Abstract
Like most metalloproteases, matrix metalloprotease 2 (MMP-2) is synthesized as a zymogen. MMP-2 propeptide plays a role in inhibition of catalytic activity through a cysteine-zinc ion pairing, disruption of which results in full enzyme activation. A variety of proteases have been shown to be involved in the activation of pro-MMP-2, including metalloproteases and serine proteases. In the previous study we showed that MMP-2 activation occurred via specific cleavages of the propeptide by thrombin followed by intermolecular autoproteolytic processing for full enzymatic activity. Thrombin also degraded MMP-2, but this degradation was reduced greatly under cell-associated conditions with a concomitant increase in activation, prompting us to elucidate the molecular mechanisms underlying thrombin-mediated MMP-2 activation. In the present study we demonstrate that heparan sulfate is essential for thrombin-mediated activation of pro-MMP-2. Binding of heparan sulfate to thrombin is primarily responsible for this activation process, presumably through conformational changes at the active site. Furthermore, interaction of MMP-2 with exosites 1 and 2 of thrombin is crucial for thrombin-mediated MMP-2 degradation, and inhibition of this interaction by heparan sulfate or hirudin fragment results in a decrease in MMP-2 degradation. Finally, we demonstrated interaction between exosite 1 and hemopexin-like domain of MMP-2, suggesting a regulatory role of hemopexin-like domain in MMP-2 degradation. Taken together, our experimental data suggest a novel regulatory mechanism of thrombin-dependent MMP-2 enzymatic activity by heparan sulfate proteoglycans.
Keywords: Enzyme Inactivation, Enzyme Processing, Heparan Sulfate, Matrix Metalloproteinase, Thrombin
Introduction
Matrix metalloprotease 2 (MMP-2)3 is widely expressed and is closely associated with diverse biological processes through the degradation of extracellular and non-extracellular matrix molecules (1, 2). Its activity is regulated at multiple levels including gene expression, compartmentalization, zymogen activation, and enzyme inactivation by extracellular inhibitors such as tissue inhibitors of metalloproteases (TIMPs) (1). Like most metalloproteases, MMP-2 is synthesized as a zymogen that is activated by conformational change (3) or proteolytic cleavage of the propeptide. Propeptides of protease zymogens have been shown to have important functions in the inhibition of catalytic activity (4); thus, removal of the propeptide is an important prerequisite for enzymatic activity. A variety of proteases have been shown to be involved in the activation of pro-MMP-2, including metalloproteases (e.g. MMP-7 and MT-MMPs) (5–10), serine proteases (e.g. thrombin, factor Xa, activated protein C, and plasmin) (11–14), and a cysteine protease (legumain) (15).
Thrombin is involved in the coagulation cascade and multiple cellular processes, including mitogenesis of fibroblasts, lymphocytes, mesenchymal cells, and smooth muscle cells, via proteolytic activation of protease activated receptors (16–20). It also plays a role in cell migration and invasion through MMP-2 activation (21, 22).
Thrombin-mediated activation of pro-MMP-2 has been well studied, with some reports indicating the involvement of MT1-MMP (12, 13, 22–25), although this remains controversial. In our previous study using metalloprotease inhibitors and pro-MMP-2 mutants incapable of cleavage by MT1-MMP, we showed MT1-MMP-independent activation of pro-MMP-2 by thrombin (26). We further demonstrated that thrombin cleaved the propeptide on the C-terminal side of Arg98 and Arg101 followed by intermolecular autoproteolytic cleavage at the Asn109-Tyr peptide bond for full enzymatic activity (26). In addition to its role in activation, thrombin also degraded MMP-2, but this degradation was reduced greatly under cell-associated conditions with a concomitant increase in activation (26). In the present study we sought to elucidate the underlying molecular mechanisms through identification of cellular molecules that associate with MMP-2.
EXPERIMENTAL PROCEDURES
Reagents
Human thrombin was purchased from Hematologic Technologies (Essex Junction, VT). Monoclonal antibodies specific to MMP-2 (sc-13595), TIMP-2 (sc-21735), and fibronectin (sc-8422), rabbit polyclonal MT1-MMP antibody (sc-30074), rabbit polyclonal collagen I (sc-8784) and IV (sc-70246) antibodies, and fibronectin siRNA (sc-29315) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and monoclonal anti-MMP-2 antibody (CA-4001) was from Calbiochem. Monoclonal anti-integrin αvβ3, polyclonal anti-integrin αv, monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and human TIMP-2 protein were purchased from Chemicon (Temecula, CA), and anti-myc antibody (clone 9E10) was from Invitrogen. Collagenase, human fibronectin, fibrinogen, hirudin fragment (Hir54–65(SO3H)63), heparan sulfate, and streptavidin-agarose were purchased from Sigma, and recombinant MMP-2 hemopexin-like domain was obtained from ProSpec (Rehovot, Israel). Monoclonal anti-heparan sulfate antibody (10E4) was purchased from Cape Cod, Inc. (East Falmouth, MA), and rabbit polyclonal anti-prothrombin antibody was from Hypen BioMed. siGENOME SMARTpool siRNA against integrin αv was purchased from Dharmacon (Lafayette, CO).
Expression Plasmids and Site-directed Mutagenesis
Plasmids for expression of full-length MMP-2 or MMP-2 with C-terminal Myc and His tags were described previously (26). cDNA encoding hemopexin-like domain (Tyr-445 to Cys-660) was amplified using MMP-2 as a template with primers 5′-AAGCTTGAGATCTGCAAACAGGACATTG-3′ (with the HindIII site underlined) and 5′-GATATCCGCAGCCTAGCCAGTCGGATT-3′ (the EcoRV site is underlined) and cloned into pSecTag2/Hygro (Invitrogen) for expression with an Ig κ-chain leader sequence and C-terminal Myc and His tags. Expression plasmids for human MT1-MMP and mouse syndecan-1 were kindly provided by Dr. Suneel Apte.
Cell Culture, Transfection, and Treatments
Human brain microvascular endothelial cell (HBMEC) line (27), COS-1, HCT116 (human colorectal carcinoma) (ATCC no. CCL-247), A549 (human lung carcinoma) (ATCC no. CCL-185), and LoVo (human colorectal adenocarcinoma) (ATCC no. CCL-229) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). K562 (human chronic myelogenous leukemia) (ATCC no. CCL-243) and K562 stably transfected with syndecan-1 were maintained in RPMI 1640 medium containing 10% FBS. Transfection with plasmids and siRNA was performed using Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen). For secreted MMP-2, transfected COS-1 cells were cultured and then transferred to 293 SFM-II medium (Invitrogen).
Flow Cytometric Analysis and Immunofluorescence
Cells were detached from 6-well plates using phosphate-buffered saline containing 5 mm EDTA and incubated with the respective antibodies at 4 °C for 2 h. The cells were washed and further incubated with FITC-conjugated goat anti-mouse antibody or goat anti-rabbit antibody (Chemicon) for 1 h. Flow cytometry was performed on a FACSCalibur (BD Biosciences, San Jose, CA), and data were analyzed using WinMDI software Version 2.8 (The Scripps Research Institute, La Jolla, CA). For immunocytochemical staining, cells were fixed and permeabilized with methanol at −20 °C for 10 min. After incubating with polyclonal anti-collagen I antibody, and subsequently with FITC-conjugated goat anti-rabbit antibody, optical images were acquired using a LSM 510 META confocal microscope (Carl Zeiss).
