A, flow cytometry profiles of asynchronous W303-1A (WT) and M3502 (cdc5-HK) strains are shown. B, average spindle length was quantitated in large-budded cells of the same strains, shown with the range that includes 25–75% of spindle lengths. The inset shows fraction of cells with short spindles, <2 μm. C, shown are flow cytometry profiles of W303-1A and M3502 arrested in G2 with nocodazole for 3 h (t = 0) and after release at 30 °C. D, quantitation of spindle length at the indicated times after nocodazole release. S.E. were all less than 1%. E, shown is tubulin staining of representative photomicrographs of cells at 40 min after nocodazole release.
DISCUSSION
In this study we determined the Dbf4 residues required for a physical interaction with Cdc5. This analysis revealed a novel Dbf4 sequence (83RSIEGA88) directly binds the PBD. Unlike most identified PBD binding sites, the Dbf4-PBD interaction did not require Ser/Thr phosphorylation and, notably, bound to a distinct binding surface within the PBD. Our results establish that Dbf4 residues 83–88 are critical for binding the polo-box domain and mediate an inhibition of Cdc5 function. Surprisingly, the ability of Cdc5 (PBD) to interact with phosphorylated substrates is apparently not required for normal yeast growth but is required for full MEN activation and for regulating spindle dynamics during mitosis.
An Alternative Mode of PBD Binding
The polo-box domains of human Plk1 and yeast Cdc5 bind proteins containing phosphoserine or phosphothreonine consensus sites. In the co-crystal structure of the Plk1-PBD with a phosphorylated peptide, the peptide binds to a shallow pocket at the interface between two polo-box motifs, called PB1 and PB2 (24, 25). Two highly conserved residues, His-538 and Lys-540 in PB2, directly contact the phosphate group. Further mutational and biological studies have confirmed that the PBD binds phosphorylated substrates in vivo and that the PBD is required for Plk1 function in human cells and in yeast, where it complements CDC5 activity (32, 33).
The optimal phosphopeptide binding motif containing S(pS/pT)(P/X) was described for Cdc5 substrates, but only a few of these binding sequences have been characterized or mapped in detail (26, 36, 57). Although a subset of Cdc5 substrates examined in one study were found to be phosphorylated by cyclin-dependent kinases (13), additional Cdc5 substrates might be primed by other kinases or by Cdc5 itself, as has been recently shown for Plk1 (58). In contrast, the PBD of Drosophila Polo was recently shown to mediate an interaction with the microtubule-associated protein Map205 without a requirement for Map205 phosphorylation (40). In addition, the Cdc5 PBD can bind to Cdc14 independently of a consensus PBD recognition site (51).
We defined a unique Dbf4 sequence (83RSIEGA88) that directly interacts with the PBD. This Dbf4 sequence differs from the PBD consensus binding sequence in two critical regards. First, this sequence does not contain the absolutely conserved serine preceding a potential phosphoserine or phosphothreonine residue. More importantly, serine phosphorylation is not required for the PBD interaction as mutation of Ser-84 to alanine had little effect on PBD binding in vitro or in vivo. In fact, a peptide containing phosphorylated Ser-84 lost the ability to compete the Dbf4-PBD interaction in vitro (Fig. 2D). Therefore, Ser-84 phosphorylation actually inhibited interaction with the PBD. If Ser-84 phosphorylation occurs in vivo, this might negatively regulate the DDK-Cdc5 interaction. Whether a similar RSIEGA sequence exists in Cdc5 substrates remains to be determined, but our data strongly suggest that the PBD utilizes a second mode of interaction to bind non-phosphorylated proteins. The PBD-HK mutant protein bound Dbf4 as the wild type but was defective for interaction with the consensus (SpSP) Spc72 peptide. Mutation of six additional PBD residues (three of which mediate Plk1::phosphopeptide contacts in the co-crystal structure) disrupted the PBD-Spc72 canonical interaction but had no effect on the PBD-Dbf4 interaction (supplemental Fig. S6). Furthermore, the Dbf4-PBD interaction was not competed by a phosphorylated consensus peptide (Fig. 4B). These data indicate that the Cdc5 PBD can interact with proteins using two different binding surfaces, one that recognizes phosphorylated substrates and one that recognizes Dbf4.
