Abstract
In CHO cells, CDK1/2-dependent phosphorylation of Ubc2/Rad6 at Ser120 stimulates its ubiquitin conjugating activity and can be replicated by a S120D point mutant (Sarcevic, B., Mawson, A., Baker, R. T., and Sutherland, R. L. (2002) EMBO J. 21, 2009–2018). In contrast, we find that ectopic expression of wild type Ubc2b but not Ubc2bS120D or Ubc2bS120A in T47D human breast cancer cells specifically stimulates N-end rule-dependent degradation but not the Ubc2-independent unfolded protein response pathway, indicating that the former is E2 limiting in vivo and likely down-regulated by Ser120 phosphorylation, as modeled by the S120D point mutation. In vitro kinetic analysis shows the in vivo phenotype of Ubc2bS120D and Ubc2bS120A is not due to differences in activating enzyme-catalyzed E2 transthiolation. However, the Ser120 mutants possess marked differences in their abilities to support in vitro conjugation by the N-end rule-specific E3α/Ubr1 ligase that presumably accounts for their in vivo effects. Initial rate kinetics of human E3α-catalyzed conjugation of the human α-lactalbumin N-end rule substrate shows Ubc2bS120D is 20-fold less active than wild type E2, resulting from an 8-fold increase in Km and a 2.5-fold decrease in Vmax, the latter reflecting a decreased ability to support the initial step in target protein conjugation; Ubc2bS120A is 8-fold less active than wild type E2 due almost exclusively to a decrease in Vmax, reflecting a defect in polyubiquitin chain elongation. These studies suggest a mechanism for the integrated regulation of diverse ubiquitin-dependent signaling pathways through E2 phosphorylation that yields differential effects on its cognate ligases.
Keywords: Enzyme Kinetics, Proteasome, Protein Degradation, Protein Kinases, Ubiquitin, E1, N-end Rule, Ubc2, Ubr1
Introduction
The modification of specific target proteins with ubiquitin is a fundamental regulatory strategy within eukaryotes (1, 2), the principal consequence of which is commitment to 26 S proteasome-mediated degradation by assembly on the target protein of polyubiquitin chains linked principally though Lys48 (3). In contrast, monomeric ubiquitin or polymeric ubiquitin assembled from Lys63 linkages serve as transposable binding elements that direct functional consequences that are independent of the 26 S proteasome (for review, see Ref. 1). The diversity of these cellular roles arising from ubiquitin conjugation reflects the intrinsically greater information content represented by the constellation of surface residues present on the polypeptide and the combinatorial contributions of polyubiquitin chain formation through each of the seven lysines contained within the polypeptide. For these reasons there is considerable interest in understanding the mechanism of ubiquitin conjugation and the processes controlling this subset of post-translational modifications.
Conjugation of ubiquitin to a substrate protein requires the sequential action of three enzymes catalyzing the two half-reactions characteristic of all ligases (for review, see Ref. 4 and 5). In the first half-reaction, ubiquitin-activating enzyme (Uba1)2 catalyzes an ATP-dependent reaction that yields a ternary complex containing both a tightly enzyme bound ubiquitin adenylate and a covalently enzyme bound ubiquitin thiolester (6, 7). The latter high energy ubiquitin intermediate is transferred to members of a superfamily of enzymes, the ubiquitin carrier proteins (E2/Ubc (generic name for ubiquitin carrier protein/ubiquitin conjugating enzyme)), to form a high energy E2-ubiquitin thiolester intermediate in a process termed transthiolation (8, 9). In the second half-reaction, aminolysis of the high energy E2-ubiquitin thiolester bond is coupled to formation of an isopeptide bond between Gly76 of ubiquitin and lysines present on the substrate protein by a second superfamily of enzymes termed ubiquitin isopeptide ligases (E3), which are also responsible for assembling linkage-specific polyubiquitin chains (for review, see Ref. 4).