Immunoblotting, Immunoprecipitation, and Protein Purification
Cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Roche Diagnostics) for 1 h at 4 °C and then centrifuged. The soluble portion of the lysate was used for Western blotting, which was performed by separation of reduced samples on SDS-PAGE followed by electroblotting to polyvinylidene difluoride membrane and detection of bound antibody by enhanced chemiluminescence (Amersham Biosciences). Medium containing thrombin and MMP-2 was immunoprecipitated with anti-MMP-2 (CA-4001) followed by reducing SDS-PAGE and immunoblotting with an anti-prothrombin antibody. MMP-2 was purified from the conditioned medium using gelatin-Sepharose according to the manufacturer's recommendations (Amersham Biosciences). The signal intensity of relevant bands from the Western blot was quantitated using ImageJ software (National Institutes of Health, Bethesda, MD).
Semiquantitative RT-PCR
Total RNA was prepared using the RNeasy Mini kit (Qiagen, Chatsworth, CA). Five micrograms of mRNA was used for synthesis of first strand cDNA with the Superscript III First-Strand kit (Invitrogen). One microliter of cDNA was used as the template for PCR. Primer sets were as follows: collagen I, forward (5′-ACGTCCTGGTGAAGTTGGTC-3′) and reverse (5′-ACCAGGGAAGCCTCTCTCTC-3′); collagen IV, forward (5′-AGAGCATCCAGCCATTCATT-3′) and reverse (5′-TTCAGCGTTTCTGACTGAGG-3′); GAPDH, forward (5′-GAAGCTCACTGGCATGGCCTT-3′) and primer (5′-CTCTCTTGCTCAGTGTCCTTGCT-3′).
Biotinylation of Heparan Sulfate and Capturing the Complex
Biotinylation of heparan sulfate was done using Sulfo-NHS-LC-Biotin according to the manufacturer's recommendations (Pierce). After biotinylation, the labeled heparan sulfate was dialyzed against 50 mm Tris-HCl, pH 7.5, containing 0.15 m NaCl. Biotinylated heparan sulfate was incubated with pro-MMP-2 at 4 °C for 2 h and captured using streptavidin-agarose. The bound protein was eluted by boiling in Laemmli sample buffer followed by SDS-PAGE and Western blotting with an anti-MMP-2 antibody.
Statistical Analysis
Data are represented as the mean and S.D. of n experiments. Statistical analysis was performed using an unpaired Student's t test. A p value less than 0.05 was considered statistically significant.
RESULTS
Integrin αvβ3, MT1-MMP, and TIMP-2 Are Not Involved in Thrombin-mediated Activation of Pro-MMP-2
In the previous study we showed an increase in thrombin-mediated MMP-2 activation and a concomitant decrease in degradation by the cell surface. Moreover, several studies have demonstrated interaction of MMP-2 with the cell surface via integrin αvβ3 and MT1-MMP·TIMP-2 complex (28, 29). Thus, before investigating the role of these molecules in thrombin-mediated activation of pro-MMP-2, the expression of integrin αvβ3, MT1-MMP, and TIMP-2 was characterized in HBMEC, COS-1, HCT116, and A549 cells. Cell surface expression of integrin αvβ3 was observed by flow cytometric analysis in HBMEC and COS-1 cells, whereas its expression was undetectable in HCT116 and A549 cells (supplemental Fig. S1A). MT1-MMP protein was not detected by immunoblotting with an anti-MT1-MMP antibody in any of the cell types (supplemental Fig. S1B). Immunoblotting with an anti-TIMP-2 antibody showed that COS-1 cells possessed the highest levels of TIMP-2, whereas its expression was absent or lower in HBMEC, HCT116, and A549 cells (supplemental Fig. S1C). Next, we examined thrombin-mediated activation and degradation of pro-MMP-2 in these cells. When pro-MMP-2-myc/His was incubated with thrombin and the incubation mixture was analyzed by immunoblotting with an anti-myc antibody, the Western blot data revealed robust increases in thrombin-mediated MMP-2 activation and decreases in degradation (supplemental Fig. S2, A, right panel, and B). In contrast, activation was only marginally detected with a significant increase in degradation under cell-free conditions (supplemental Fig. S2A, left panel). Thus, these data together with the lack of integrin αvβ3, MT1-MMP, and TIMP-2 expression in some cell types suggest that thrombin-mediated activation of pro-MMP-2 occurs independently of cell surface integrin αvβ3, MT1-MMP, and TIMP-2. We further investigated the role of integrin αvβ3, MT1-MMP, and TIMP-2 in thrombin-mediated activation of pro-MMP-2. To examine the role of integrin αvβ3 in this activation, pro-MMP-2 was incubated with thrombin in HBMEC cells transfected with control and integrin αv-specific siRNA. Although integrin αv expression was significantly repressed (supplemental Fig. S3A, right panel), pro-MMP-2 activation was not altered in the integrin αv siRNA-treated cells (supplemental Fig. S3A, left panel). Previously, we demonstrated that the increase in MMP-2 activation under cell-associated conditions was not due to the proteolytic activity of MT1-MMP (26). Furthermore, it has been shown that MMP-2 binds MT1-MMP using TIMP-2 as an adaptor (29). Therefore, we tested whether MT1-MMP could act as an MMP-2 receptor for thrombin-mediated activation of pro-MMP-2. Incubation of pro-MMP-2 with thrombin in COS-1 cells (which do not express endogenous MT1-MMP) transfected with catalytically inactive MT1-MMP E240A (supplemental Fig. S3B, right panel) did not affect activation (supplemental Fig. S3B, left panel). Furthermore, supplemental Fig. S3C shows that incubation with TIMP-2 did not have any influence on the activation in A549 cells that expressed a negligible level of endogenous TIMP-2 (supplemental Fig. S1C). Taken together, these results suggest that integrin αvβ3, MT1-MMP, and TIMP-2 are not involved in the thrombin-mediated activation of pro-MMP-2.