Although the Plk1-HK mutant is defective for phosphopeptide binding in vitro and Plk1 activity in vivo, we found that the analogous cdc5-H641A, K643M mutant in budding yeast had a wild type growth rate and exhibited no temperature sensitivity (Fig. 5, A and B). This mutant instead exhibited a G2/M delay by flow cytometry, increased resistance to benomyl, and had an elongated spindle phenotype. These data suggest that Cdc5-HK protein is defective for interactions that restrain spindle elongation or that promote spindle disassembly. Because spindle disassembly follows Cdc5 activation of the MEN (55), a defect in MEN activation could account for the longer spindle length in the mutant. The synthetic lethality of cdc5-HK with cdc15 or dbf2 ts mutants supported the idea that cdc5-HK has a defect in MEN activation. However, in arguing against the MEN defect per se causing increased resistance to benomyl, we found that the cdc5-5 and cdc15-4 MEN mutants had similar or greater sensitivity to benomyl than wild type. The cdc5-1 and cdc5-2 mutants also exhibited a greater sensitivity to benomyl than wild type (data not shown). The wild type growth rate and unique resistance to benomyl argue that the HK mutation caused another defect in Cdc5 activity and that cdc5-HK was not simply another hypomorphic MEN mutant. The increased rate of spindle elongation in the mutant compared with the wild type after release of arrested G2/M cells strongly suggests that the cdc5-HK mutant has altered spindle dynamics. These data indicate that the pincer residues are not required for the essential function of Cdc5 and, because the PBD-HK mutant does not bind phosphopeptides in vitro, strongly suggest that PBD interactions with phosphoproteins are not essential in yeast.
Dbf4 Is a Scaffold for Cdc5 Inhibition
When a Cdc5-Dbf4 two-hybrid interaction was first described it was proposed that Cdc5 might have a novel role in DNA replication and that Dbf4 possibly functioned as a scaffold between these two essential kinases (16). Although Cdc5 is absent in G1 and early S-phase phase (59–61), it could potentially influence DNA replication during the preceding mitosis. However, when cells are released from a G1 block in the absence of Cdc5 expression, DNA synthesis occurs on schedule, and cells arrest in telophase with segregated chromatids (57, 62, 63). The Xenopus Plk1 ortholog, Plx1, was recently shown to influence DNA replication in response to DNA damage, raising the possibility that additional Polo orthologs might have a similar role (12). We recently proposed that DDK inhibits Cdc5 function during mitotic exit through a direct Dbf4-Cdc5 interaction (2). Here we show that multiple dbf4 mutants defective in the Dbf4-Cdc5 interaction suppress the cdc5-1 temperature sensitivity. Furthermore, increased Dbf4 expression is lethal to cdc5-1 cells at the permissive temperature, but only if Dbf4 can bind to Cdc5 (Fig. 3D). These data indicate that Dbf4 inhibits Cdc5 function by direct association with the PBD. Because DDK phosphorylates Cdc5 in vitro (2), these findings suggest that Dbf4 may serve as the scaffold in late S-phase so that Cdc7 kinase can inhibit Cdc5 by phosphorylating Cdc5 or an essential Cdc5 substrate. It is also possible that Dbf4 inhibits Cdc5 simply by binding to the PBD to prevent access to essential mitotic substrates, as overproduction of a Dbf4 N-terminal peptide is lethal to cdc5-1 (Fig. 3E and Ref. 2).
The DDK regulation of Cdc5 is not required for cell division control under normal conditions, as cell cycle progression occurs normally in dbf4 mutants defective for the Cdc5 interaction in otherwise wild type cells (supplemental Fig. S5 and Ref. 41). This suggests redundant mechanisms to delay Cdc5 activation until anaphase onset. For instance, Kin4 kinase antagonizes Cdc5 function when mitotic spindle positioning errors occur, although this may occur through inhibitory phosphorylation of Cdc5 substrates and not direct Cdc5 phosphorylation (64, 65). DNA damage and the Rad53 checkpoint kinase also block mitotic exit by directly or indirectly affecting Cdc5 activity (63, 66). Interestingly, the dbf4-NΔ109 deletion mutant that lacks the Polo binding site is synthetically lethal with rad53-1 (41), raising the possibility that Dbf4 and Rad53 have redundant, but together essential, roles to inhibit Cdc5 during an unperturbed cell cycle.
In summary, we have uncovered a novel PBD binding motif in Dbf4 that may be conserved in other PBD-binding proteins. We suggest that DDK uses this unique motif to bind Cdc5 and inhibit its essential function in the mitotic cell cycle. Presumably the Dbf4-PBD interaction does not preclude binding to substrates containing a phosphorylated consensus site based on our peptide competition studies. Therefore, the DDK-Cdc5 ternary complex could in principal interact with Cdc5 substrates containing a phospho-PBD consensus sequence. This model agrees with the finding that DDK and Cdc5 interact during meiosis and that both proteins phosphorylate the Cdc5 substrate Mam1 to promote monopolar spindle orientation during meiosis I (1). Defining the Dbf4-PBD physical interaction allows further investigation of how DDK regulates mitotic and meiotic events.