Cells utilize multiple strategies to regulate protein degradation through the ubiquitin-proteasome system. Increases in overall protein degradation, termed general enhanced degradation, results from the coordinated induction of ubiquitin and specific ligation components, as occurs in the terminal maturation of erythrocytes (10), the developmentally programmed post-eclosion degeneration of Manduca sexta intersegmental muscles during insect metamorphosis (11, 12), and skeletal muscle atrophy accompanying starvation or denervation (13). In contrast, specific enhanced degradation of a single protein or subset of proteins within the larger population of intracellular constituents is largely regulated by substrate availability. In such cases, proteins generally are not targeted by induction of the responsible ligation pathway but by increasing specific substrate availability for conjugation by regulating the rate of a post-translational modification(s) that predisposes the resulting protein for conjugation, such as occurs after phosphorylation of IκB and proline hydroxylation of Hif1α (14, 15). Thus, substrate availability reflects the combinatorial regulation of spatial and/or temporal signals from disparate signaling pathways that culminate in specific enhanced degradation. Control of protein targeting by modification of components within the ubiquitination machinery also regulates substrate conjugation independent of substrate availability by altering protein-protein interactions or by activating specific ligases. Among many examples, the anaphase promoting complex required for ubiquitination of mitotic cyclins and other regulatory proteins that are targeted for destruction during cell division is activated in part by Cdk1-dependent phosphorylation (16). Other studies suggest that phosphorylation of Cdc34/Ubc3 by Cdk2 also regulates its activity as an E2 in supporting anaphase promoting complex-dependent ubiquitination (17).
Sarcevic et al. (18) have demonstrated a role for Cdk1/2-dependent phosphorylation of Ubc2/Rad6 at Ser120 in regulating histone ubiquitination and cell cycle progression in Chinese hamster ovary cells and Saccharomyces cerevisiae. In vivo studies additionally demonstrate a significantly reduced growth rate and extended cell cycle for S. cerevisiae conditional rad6 deletion mutants complemented with HsUbc2aS120A or HsUbc2aS120T point mutants; in contrast, complementation with a HsUbc2aS120D mutant results in yeast exhibiting wild type growth rates, indicating that substitution with aspartate mimics the effect of phosphorylation (18). These studies did not address the effect of Ser120 phosphorylation on Uba1-catalyzed ubiquitin transthiolation or other targeting pathways utilizing Ubc2. The Ubc2 protein is required for post-replicative DNA repair in eukaryotes and is recruited to chromatin lesions by the Rad5 and Rad18 Ring-finger proteins to form a complex with Mms2 and Ubc13 that catalyzes Lys63-linked polyubiquitin chain formation (19, 20). In addition, Ubc2 supports the N-end rule targeting pathway through Ubr1/E3α ubiquitin ligase-catalyzed Lys48-linked polyubiquitin chain formation that is required for degrading incorrectly trafficked cellular proteins, regulatory components of several key signaling pathways, and unfolded proteins (21–24) as well as the monoubiquitination of histones during transcriptional regulation (25–27). The Ubr1 ligase also serves as an intracellular leucine sensor in regulating steady state levels of mTOR and its consequent signaling pathways (28).
In the present studies, we have used site directed mutagenesis and in vitro kinetic assays to examine the function(s) of Ser120 within human Ubc2 and the paralogous position of an unrelated E2 family (Ubc5). The results indicate that position 120 represents a key binding residue for interaction of E2 paralogs with the Uba1 and accounts in part for the family-specific difference in binding affinity for Ubc2 versus Ubc5. More important, mimicking phosphorylation by mutation of Ser120 of Ubc2 to aspartate functionally abrogates its ability to support E3α-dependent N-end rule targeting through specific effects on the mechanism of ligase-catalyzed Lys48 polyubiquitin chain formation. The latter observations suggest that phosphorylation at Ser120 may serve as a regulatory switch, shifting the flux of Ubc2-dependent conjugation among different regulatory pathways.