Thrombin-mediated Activation of Pro-MMP-2 Is Not Associated with Cellular Expression of Fibronectin and Collagens
Because fibronectin and collagens have been shown to interact with MMP-2 (30–33), we examined the role of these molecules in thrombin-mediated activation of pro-MMP-2. First, Western blot analysis was performed to determine the expression of fibronectin in HBMEC, COS-1, HCT116, and A549 cells. Although an increase in thrombin-mediated activation of pro-MMP-2 was observed in COS-1 and HCT116 cells (supplemental Fig. S2B), the expression of fibronectin was negligible in these cells (supplemental Fig. S4A), suggesting that the activation occurs independently of fibronectin. This was further verified in HBMEC cells transfected with fibronectin siRNA. Although fibronectin expression was greatly repressed in the siRNA-treated cells (supplemental Fig. S4B, right panel), thrombin-mediated activation of pro-MMP-2 was not affected (supplemental Fig. S4B, left panel). Likewise, in HCT116 cells pro-MMP-2 activation was unaltered by incubation with purified fibronectin (supplemental Fig. S4C). Thus, these results indicate that interaction of pro-MMP-2 with fibronectin does not influence thrombin-mediated activation of pro-MMP-2 on the cell surface. Next, we examined the role of collagen I and IV in thrombin-mediated activation of pro-MMM-2. RT-PCR analysis showed variable mRNA expression of collagens I and IV in HBMEC, HCT116, A549, and LoVo cells (supplemental Fig. S5A). However, protein expression of collagen I was not detected in HBMEC and A549 cells by immunocytochemistry (data not shown), although an increase in activated MMP-2 was observed in these cells (supplemental Fig. S2). These data indicate that the activation does not depend on collagen I expression in these cells. The lack of collagen IV expression in LoVo cells had been previously reported (34) and was confirmed in our study (supplemental Fig. S5A). When pro-MMP-2 was incubated with thrombin in LoVo cells, thrombin-mediated activation occurred normally (supplemental Fig. S5B), indicating that the activation was collagen IV-independent. Collagen-independent activation was further verified in HBMEC cells that had been pretreated with collagenase. Although removal of collagen IV was confirmed by flow cytometric analysis (supplemental Fig. S5C), thrombin-mediated activation of pro-MMP-2 was not influenced by collagenase treatment (supplemental Fig. S5D). These results further demonstrate that the activation is not related to collagen expression.
Heparan Sulfate Proteoglycans Are Primarily Responsible for Thrombin-mediated Activation of Pro-MMP-2
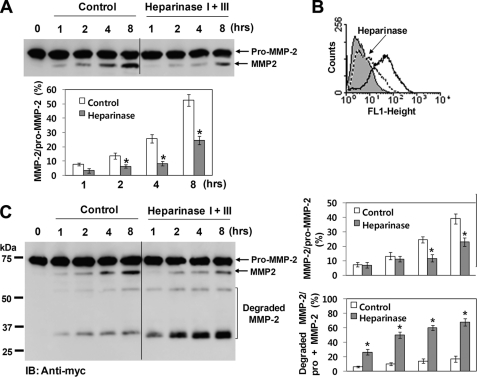
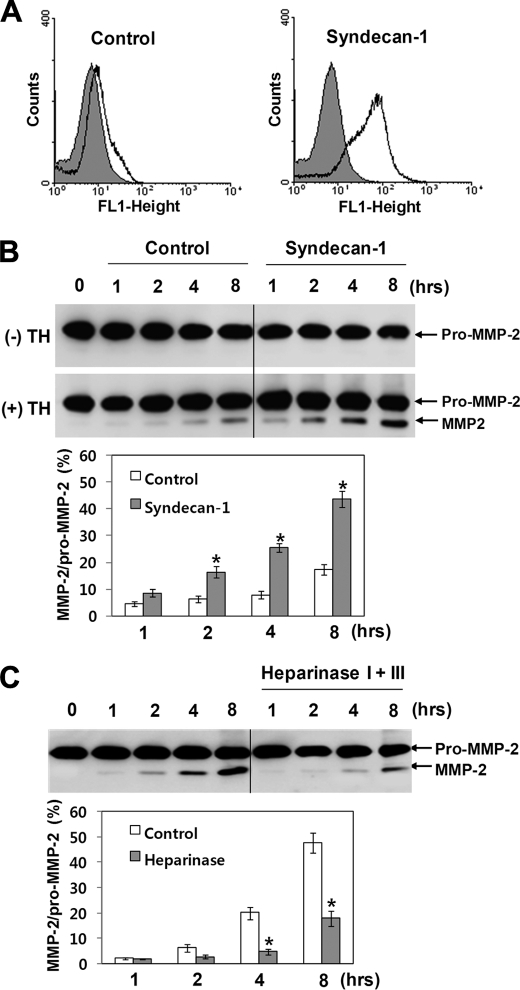
MMP-2 is localized at the cell surface by binding to the heparan sulfate proteoglycans (35). Therefore, we investigated the role of cell surface heparan sulfate proteoglycans in thrombin-mediated activation of pro-MMP-2. After removal of heparan sulfate from the cell surface by treatment with heparinase I and III (Fig. 1B), pro-MMP-2 was incubated with thrombin in these cells, and the reaction mixture was analyzed by immunoblotting with an anti-MMP-2 antibody. As shown in Fig. 1A, thrombin-mediated activation was significantly decreased in the heparinase-treated cells. Similar experiments were performed using pro-MMP-2-myc/His. Analysis of the incubation mixture by immunoblotting with an anti-myc antibody showed a decrease in MMP-2 activation and a corresponding increase in degradation after heparinase treatment (Fig. 1C). To further confirm the role of heparan sulfate in this activation process, we took advantage of the deficient expression of heparan sulfate in K562 cells, which was confirmed by flow cytometric analysis (Fig. 2A, left panel). When pro-MMP-2 was incubated with thrombin in these cells, immunoblotting with an anti-MMP-2 antibody showed a slight increase in MMP-2 activation (Fig. 2B, lower panel). In contrast, thrombin-mediated activation was significantly increased in cells expressing syndecan-1, which showed high expression of cell surface heparan sulfate (Fig. 2, A, right panel, and B, lower panel). Pro-MMP-2 in these cells remained unaffected in the absence of thrombin (Fig. 2B, upper panel). A control experiment was undertaken to show that heparan sulfate was specifically involved in this activation process by treating the transfected cells with heparinases. When heparan sulfate was removed from the cell surface by treatment with heparinases (data not shown) and pro-MMP-2 was incubated with thrombin in these cells, immunoblotting with an anti-MMP-2 antibody revealed that thrombin-mediated activation of pro-MMP-2 was significantly decreased in the heparinase-treated cells (Fig. 2C), confirming the specific role of heparan sulfate in this activation. Taken together, these data suggest that heparan sulfate proteoglycans are primarily responsible for thrombin-mediated activation of pro-MMP-2 on the cell surface.
FIGURE 1.
Heparan sulfate proteoglycans are required for thrombin-mediated activation of pro-MMP-2. A, Western blot analysis using an anti-MMP-2 antibody of 20 μg/ml pro-MMP-was 2 incubated with 50 nm thrombin in HBMEC cells for the indicated times. Cells were pretreated with or without heparinase I (5 units/ml) and III (2 units/ml) for 2 h to remove cell surface heparan sulfate proteoglycans. These experiments were performed in serum-free medium. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2. Data represent the mean and S.D. of three experiments. *, p < 0.05 versus untreated control. B, flow cytometric analysis of cell surface expression of heparan sulfate in HBMEC cells treated with or without heparinase I and III is shown. A sample lacking primary antibody was used as a control. Cell surface heparan sulfate (solid line) is deficient in the cells treated with heparinases (dotted line) (n = 3). C, Western blot (IB) analysis using an anti-myc antibody of 20 μg/ml pro-MMP-2-myc/His incubated with 50 nm thrombin in HBMEC cells for the indicated times is shown. Cells were pretreated with or without heparinase I (5 units/ml) and III (2 units/ml) for 2 h. Degradation products are indicated. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2 or the ratio of the degraded MMP-2 to total MMP-2 (pro-MMP-2 + MMP-2). Data represent the mean and S.D. of three experiments. *, p < 0.05 versus untreated control.