Supplementary Material
Acknowledgments
We thank the Flow Cytometry laboratory for technical assistance and FuJung Chang, Carrie Gabrielse, and Jessica Kenworthy for technical help. We also thank Angelika Amon and Wolfgang Zacharaie for strains and helpful discussions.
This work was supported by the Van Andel Research Institute and American Cancer Society Grant RSG-0506301GMC.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–6.
- DDK
- Dbf4-dependent kinase
- SCM
- Synthetic Complete medium
- ts
- temperature sensitivity
- MEN
- mitotic exit network
- Plk
- Polo-like kinase
- PBD
- polo-box domain.
REFERENCES
- 1.Matos J., Lipp J. J., Bogdanova A., Guillot S., Okaz E., Junqueira M., Shevchenko A., Zachariae W. (2008) Cell 135, 662–678 [DOI] [PubMed] [Google Scholar]
- 2.Miller C. T., Gabrielse C., Chen Y. C., Weinreich M. (2009) PLoS Genet. 5, e1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blow J. J., Tanaka T. U. (2005) EMBO Rep. 6, 1028–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jares P., Donaldson A., Blow J. J. (2000) EMBO Rep. 1, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston L. H., Masai H., Sugino A. (1999) Trends Cell Biol. 9, 249–252 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi T. S., Basu A., Bermudez V., Hurwitz J., Walter J. C. (2008) Genes Dev. 22, 1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasanuma H., Hirota K., Fukuda T., Kakusho N., Kugou K., Kawasaki Y., Shibata T., Masai H., Ohta K. (2008) Genes Dev. 22, 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan L., Niu H., Futcher B., Zhang C., Shokat K. M., Boulton S. J., Hollingsworth N. M. (2008) Genes Dev. 22, 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo H. C., Wan L., Rosebrock A., Futcher B., Hollingsworth N. M. (2008) Mol. Biol. Cell 19, 4956–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan M., Holt L., Morgan D. O. (2008) Mol. Cell. Biol. 28, 5328–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marston A. L. (2009) Curr. Biol. 19, R74–R76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trenz K., Errico A., Costanzo V. (2008) EMBO J. 27, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K. S., Park J. E., Asano S., Park C. J. (2005) Oncogene 24, 217–229 [DOI] [PubMed] [Google Scholar]
- 14.Petronczki M., Lénárt P., Peters J. M. (2008) Dev. Cell 14, 646–659 [DOI] [PubMed] [Google Scholar]
- 15.Kitada K., Johnson A. L., Johnston L. H., Sugino A. (1993) Mol. Cell. Biol. 13, 4445–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy C. F., Pautz A. (1996) Mol. Cell. Biol. 16, 6775–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunkel C. E., Glover D. M. (1988) J. Cell Sci. 89, 25–38 [DOI] [PubMed] [Google Scholar]
- 18.Archambault V., Glover D. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 19.Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 20.Lowery D. M., Lim D., Yaffe M. B. (2005) Oncogene 24, 248–259 [DOI] [PubMed] [Google Scholar]
- 21.Lee K. S., Grenfell T. Z., Yarm F. R., Erikson R. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seong Y. S., Kamijo K., Lee J. S., Fernandez E., Kuriyama R., Miki T., Lee K. S. (2002) J. Biol. Chem. 277, 32282–32293 [DOI] [PubMed] [Google Scholar]
- 23.Yaffe M. B., Smerdon S. J. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 225–244 [DOI] [PubMed] [Google Scholar]
- 24.Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N. (2003) EMBO J. 22, 5757–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 26.Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strebhardt K., Ullrich A. (2006) Nat. Rev. Cancer 6, 321–330 [DOI] [PubMed] [Google Scholar]
- 28.Reindl W., Yuan J., Krämer A., Strebhardt K., Berg T. (2008) Chem. Biol. 15, 459–466 [DOI] [PubMed] [Google Scholar]
- 29.Watanabe N., Sekine T., Takagi M., Iwasaki J., Imamoto N., Kawasaki H., Osada H. (2009) J. Biol. Chem. 284, 2344–2353 [DOI] [PubMed] [Google Scholar]
- 30.de Cárcer G., Pérez de Castro I., Malumbres M. (2007) Curr. Med. Chem. 14, 969–985 [DOI] [PubMed] [Google Scholar]
- 31.Hartwell L. H., Culotti J., Reid B. (1970) Proc. Natl. Acad. Sci. U.S.A. 66, 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K. S., Erikson R. L. (1997) Mol. Cell. Biol. 17, 3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang B., Pan H., Lu L., Li J., Stambrook P., Li B., Dai W. (1997) J. Biol. Chem. 272, 28646–28651 [DOI] [PubMed] [Google Scholar]