MATERIALS AND METHODS
Bovine ubiquitin, creatine phosphokinase, yeast inorganic pyrophosphatase, and human α-lactalbumin were purchased from Sigma. Ubiquitin and α-lactalbumin were further purified to apparent homogeneity (29, 30). Recombinant UbK48R was that described previously (29). Wild type and mutant ubiquitin were radioiodinated using carrier free Na[125I] (Perkin Elmer Life Sciences) by the chloramine-T method (31) and generally yielded specific activities of 15,000–20,000 cpm/pmol (32). Human ubiquitin-activating enzyme (HsUba1) was prepared from outdated red blood cells and was quantitated by the stoichiometric formation of HsUba1-125I-ubiquitin thiolester (33). The N-end rule E3α ligase was purified from human erythrocyte fraction II by E2 ligand affinity chromatography (34).
Generation of HsUbc2b and HsUbc5B Point Mutants
Point mutants of HsUbc2b/E214kb were generated by overlap extension PCR from pGEX-HsUbc2b to yield pGEX-HsUbc2bS120A, pGEX-HsUbc2bS120D, and pGEX-HsUbc2bS120E expression plasmids; an identical approach was used to generate pGEX-HsUbc5BD116N and pGEX-HsUbc5BD117S point mutants from pGEX-HsUbc5B (30, 35). The HsUbc5BD116N/D117S double mutant was similarly generated by sequential introduction of single mutations into HsUbc5B. The complete coding region of each mutant was sequenced to confirm the desired modification and to preclude secondary mutations. The corresponding GST fusion proteins were expressed in Escherichia coli BL21 cells by 0.4 mm isopropyl-β-d-thiogalactopyranoside induction for 2 h then purified by glutathione-Sepharose affinity column chromatography (30). The GST moiety was processed from the resulting fusion proteins using thrombin at a final concentration of 50 IU/ml. Quantitative processing and production of the intact recombinant E2 proteins were confirmed by MALDI-TOF mass spectroscopy. Processed GST and residual GST-E2 fusion protein were removed by passing the sample through a second glutathione-Sepharose affinity column (30). Processed recombinant E2 contained in the resulting unadsorbed fraction was purified to apparent homogeneity by Mono Q anion exchange FPLC (9). The E2 mutants were quantitated by their HsUba1-catalyzed stoichiometric formation of 125I-ubiquitin thiolesters (30). The E2 proteins were typically greater than 90% active when compared with protein content spectrophotometrically quantitated using empirical 280-nm extinction coefficients for HsUbc2b (1.37 ml·mg−1) or HsUbc5B (1.52 ml·mg−1). No consistent difference in fraction of active protein was observed between wild type and mutant HsUbc2B or HsUbc5B, precluding destabilization of the E2 proteins by the residue changes.
Transthiolation Kinetics For HsUbc2b Mutants
Initial rates of E2 transthiolation were measured by the same method employed for the stoichiometric end point assays except that HsUba1 was present at a catalytic concentration (30). Reactions of a 50-μl final volume contained 50 mm Tris-HCl (pH 7.5), 1 mm ATP, 10 mm MgCl2, 1 mm DTT, 0.5 nm HsUba1, 5 μm 125I-ubiquitin, and the indicated concentrations of recombinant wild type or mutant HsUbc2b or HsUbc5B (30). Reactions were initiated by the addition of radiolabeled ubiquitin. After 1 min of incubation at 37°C, the assays were quenched by the addition of 2× SDS sample buffer from which β-mercaptoethanol had been omitted. Samples were immediately resolved by non-reducing SDS-PAGE without boiling in order to stoichiometrically preserve the thiolesters formed for subsequent autoradiography and quantitation by γ counting (9, 30). Preliminary experiments confirmed that 1-min incubations were within the initial velocity region for HsUba1-catalyzed transthiolation (30). Kinetic parameters were determined by nonlinear hyperbolic regression analysis using the Grafit suite of programs to obviate weighting errors (30).