FIGURE 2.
Expression of syndecan-1 increases thrombin-mediated activation of pro-MMP-2 in K562 cells. A, flow cytometric analysis shows cell surface expression of heparan sulfate in K562 cells stably transfected with syndecan-1 is shown. A sample lacking primary antibody was used as a control. B, Western blot analysis using an anti-MMP-2 antibody of 20 μg/ml pro-MMP-2 incubated for the indicated times with or without 50 nm thrombin in K562 and K562 cells expressing syndecan-1 is shown. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2. Data represent the mean and S.D. of three experiments. *, p < 0.05 versus control. C, Western blot analysis using an anti-MMP-2 antibody of 20 μg/ml pro-MMP-2 incubated with 50 nm thrombin in K562 cells expressing syndecan-1 for the indicated times is shown. Cells were pretreated with or without heparinase I (5 units/ml) and III (2 units/ml) for 2 h. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2. Data represent the mean and S.D. of three experiments. *, p < 0.05 versus untreated control.
Heparan Sulfate Increases Thrombin-mediated Activation of Pro-MMP-2 under Cell-free Conditions
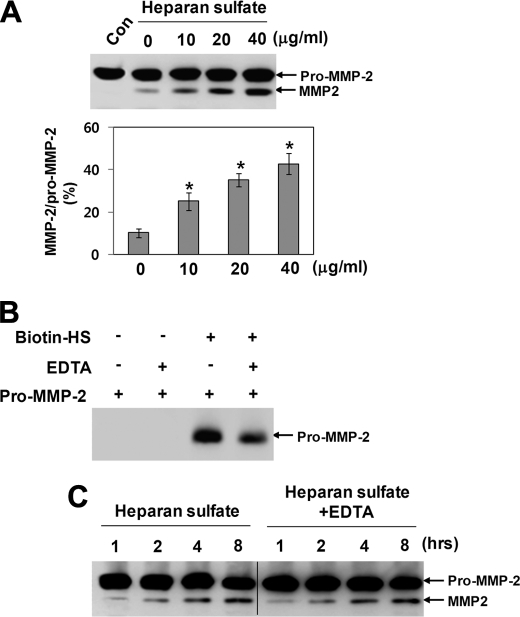
Heparan sulfate was used to corroborate its functional role in thrombin-mediated activation of pro-MMP-2 under cell-free conditions. Pro-MMP-2 was incubated with thrombin in the presence of heparan sulfate under cell-free conditions, and the reaction mixture was analyzed by immunoblotting with an anti-MMP-2 antibody. The Western blot data showed a significant increase in thrombin-mediated activation in the presence of heparan sulfate (Fig. 3A). MMP-2 has been reported to bind to heparan sulfate, possibly via its hemopexin-like domain, in a calcium-dependent manner (30, 36). Therefore, we tested whether a direct interaction between pro-MMP-2 and heparan sulfate was responsible for this activation process. When metal ions were removed from pro-MMP-2 by treatment with EDTA and dialysis, most of the pro-MMP-2 did not bind to biotinylated heparan sulfate at 0.15 m NaCl, whereas significant binding of non-treated pro-MMP-2 to the heparan sulfate was observed at the same concentration of NaCl (Fig. 3B). Next, pro-MMP-2 was incubated with thrombin in the presence of heparan sulfate, and the reaction mixture was analyzed by immunoblotting with an anti-MMP-2 antibody. The results showed that removal of the metal ions did not affect thrombin-mediated activation of pro-MMP-2 in the presence of heparan sulfate (Fig. 3C). Thus, these data suggest that a direct interaction of pro-MMP-2 with heparan sulfate is not involved in this activation process.
FIGURE 3.
Heparan sulfate increases thrombin-mediated activation of pro-MMP-2 under cell-free conditions. A, Western blotting using an anti-MMP-2 antibody of 20 μg/ml pro-MMP-2 incubated with 50 nm thrombin in the presence of heparan sulfate under cell-free conditions for 8 h is shown. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2. Data represent the mean and S.D. of three experiments. *, p < 0.05 versus 0 μg/ml heparan sulfate. B, Western blot analysis of the eluates from streptavidin-agarose using an anti-MMP-2 antibody is shown. Biotinylated heparan sulfate (Biotin-HS) was incubated with pro-MMP-2 and captured using streptavidin-agarose. Metal ions were removed from pro-MMP-2 by treatment with 50 mm EDTA and dialysis against 50 mm Tris-HCl, pH 7.5, containing 0.15 m NaCl at 4 °C for 24 h (n = 2). C, Western blotting using an anti-MMP-2 antibody of 20 μg/ml pro-MMP-2 incubated with 50 nm thrombin in the presence of heparan sulfate (20 μg/ml) under cell-free conditions for the indicated times is shown. Metal ions were removed from pro-MMP-2 as described above (n = 3).