- 34.Snead J. L., Sullivan M., Lowery D. M., Cohen M. S., Zhang C., Randle D. H., Taunton J., Yaffe M. B., Morgan D. O., Shokat K. M. (2007) Chem. Biol. 14, 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asano S., Park J. E., Sakchaisri K., Yu L. R., Song S., Supavilai P., Veenstra T. D., Lee K. S. (2005) EMBO J. 24, 2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crasta K., Lim H. H., Giddings T. H., Jr., Winey M., Surana U. (2008) Nat. Cell Biol. 10, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geymonat M., Spanos A., Walker P. A., Johnston L. H., Sedgwick S. G. (2003) J. Biol. Chem. 278, 14591–14594 [DOI] [PubMed] [Google Scholar]
- 38.Hornig N. C., Uhlmann F. (2004) EMBO J. 23, 3144–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowery D. M., Mohammad D. H., Elia A. E., Yaffe M. B. (2004) Cell Cycle 3, 128–131 [PubMed] [Google Scholar]
- 40.Archambault V., D'Avino P. P., Deery M. J., Lilley K. S., Glover D. M. (2008) Genes Dev. 22, 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrielse C., Miller C. T., McConnell K. H., DeWard A., Fox C. A., Weinreich M. (2006) Genetics 173, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 43.Sherman F., Fink G. R., Hicks J. B. (1986) Methods in Yeast Genetics, pp. 523–585, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44.Weinreich M., Stillman B. (1999) EMBO J. 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Souès S., Adams I. R. (1998) J. Cell Sci. 111, 2809–2818 [DOI] [PubMed] [Google Scholar]
- 46.Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. (1994) Mol. Cell. Biol. 14, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Zhang J., Schopfer F. J., Martynowski D., Garcia-Barrio M. T., Kovach A., Suino-Powell K., Baker P. R., Freeman B. A., Chen Y. E., Xu H. E. (2008) Nat. Struct. Mol. Biol. 15, 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 49.Kang Y. H., Park J. E., Yu L. R., Soung N. K., Yun S. M., Bang J. K., Seong Y. S., Yu H., Garfield S., Veenstra T. D., Lee K. S. (2006) Mol. Cell 24, 409–422 [DOI] [PubMed] [Google Scholar]
- 50.García-Alvarez B., de Cárcer G., Ibañez S., Bragado-Nilsson E., Montoya G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahal R., Amon A. (2008) Cell Cycle 7, 3262–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ullman E. F., Kirakossian H., Singh S., Wu Z. P., Irvin B. R., Pease J. S., Switchenko A. C., Irvine J. D., Dafforn A., Skold C. N. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5426–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pintard L., Peter M. (2001) Mol. Cell 8, 1155–1156 [DOI] [PubMed] [Google Scholar]
- 54.Park C. J., Song S., Lee P. R., Shou W., Deshaies R. J., Lee K. S. (2003) Genetics 163, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stegmeier F., Amon A. (2004) Annu. Rev. Genet. 38, 203–232 [DOI] [PubMed] [Google Scholar]
- 56.Shou W., Deshaies R. J. (2002) BMC Genet 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida S., Kono K., Lowery D. M., Bartolini S., Yaffe M. B., Ohya Y., Pellman D. (2006) Science 313, 108–111 [DOI] [PubMed] [Google Scholar]
- 58.Lee K. S., Park J. E., Kang Y. H., Zimmerman W., Soung N. K., Seong Y. S., Kwak S. J., Erikson R. L. (2008) Cell Cycle 7, 141–145 [DOI] [PubMed] [Google Scholar]
- 59.Charles J. F., Jaspersen S. L., Tinker-Kulberg R. L., Hwang L., Szidon A., Morgan D. O. (1998) Curr. Biol. 8, 497–507 [DOI] [PubMed] [Google Scholar]
- 60.Cheng L., Hunke L., Hardy C. F. (1998) Mol. Cell. Biol. 18, 7360–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirayama M., Zachariae W., Ciosk R., Nasmyth K. (1998) EMBO J. 17, 1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S. J. (2001) Cell 107, 655–665 [DOI] [PubMed] [Google Scholar]
- 63.Liang F., Wang Y. (2007) Mol. Cell. Biol. 27, 5067–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Aquino K. E., Monje-Casas F., Paulson J., Reiser V., Charles G. M., Lai L., Shokat K. M., Amon A. (2005) Mol. Cell 19, 223–234 [DOI] [PubMed] [Google Scholar]
- 65.Pereira G., Schiebel E. (2005) Mol. Cell 19, 209–221 [DOI] [PubMed] [Google Scholar]
- 66.Sanchez Y., Bachant J., Wang H., Hu F., Liu D., Tetzlaff M., Elledge S. J. (1999) Science 286, 1166–1171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.