E3α-catalyzed Conjugation Assays
Initial rates of E3α-catalyzed 125I-ubiquitin conjugation were measured in coupled assays as described previously (30, 34). Incubations of a 25-μl final volume contained 50 mm Tris-HCl (pH 7.5), 1 mm ATP, 10 mm MgCl2, 10 mm creatine phosphate, 1 IU/ml creatine phosphokinase, 1 mm DTT, 1 IU/ml HPLC-purified yeast inorganic pyrophosphatase, 5 μm 125I-ubiquitin, 25 μm human α-lactalbumin, 30 nm HsUba1, 1 μg of human E3α, and the indicated concentration of wild type or mutant HsUbc2b. After the indicated time at 37 °C, reactions were quenched by the addition of 2× SDS sample buffer containing 1% (v/v) β-mercaptoethanol then incubated for 5 min at 100 °C. Ubiquitin adducts were resolved by reducing SDS-PAGE and identified by autoradiography of the dried gel (30, 34). Conjugated 125I-ubiquitin was determined by γ-counting of excised lanes, and initial velocities were calculated from the specific radioactivity of the 125I-ubiquitin (30, 34). Where indicated, 125I-UbK48R was substituted for wild type radioiodinated ubiquitin. Control experiments confirmed that the assays were E3α limiting by the absence of an effect on initial velocity of doubling the concentration of HsUba1.
Transfection Experiments
The ubiquitin-proteasome reporter constructs pUbRYFP and pYFPCL1 were the generous gift of Dr. Nico Dantuma (Karolinska Institute) and are described elsewhere (36–38). The pTriEX-HsUbc2b, pTriEX-HsUbc2bS120A, and pTriEX-HsUbc2bS120D plasmids were generated by subcloning the corresponding EcoR1/HindIII full-length coding regions from the respective pGEX bacterial expression plasmids into pTriEx-3, then the DNA was sequenced to obviate secondary mutations. The latter strategy resulted in appending of an amino-terminal MAISRELVDPN sequence to the HsUbc2b proteins, allowing one to distinguish the ectopically expressed E2 from endogenous protein as a band migrating at a 1200-Da greater relative molecular weight. A similar cloning strategy was used to generate pTriEx-YFP from pUbRYFP but without the appended amino-terminal peptide.
Human T47D breast cancer cells were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum at 37 °C in a 5% (v/v) CO2 atmosphere. For each experimental condition, triplicate 60-mm plates at 80% confluency were transfected for 3 h at 37 °C with 4 μg of individual plasmid DNA using Lipofectamine Plus according to the manufacturer's protocol. After 3 h, cultures were transferred to fresh medium. After 24 h, cultures were rinsed twice with 25 mm phosphate-buffered saline and harvested directly into 0.5 ml of 1× SDS sample buffer then incubated at 100 °C for 5 min (39). Aliquots of each replicate were analyzed by SDS-PAGE and Western blot, as described in the accompanying legend to Fig. 6. (40). The resulting fluorescence was directly quantitated using a Kodak Image Station 2000R instrument under linear sampling conditions. Aliquots of the replicate samples were separately pooled, resolved by SDS-PAGE, and analyzed by Western blotting using anti-HsUbc2b antibody to quantitate endogenous versus ectopically expressed wild type and mutant HsUbc2b protein (40).
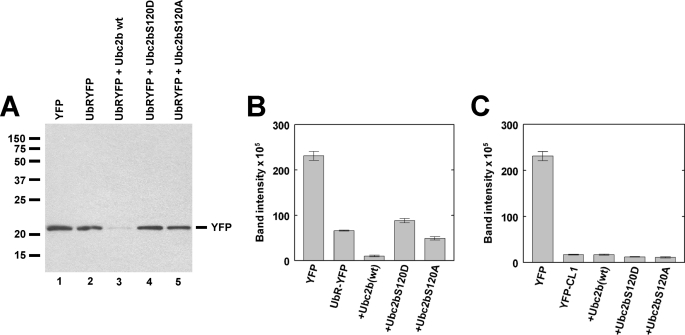
FIGURE 6.
Ser120 mutation inhibits degradation of a model N-end rule substrate. A, shown is a Western blot stained for YFP and visualized by chemiluminescence for pooled samples from triplicate plates of T47D human breast cancer cells transfected as described under “Materials and Methods” to ectopically express YFP or the model N-end rule substrate UbRYFP in the absence or presence of wild type or mutant HsUbc2b as indicated. Relative molecular weights are indicated to the left, and the migration position for free YFP is indicated to the right. B, shown is fluorescence quantitation of the Western blot of panel A. C, shown is fluorescence quantitation of the Western blot derived from a parallel experiment identical to that of panels A and B but expressing the model unfolded protein response substrate YFP-CL1. The YFP alone control is identical for panels A and B. Error bars correspond to the S.D. for triplicate samples.