Binding of Heparan Sulfate to Thrombin Leads to a Decrease in Thrombin-mediated MMP-2 Degradation and a Concomitant Increase in Activation
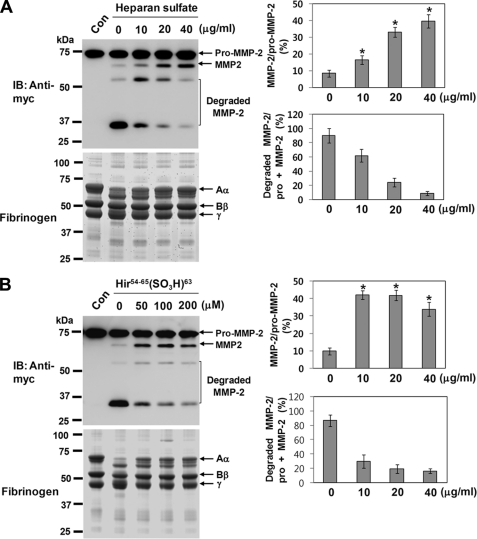
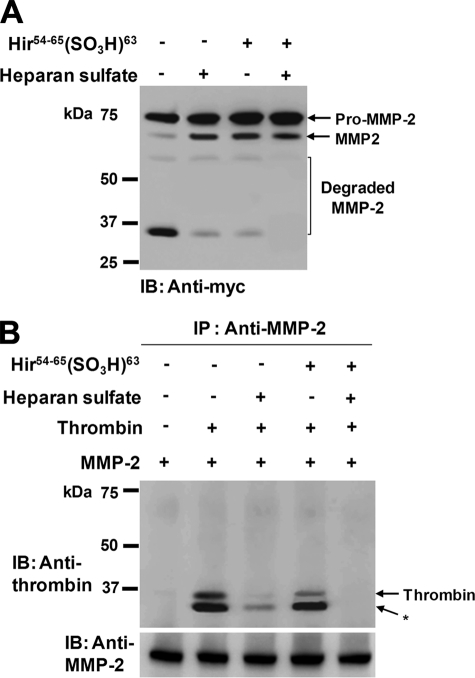
Thrombin has been shown to possess two positively charged domains (37, 38). These sites (termed exosites 1 and 2) contribute to the specificity of thrombin by serving as binding sites for substrates, cofactors, and other ligands. Exosite 1 is known to be the binding site for fibrinogen, hirudin, heparin cofactor II, and thrombin receptor, whereas exosite 2 binds heparin, other glycosaminoglycans, and prothrombin fragment 2. Therefore, we investigated the possibility that binding of heparan sulfate to exosite 2 might elicit different effects on thrombin-mediated activation and degradation of pro-MMP-2. Experiments were performed using pro-MMP-2-myc/His and immunoblotting with an anti-myc antibody. When pro-MMP-2-myc/His was incubated with thrombin in the presence of heparan sulfate under cell-free conditions, a significant decrease in MMP-2 degradation products was observed with a corresponding increase in activated MMP-2 (Fig. 4A, upper panel), suggesting that binding of heparan sulfate to thrombin may cause conformational changes at the active site with concomitant alterations in the substrate specificity of thrombin. Thus, we examined the influence of heparan sulfate on the substrate specificity of thrombin toward fibrinogen. Fibrinogen was incubated with thrombin in the presence of heparan sulfate, and the reaction mixture was analyzed by SDS-PAGE. Fibrinogen cleavage by thrombin in the presence of heparan sulfate was significantly decreased (Fig. 4A, lower panel), confirming alterations in the enzymatic activity of thrombin. The interaction of exosite 1 with substrates or inhibitors has also been reported to induce allosteric changes that alter the substrate specificity of thrombin (39, 40). Therefore, the cleavage experiments were also performed in the presence of hirudin fragment that binds specifically to the exosite 1 of thrombin (39), yielding similar results with pro-MMP-2-myc/His and fibrinogen (Fig. 4B). Under these conditions we noticed that MMP-2 degradation was not completely inhibited, even by increasing the concentration of heparan sulfate or hirudin fragment (Fig. 4, A and B). Thus, we assumed that simultaneous binding of each ligand to the exosites is necessary for the complete inhibition of MMP-2 degradation. To test this, thrombin cleavage of pro-MMP-2-myc/His was performed in the combined presence of heparan sulfate and hirudin fragment. Western blot analysis of the incubation mixture using an anti-myc antibody showed that MMP-2 degradation was almost completely inhibited by combined treatment with heparan sulfate and hirudin fragment, whereas activation was only slightly affected (Fig. 5A). Overall, these data suggest that the interaction of exosite 1 or 2 with its ligand may induce conformational changes at the active site of thrombin to facilitate pro-MMP-2 activation. Moreover, binding of MMP-2 to both exosites of thrombin is likely to be required for its degradation. Thus, inhibition of MMP-2 binding to exosite 1 or 2 may prevent MMP-2 degradation. To determine whether MMP-2 can interact with thrombin in the presence of heparan sulfate and hirudin fragment, co-immunoprecipitation experiments were performed. Immunoprecipitation with an anti-MMP-2 antibody followed by immunoblotting with an anti-thrombin antibody revealed interaction between thrombin and MMP-2 (Fig. 5B). This binding was mostly abolished in the presence of heparan sulfate but was marginally affected by hirudin fragment, indicating that the stronger interaction of MMP-2 with thrombin occurs via exosite 2. Moreover, the interaction between these two proteins completely disappeared in the combined presence of heparan sulfate and hirudin fragment (Fig. 5B), which corresponds to the absence of MMP-2 degradation (Fig. 5A). Thus, these data suggest that binding of MMP-2 to both exosites is essential for its degradation by thrombin but not for its activation.
FIGURE 4.
Binding of heparan sulfate to thrombin elicits a decrease in thrombin-mediated MMP-2 degradation and a concomitant increase in activation. A, shown is the effect of heparan sulfate on the activation and degradation of MMP-2 and fibrinogen cleavage by thrombin. Pro-MMP-2-myc/His (20 μg/ml) or fibrinogen (50 μg/ml) was incubated with 50 nm thrombin in the presence of heparan sulfate under cell-free conditions. After 8 h the incubation mixtures were analyzed by Western blotting (IB)with an anti-myc antibody or by reducing SDS-PAGE. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2 or the ratio of the degraded MMP-2 to total MMP-2 (pro-MMP-2 + MMP-2). Data represent the mean and S.D. of three experiments. *, p < 0.05 versus 0 μg/ml heparan sulfate. Arrows indicate Aα, Bβ, and γ chains of fibrinogen (lower panel) (n = 2). B, shown is the effect of hirudin fragment on the activation and degradation of MMP-2 and fibrinogen cleavage by thrombin. Pro-MMP-2-myc/His (20 μg/ml) or fibrinogen (50 μg/ml) was incubated with 50 nm thrombin in the presence of hirudin fragment under cell-free conditions. After 8 h the incubation mixtures were analyzed as described above. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2 or the ratio of the degraded MMP-2 to total MMP-2 (pro-MMP-2 + MMP-2). Data represent the mean and S.D. of three experiments. *, p < 0.05 versus 0 μg/ml hirudin fragment.
FIGURE 5.
Simultaneous binding of MMP-2 to exosite 1 and 2 is essential for its degradation by thrombin. A, Western blot (IB) analysis using an anti-myc antibody of 20 μg/ml pro-MMP-2 incubated with 50 nm thrombin in the presence of heparan sulfate (40 μg/ml) and/or hirudin fragment (200 μm) for 8 h (n = 2) is shown. B, co-immunoprecipitation of MMP-2 and thrombin is shown. DMEM containing thrombin (200 nm) and MMP-2 (80 μg/ml) was immunoprecipitated in the presence of heparan sulfate (40 μg/ml) and/or hirudin fragment (200 μm) with anti-MMP-2 (CA-4001) followed by reducing SDS-PAGE and Western blotting with an anti-prothrombin antibody. Note that an autolytic cleavage product of thrombin (*) is seen. The panel at the bottom shows that MMP-2 is present in the immunoprecipitates (n = 3).
Hemopexin-like Domain Regulates Thrombin-mediated Degradation of MMP-2 through Binding to Exosite 1
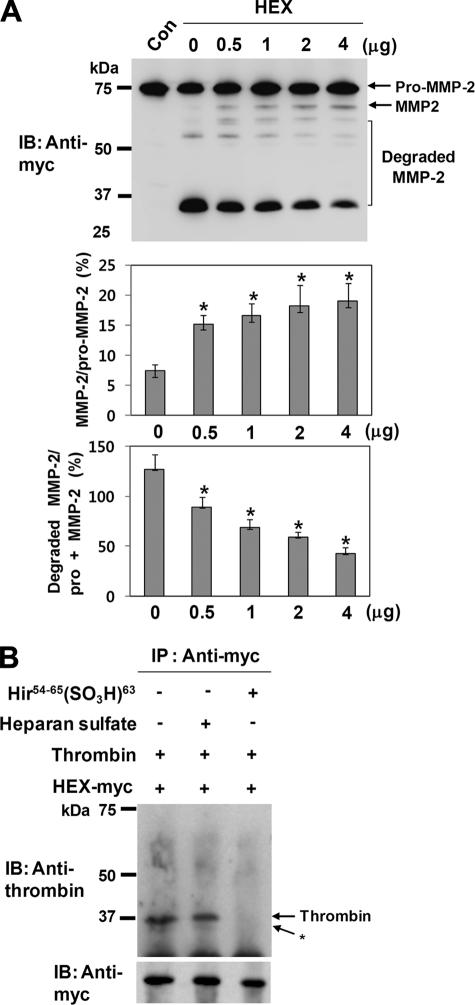
It has been shown that the hemopexin-like domain retains regulatory functions for MMP-2 enzymatic activity (6, 41, 42). Therefore, we investigated the functional role of the hemopexin-like domain in thrombin-mediated activation and degradation of pro-MMP-2. Pro-MMP-2-myc/His was incubated with thrombin in the presence of recombinant hemopexin-like domain under cell-free conditions, and the incubation mixture was analyzed by immunoblotting with an anti-myc antibody, revealing a decrease in MMP-2 degradation and a slight increase in activation (Fig. 6A). These data suggest that the interaction of thrombin with the recombinant hemopexin-like domain may disturb its binding to MMP-2 but result in minor conformational changes at the active site. Thus, to examine the interaction between thrombin and MMP-2 hemopexin-like domain, a plasmid expressing C-terminal myc/His-tagged hemopexin-like domain was constructed, and co-immunoprecipitation experiments were performed. Immunoprecipitation with an anti-myc antibody followed by immunoblotting with an anti-thrombin antibody showed binding of thrombin to the hemopexin-like domain (Fig. 6B). Moreover, the binding between these two proteins was completely prevented in the presence of hirudin fragment, whereas heparan sulfate did not affect the binding (Fig. 6B). Therefore, these data suggest that the hemopexin-like domain and hirudin fragment share exosite 1 for their binding to thrombin.
FIGURE 6.
Degradation of MMP-2 by thrombin is dependent on the hemopexin-like domain. A, Western blot (IB) analysis using an anti-myc antibody of 20 μg/ml pro-MMP-2 incubated with 50 nm thrombin in the presence of recombinant hemopexin-like domain of MMP-2 (HEX) for 12 h is shown. The bar graph shows the ratio of activated MMP-2 to pro-MMP-2 or the ratio of the degraded MMP-2 to total MMP-2 (pro-MMP-2 + MMP-2). Data represent the mean and S.D. of three experiments. *, p < 0.05 versus 0 μg/ml HEX. B, co-immunoprecipitation (IP) of MMP-2 hemopexin-like domain and thrombin is shown. DMEM containing thrombin (100 nm) and C-terminal myc/His-tagged hemopexin-like domain (HEX-myc) (20 μg/ml) was immunoprecipitated in the presence of heparan sulfate (40 μg/ml) or hirudin fragment (200 μm) with an anti-myc antibody followed by Western blotting with an anti-prothrombin antibody. Myc/His-tagged hemopexin-like domain was expressed in COS-1 cells and purified using anti-c-myc-agarose according to the manufacturer's recommendations (Sigma) (n = 3).
DISCUSSION
Although studies on the regulation of MMP-2 enzymatic activity have focused mainly on its activation by proteolytic processing of the propeptide (43), negative regulation of its enzymatic activity through degradation has also been reported (23, 26, 44). This indicates that MMP-2 enzymatic activity is regulated through both activation and degradation, thus, restricting excessive enzymatic activity. Moreover, previous studies have shown that plasmin is involved in MMP-2 degradation under cell-free conditions but that degradation is reduced greatly by the cell surface with a concomitant activation of pro-MMP-2 (23). In previous work we also showed that treatment with thrombin results in an increase in MMP-2 activation and a corresponding decrease in degradation under cell-associated conditions (26). Therefore, by seeking cellular molecules that associate with MMP-2, we hoped to elucidate the underlying molecular mechanisms of MMP-2 regulation.
The data provided here include several novel observations. First, heparan sulfate is a unique requirement for thrombin-mediated activation of pro-MMP-2. Second, contrary to our previous predictions, integrin αvβ3, MT1-MMP, TIMP-2, fibronectin, and collagens are not necessary for this activation. Third, binding of heparan sulfate to thrombin is primarily responsible for this activation, presumably through conformational changes at the active site of thrombin. Fourth, binding of MMP-2 to both exosites of thrombin is crucial for thrombin-mediated MMP-2 degradation. Thus, disruption of this interaction by exosite ligands results in a decrease in MMP-2 degradation. Finally, hemopexin-like domain regulates MMP-2 degradation through binding to exosite 1 of thrombin.
The essential role of heparan sulfate in thrombin-mediated activation of pro-MMP-2 suggests a novel mechanism for the regulation of MMP-2 enzymatic activity by cell surface molecules. There is one example in the literature in which MMP-2 activation is regulated by a heparan sulfate proteoglycan (synedican-2), but in this case MMP-2 activation was suppressed by the heparan sulfate proteoglycan, whereas activation was initiated by MT1-MMP (45). Moreover, syndecan-2, but not other syndecans, was specifically involved in this regulation process. In contrast, all heparan sulfate proteoglycans seem to be involved in thrombin-mediated activation because a similar increase in thrombin-mediated activation was observed in the presence of heparan sulfate under cell-free conditions. It was also shown that a glycosylphosphatidylinositol-anchored glycoprotein, RECK (reversion-inducing cysteine-rich protein with Kazal motifs), negatively regulated MT1-MMP-mediated activation of pro-MMP-2 on the cell surface (46). These data together with ours demonstrate a regulatory mechanism for MMP-2 enzymatic activity by cell surface molecules.
Various MMPs, including MMP-2, -7, -9, and -13, bind to heparan sulfate proteoglycans (35). It has been suggested that this binding might position the enzyme for directed proteolytic attack for activation (35). Bone sialoprotein, an acidic phosphoprotein, has also been shown to bind pro-MMP-2 (47, 48). Binding of sialoprotein to pro-MMP-2 is associated with structural changes and increased susceptibility to plasmin cleavage, leading to an increase in plasmin-mediated MMP-2 activation. Based on these data, we postulated that binding of heparan sulfate to MMP-2 may induce conformational changes or optimal orientation of the enzyme for proteolytic activation. Contrary to our expectations, the experimental evidence excludes this possibility, as disruption of the interaction between pro-MMP-2 and heparan sulfate does not affect MMP-2 activation. However, although the direct interaction of pro-MMP-2 with heparan sulfate was not required for thrombin-mediated activation under cell-free conditions, binding of pro-MMP-2 and thrombin to cell surface heparan sulfate proteoglycans (49) would bring them together in an organized fashion and in high concentration, leading to proteolysis at the cell surface even in the presence of high concentrations of inhibitors.