RESULTS
Ser120 Contributes to HsUba1 Binding
The in vivo phenotypes reported by Sarcevic et al. (18) that are associated with mutating or phosphorylating Ser120 of Ubc2 could result from the effect(s) on the step of Uba1-catalyzed transthiolation to charge the E2 with activated ubiquitin or downstream effect(s) on subsequent steps of Ubc2-ubiquitin thiolester-supported conjugation by its cognate ligase(s).3 We have shown that the initial rate for E2 transthiolation from E1 ternary complex serves as a sensitive and quantitative reporter function for monitoring E1-E2 binding interactions within the pathways for conjugation of ubiquitin and other Class 1 ubiquitin-like proteins (30, 41, 42). Therefore, to test the role of Ser120 on binding to HsUba1, we examined the concentration dependence of wild type or mutant HsUbc2b on the initial rate of HsUba1-catalyzed transthiolation. The autoradiogram of Fig. 1A obtained by non-reducing SDS-PAGE to preserve the HsUbc2b-125I-ubiquitin thiolesters formed during the incubations demonstrates formation of increasing amounts of the high energy intermediate within 1 min with increasing concentrations of the carrier protein under HsUba1 limiting conditions. In contrast, much less 125I-ubiquitin thiolester is formed with a HsUbc2bS120A point mutant over a similar concentration range (Fig. 1A). The incubation time and HsUba1 concentration used in Fig. 1A were empirically chosen to fall within the initial velocity range for HsUbc2b transthiolation; therefore, the autoradiographic intensities of the resulting HsUbc2b-125I-ubiquitin thiolester bands are proportional to the respective initial rates for 125I-ubiquitin thiolester transfer from HsUba1 ternary complex to HsUbc2b (30). Excision and γ counting of the resulting thiolester bands allowed absolute quantitation of the HsUbc2b-125I-ubiquitin thiolesters (9, 30). The linearity of a double-reciprocal plot obtained from a parallel experiment at slightly different concentrations of HsUbc2b confirms that the mechanism follows hyperbolic Michaelis-Menten kinetics from which Km and kcat (determined as Vmax/[HsUba1]o) could be calculated (Fig. 1B). To avoid errors associated with weighting of the datum points, values of Km and Vmax were determined by nonlinear hyperbolic regression analysis (30).
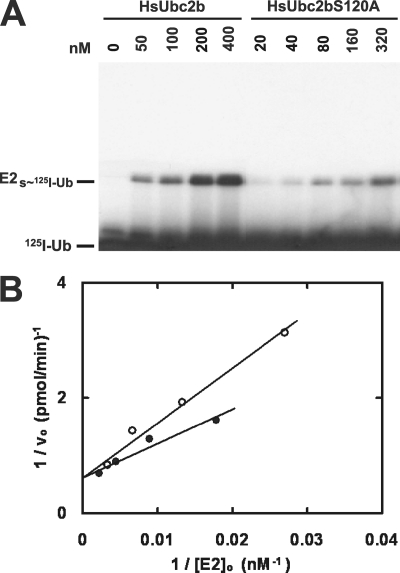
FIGURE 1.
Ser120 contributes to HsUba1 binding. A, shown is an autoradiogram of a non-reducing SDS-PAGE gel illustrating the [HsUbc2b]o-dependent formation of the resulting 125I-ubiquitin thiolester catalyzed by HsUba1 under initial velocity conditions, as described under “Materials and Methods.” B, shown is a double-reciprocal plot of the corresponding initial rates calculated after excising the E2-125I-ubiquitin thiolester bands from the gel of panel A and determining associated radioactivity by γ counting. Closed circles, wild type HsUbc2b; open circles, HsUbc2bS120A.