Our observation that binding of heparan sulfate to thrombin elicits an increase in thrombin-mediated MMP-2 activation and a concomitant decrease in degradation suggests two different mechanisms for the regulation of MMP-2 enzymatic activity through its activation and degradation. Just as multifunctional enzymes acquire specificity by exosite-dependent interactions, thrombin has two positively charged patches (exosites 1 and 2) that contribute to its specificity by binding substrates, cofactors, and other ligands (37, 38). Consistent with this, the present study showed that interaction of exosite 2 with heparan sulfate resulted in an increase in activated MMP-2, suggesting that ligand binding to exosite 2 of thrombin may induce allosteric changes of the active site with concomitant alterations in the substrate specificity of thrombin. Similar results were obtained with hirudin fragment, which is known to bind to exosite 1, although it was reported that ligand binding to exosites 1 and 2 differentially affected the conformation of the active site (39, 40). This demonstrates another mechanism by which thrombin utilizes exosite 1 and 2 to regulate its substrate specificity and reveals that different ligands play the same role in the regulation of thrombin activity.
The precise mechanism by which binding of heparan sulfate to thrombin induces a decrease in MMP-2 degradation is presently unclear. Potential mechanisms are suggested by our observation that the interaction between thrombin and MMP-2 is mostly abolished in the presence of heparan sulfate. Furthermore, the complete inhibition of the interaction between these two proteins in the combined presence of heparan sulfate and hirudin fragment suggests the involvement of both exosites in binding to MMP-2. Thus, we speculate that the two exosites may cooperatively bind MMP-2, thus, positioning MMP-2 for proteolytic degradation by thrombin. Inhibition of this binding prevents optimal orientation of MMP-2 for degradation and, thus, hinders MMP-2 degradation without affecting its activation. Another possible mechanism is that binding of both exosites to MMP-2 may induce conformational changes in MMP-2, which expose new cleavage sites for proteolytic degradation. Cooperative binding of exosites 1 and 2 was also reported for fibrin, factor V, factor VIII, and β2-glycoprotein I (50–53). Although none of these showed involvement in the degradation of the substrates, a role of the exosites in the activation of factor V and factor VIII was demonstrated (51, 52). These studies showed that recognition of factor V and factor VIII by both exosites was important for their activation by thrombin despite differential influences on the cleavage site specificity. The importance of exosite 1 in the proteolytic degradation of the substrates has been shown for ADAMTS13. In this study, preincubation of thrombin with an exosite 1 ligand, but not with an exosite 2 ligand, inhibited the degradation of ADAMTS13, suggesting the involvement of exosite 1 in this proteolytic process (54). Therefore, these data combined with ours indicate that binding to exosites 1 and/or 2 plays a role in the proteolytic cleavage of the substrates by thrombin, resulting in activation or degradation of the substrates. The present study demonstrated binding of exosite 1 to the hemopexin-like domain of MMP-2, but the binding site for exosite 2 remains to be elucidated. We suggest that exosite 2 may bind to the MMP-2 C-terminal domain because the hemopexin-like domain did not bind to exosite 2 and the interaction between thrombin and MMP-2 completely disappeared only in the combined presence of heparan sulfate (an exosite 2 ligand) and hirudin fragment (an exosite 1 ligand).
In conclusion, we propose a novel regulatory mechanism for thrombin-dependent MMP-2 enzymatic activity that is modulated by heparan sulfate proteoglycans. Interaction with heparan sulfate may induce allosteric changes of the active site with concomitant alterations in the substrate specificity of thrombin. This converts thrombin from a procoagulant to a potent extracellular matrix degradation enzyme by virtue of its ability to activate pro-MMP-2.
Supplementary Material
This work was supported by Future-based Technology Development Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (no. 2009-0081760).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MMP-2
- matrix metalloprotease 2
- MT
- membrane type
- TIMP
- tissue inhibitors of metalloproteases
- HBMEC
- human brain microvascular endothelial cell.
REFERENCES
- 1.Somerville R. P., Oblander S. A., Apte S. S. (2003) Genome Biol. 4, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woessner J. F., Jr. (1994) Ann. N.Y. Acad. Sci. 732, 11–21 [DOI] [PubMed] [Google Scholar]
- 3.Bescond A., Augier T., Chareyre C., Garçon D., Hornebeck W., Charpiot P. (1999) Biochem. Biophys. Res. Commun. 263, 498–503 [DOI] [PubMed] [Google Scholar]
- 4.Lazure C. (2002) Curr. Pharm. Des. 8, 511–531 [DOI] [PubMed] [Google Scholar]
- 5.Crabbe T., Smith B., O'Connell J., Docherty A. (1994) FEBS Lett. 345, 14–16 [DOI] [PubMed] [Google Scholar]
- 6.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995) J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 7.Morrison C. J., Butler G. S., Bigg H. F., Roberts C. R., Soloway P. D., Overall C. M. (2001) J. Biol. Chem. 276, 47402–47410 [DOI] [PubMed] [Google Scholar]
- 8.Nakada M., Yamada A., Takino T., Miyamori H., Takahashi T., Yamashita J., Sato H. (2001) Cancer Res. 61, 8896–8902 [PubMed] [Google Scholar]
- 9.Pei D. (1999) J. Biol. Chem. 274, 8925–8932 [DOI] [PubMed] [Google Scholar]
- 10.Nie J., Pei D. (2003) Cancer Res. 63, 6758–6762 [PubMed] [Google Scholar]
- 11.Rauch B. H., Bretschneider E., Braun M., Schrör K. (2002) Circ. Res. 90, 1122–1127 [DOI] [PubMed] [Google Scholar]
- 12.Baramova E. N., Bajou K., Remacle A., L'Hoir C., Krell H. W., Weidle U. H., Noel A., Foidart J. M. (1997) FEBS Lett. 405, 157–162 [DOI] [PubMed] [Google Scholar]