The data of Fig. 1 yielded a Km of 123 ± 19 nm and a kcat of 4.5 ± 0.3 s−1 for wild type HsUbc2b, in good agreement with previously published values (30, 41, 42), Fig. 2A and supplemental Table 1. Mutation of Ser120 to alanine resulted in a Km of 180 ± 11 nm for HsUbc2bS120A, representing a modest ΔΔGbinding of 0.2 kcal/mol, Fig. 2A; therefore, Ser120 makes a small but significant contribution to the overall binding energy of −9.4 kcal/mol for association of wild type HsUbc2b with HsUba1 ternary complex. Mutating Ser120 to aspartate results in a much larger effect on binding to HsUba1, yielding a Km of 37 ± 5 nm for HsUbcS120D (Fig. 2A) that represents a ΔΔGbinding of 0.7 kcal/mol. As found with HsUbc2bS120A, mutating Ser120 to aspartate has no significant effect on kcat (Fig. 2A). There is a modest effect of geometry associated with inserting an additional methylene carbon into the side chain of aspartate when Ser120 is mutated to glutamate (Km = 53 ± 9 nm; ΔΔGbinding = 0.5 kcal/mol) but no effect on kcat (Fig. 2A). Therefore, Ser120 lies at the binding interface between HsUba1 ternary complex and uncharged HsUbc2b. Phosphorylation of Ser120, modeled by mutation to aspartate to allow quantitative kinetic characterization, enhances binding of HsUbc2b with the ubiquitin-activating enzyme but does not alter transition state stability. These results indicate that phosphorylation of Ser120, as modeled by mutation to aspartate, should provide a modest advantage in the ability of phosphoHsUbc2 to compete with other E2 paralogs for charging by Uba1; however, the effect is probably not large enough to account for the gain-of-function phenotypes reported by Sarcevic et al. (18), as E1 is not thought to be the rate-limiting step for ubiquitin conjugation (5).
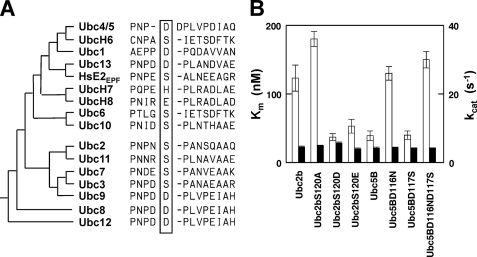
FIGURE 2.
Asp120 accounts for high affinity binding of HsUbc5B to HsUba1. A, shown is a sequence comparison of human E2 paralogs flanking Ser120 (HsUbc2b numbering). The E2 families are arranged phylogenetically and segregate into distinct families comprising the Ubc4/5- and Ubc2/Rad6-like superclades. B, shown is a summary of kinetic constants (±S.E.) for the indicated wild type or mutant HsUbc2b and HsUbc5B paralogs in HsUba1-catalyzed transthiolation, determined as described in the legend to Fig. 1 and “Materials and Methods.” Data values are summarized in supplemental Table 1. Open bars, values of Km (left axis); closed bars, values of kcat (right axis).