- 13.Lafleur M. A., Hollenberg M. D., Atkinson S. J., Knäuper V., Murphy G., Edwards D. R. (2001) Biochem. J. 357, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson M. T., Smith M. M., Smith S. M., Jackson C. J., Xue M., Little C. B. (2009) Arthritis Rheum. 60, 780–791 [DOI] [PubMed] [Google Scholar]
- 15.Chen J. M., Fortunato M., Stevens R. A., Barrett A. J. (2001) Biol. Chem. 382, 777–783 [DOI] [PubMed] [Google Scholar]
- 16.Kahan C., Seuwen K., Meloche S., Pouysségur J. (1992) J. Biol. Chem. 267, 13369–13375 [PubMed] [Google Scholar]
- 17.Chen D., Carpenter A., Abrahams J., Chambers R. C., Lechler R. I., McVey J. H., Dorling A. (2008) J. Exp. Med. 205, 1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozaki Y., Nishimura M., Sekiya K., Suehiro F., Kanawa M., Nikawa H., Hamada T., Kato Y. (2007) Stem Cells Dev. 16, 119–129 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Kong L., Kang J., Morgan J. H., 3rd, Shillcutt S. D., Robinson J. S., Jr., Nakayama D. K. (2009) Neurosci. Lett. 451, 199–203 [DOI] [PubMed] [Google Scholar]
- 20.McNamara C. A., Sarembock I. J., Gimple L. W., Fenton J. W., 2nd, Coughlin S. R., Owens G. K. (1993) J. Clin. Invest. 91, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galis Z. S., Kranzhöfer R., Fenton J. W., 2nd, Libby P. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 483–489 [DOI] [PubMed] [Google Scholar]
- 22.Henderson N., Markwick L. J., Elshaw S. R., Freyer A. M., Knox A. J., Johnson S. R. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L1030–L1038 [DOI] [PubMed] [Google Scholar]
- 23.Mazzieri R., Masiero L., Zanetta L., Monea S., Onisto M., Garbisa S., Mignatti P. (1997) EMBO J. 16, 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen M., Arkell J., Jackson C. J. (1999) Lab. Invest. 79, 467–475 [PubMed] [Google Scholar]
- 25.Monea S., Lehti K., Keski-Oja J., Mignatti P. (2002) J. Cell. Physiol. 192, 160–170 [DOI] [PubMed] [Google Scholar]
- 26.Koo B. H., Park M. Y., Jeon O. H., Kim D. S. (2009) J. Biol. Chem. 284, 23375–23385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan M. K., Williams K. A., Kivisäkk P., Pearce D., Stins M. F., Ransohoff R. M. (2004) J. Neuroimmunol. 153, 150–157 [DOI] [PubMed] [Google Scholar]
- 28.Brooks P. C., Strömblad S., Sanders L. C., von Schalscha T. L., Aimes R. T., Stetler-Stevenson W. G., Quigley J. P., Cheresh D. A. (1996) Cell 85, 683–693 [DOI] [PubMed] [Google Scholar]
- 29.Butler G. S., Butler M. J., Atkinson S. J., Will H., Tamura T., Schade van Westrum S., Crabbe T., Clements J., d'Ortho M. P., Murphy G. (1998) J. Biol. Chem. 273, 871–880 [DOI] [PubMed] [Google Scholar]
- 30.Wallon U. M., Overall C. M. (1997) J. Biol. Chem. 272, 7473–7481 [DOI] [PubMed] [Google Scholar]
- 31.Murphy G., Nguyen Q., Cockett M. I., Atkinson S. J., Allan J. A., Knight C. G., Willenbrock F., Docherty A. J. (1994) J. Biol. Chem. 269, 6632–6636 [PubMed] [Google Scholar]
- 32.Steffensen B., Wallon U. M., Overall C. M. (1995) J. Biol. Chem. 270, 11555–11566 [DOI] [PubMed] [Google Scholar]
- 33.Gioia M., Monaco S., Van Den Steen P. E., Sbardella D., Grasso G., Marini S., Overall C. M., Opdenakker G., Coletta M. (2009) J. Mol. Biol. 386, 419–434 [DOI] [PubMed] [Google Scholar]
- 34.Ikeda K., Iyama K., Ishikawa N., Egami H., Nakao M., Sado Y., Ninomiya Y., Baba H. (2006) Am. J. Pathol. 168, 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu W. H., Woessner J. F., Jr. (2000) J. Biol. Chem. 275, 4183–4191 [DOI] [PubMed] [Google Scholar]
- 36.Crabbe T., Ioannou C., Docherty A. J. (1993) Eur. J. Biochem. 218, 431–438 [DOI] [PubMed] [Google Scholar]
- 37.Huntington J. A. (2005) J. Thromb. Haemost. 3, 1861–1872 [DOI] [PubMed] [Google Scholar]
- 38.Bode W. (2006) Semin. Thromb. Hemost. 32, 16–31 [DOI] [PubMed] [Google Scholar]
- 39.Fredenburgh J. C., Stafford A. R., Weitz J. I. (1997) J. Biol. Chem. 272, 25493–25499 [DOI] [PubMed] [Google Scholar]
- 40.Verhamme I. M., Olson S. T., Tollefsen D. M., Bock P. E. (2002) J. Biol. Chem. 277, 6788–6798 [DOI] [PubMed] [Google Scholar]
- 41.Brooks P. C., Silletti S., von Schalscha T. L., Friedlander M., Cheresh D. A. (1998) Cell 92, 391–400 [DOI] [PubMed] [Google Scholar]
- 42.Morgunova E., Tuuttila A., Bergmann U., Isupov M., Lindqvist Y., Schneider G., Tryggvason K. (1999) Science 284, 1667–1670 [DOI] [PubMed] [Google Scholar]
- 43.Ra H. J., Parks W. C. (2007) Matrix Biol. 26, 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorsa T., Salo T., Koivunen E., Tyynelä J., Konttinen Y. T., Bergmann U., Tuuttila A., Niemi E., Teronen O., Heikkilä P., Tschesche H., Leinonen J., Osman S., Stenman U. H. (1997) J. Biol. Chem. 272, 21067–21074 [DOI] [PubMed] [Google Scholar]
- 45.Munesue S., Yoshitomi Y., Kusano Y., Koyama Y., Nishiyama A., Nakanishi H., Miyazaki K., Ishimaru T., Miyaura S., Okayama M., Oguri K. (2007) J. Biol. Chem. 282, 28164–28174 [DOI] [PubMed] [Google Scholar]
- 46.Oh J., Takahashi R., Kondo S., Mizoguchi A., Adachi E., Sasahara R. M., Nishimura S., Imamura Y., Kitayama H., Alexander D. B., Ide C., Horan T. P., Arakawa T., Yoshida H., Nishikawa S., Itoh Y., Seiki M., Itohara S., Takahashi C., Noda M. (2001) Cell 107, 789–800 [DOI] [PubMed] [Google Scholar]
- 47.Fedarko N. S., Jain A., Karadag A., Fisher L. W. (2004) FASEB J. 18, 734–736 [DOI] [PubMed] [Google Scholar]
- 48.Jain A., Karadag A., Fisher L. W., Fedarko N. S. (2008) Biochemistry 47, 10162–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimada K., Ozawa T. (1985) J. Clin. Invest. 75, 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pospisil C. H., Stafford A. R., Fredenburgh J. C., Weitz J. I. (2003) J. Biol. Chem. 278, 21584–21591 [DOI] [PubMed] [Google Scholar]
- 51.Esmon C. T., Lollar P. (1996) J. Biol. Chem. 271, 13882–13887 [DOI] [PubMed] [Google Scholar]
- 52.Segers K., Dahlbäck B., Bock P. E., Tans G., Rosing J., Nicolaes G. A. (2007) J. Biol. Chem. 282, 33915–33924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahgozar S., Yang Q., Giannakopoulos B., Yan X., Miyakis S., Krilis S. A. (2007) Arthritis Rheum. 56, 605–613 [DOI] [PubMed] [Google Scholar]
- 54.Crawley J. T., Lam J. K., Rance J. B., Mollica L. R., O'Donnell J. S., Lane D. A. (2005) Blood 105, 1085–1093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.