Asp120 Accounts for High Affinity Binding of HsUbc5B to HsUba1
Sequence comparison of the human E2 paralogs reveals that residue 120 or its structurally paralogous position within the core catalytic domain of the superfamily is markedly conserved for either serine or aspartate (Fig. 2A). The only exceptions are the related vertebrate-specific UbcH7 and UbcH8 isoforms that bear histidine or glutamate, respectively, at this position (Fig. 2A) based on superposition of the crystal structures for ScUbc2 (PDB code 1AYZ), HsUbcH7 (PDB code 1FBV), and HsUbcH8 (PDB code 1WZW) (results not shown). There is no consistent pattern to the preference for serine versus aspartate at position 120 between the two major E2 superclades or among the E2 families comprising the superclades (Fig. 2A). The kinetic results with HsUbc2bS120D suggest that E2 families harboring aspartate at the paralogous position should exhibit enhanced affinities for binding to HsUba1 ternary complex (Fig. 2B and supplemental Table 1. To test this directly, we examined the concentration dependence of wild type HsUbc5B on the initial rate for HsUba1-catalyzed transthiolation in a manner identical to that for HsUbc2b, as an example of an E2 family containing aspartate at this position. Initial rates of HsUba1-catalyzed HsUbc5B transthiolation versus [HsUbc5B]o yielded a Km of 39 ± 7 nm, in excellent agreement with that determined for HsUbc2bS120D (Fig. 2B) and consistent with the hypothesis that the identity of the residue at position 120 (HsUbc2b numbering) significantly influences binding of E2 paralogs to the ubiquitin activating enzyme. As further confirmation, we next examined the kinetics for HsUbc5B in which the paralogous aspartate was mutated to an uncharged group. ClustalW analysis initially predicted Asp117 of HsUbc5B as being paralogous to Ser120 of HsUbc2b, but mutation of this residue to serine failed significantly to affect the Km (40 ± 6 nm) relative to that of wild type HsUbc5b (Fig. 2B). However, a detailed comparison of structures for yeast Ubc2 (PDB code 1AYZ) versus human Ubc5B (PDB code 2ESK) indicated that Asp116 occupies the position paralogous to Ser120 of Ubc2. Subsequent mutation and kinetic analysis yielded a Km of 130 ± 10 nm and a kcat of 4.4 ± 0.1 s−1 for HsUbc5BD116N4 (Fig. 2B). The HsUbc5B double mutant exhibited Km and kcat values indistinguishable from those found by mutating Asp116 alone, indicating that the effect on affinity of binding to HsUba1 ternary complex is highly position-specific (Fig. 2B). In addition, the observation that mutation of Asp116 of HsUbc5B and its paralogous Ser120 of HsUbc2b alters Km but not kcat demonstrates that this position is exclusively a binding residue within the HsUba1 catalytic cycle.
Ser120 of HsUbc2b Is Critical For E3α-dependent Conjugation
The previous transthiolation kinetics demonstrate that position 120 is important in defining the affinity with which HsUbc2b, HsUbc5B, and presumably other E2 paralogs bind to the Uba1 ternary complex, thus, determining their ability to compete for charging by the ubiquitin-activating enzyme to support their respective downstream conjugation pathways. However, the presumed effect of phosphorylation modeled by the S120D point mutant is not sufficiently large to account for the functional phenotypes observed (18), suggesting that the principal consequences are in the step of E3-catalyzed ubiquitin conjugation. To investigate the contribution of position 120 in the subsequent step of E3-dependent substrate conjugation, we examined the HsUbc2b point mutants for their ability to support the human N-end rule ligase E3α/Ubr1 (30, 34).
Fig. 3 shows a representative autoradiogram for E3α-dependent ubiquitination of the model type 1 N-end rule substrate human α-lactalbumin (43). No conjugation was observed in the absence of ligand affinity purified human E3α (Fig. 3, lane 1); in contrast, a modest rate of HsUbc2b autoubiquitination was observed in the presence of E3α when α-lactalbumin was omitted (Fig. 3, lane 2). In the presence of E3α and substrate, a ladder of polyubiquitinated α-lactalbumin conjugates was formed that corresponds in mobility to the successive addition of single ubiquitin moieties (Fig. 3, lanes 3–5), as shown previously (34). Because the incubations were conducted under initial velocity conditions, the relative autoradiographic intensities are proportional to their rates of formation, within the linear response range of the film. When the concentration of HsUbc2b was increased from 100 to 500 nm, no qualitative difference in conjugation was observed except for an increased level of E2 autoubiquitination found also in the absence of substrate (Fig. 3, lanes 2 and 5). The relative independence of the initial rate of α-lactalbumin polyubiquitination when [HsUba1]o was doubled confirms that the assay is E3α limiting, as does the small effect of increasing [HsUbc2b]o, as the ligase is near saturation with respect to the reported Km for HsUbc2b-125I-ubiquitin thiolester binding to human E3α of 54 ± 18 nm (30). Because such measurements reflect only the rate-limiting step, the E3α-catalyzed step can, thus, be kinetically isolated under these conditions even though multiple steps are involved in the ubiquitin conjugation cascade, as has been discussed previously (30).
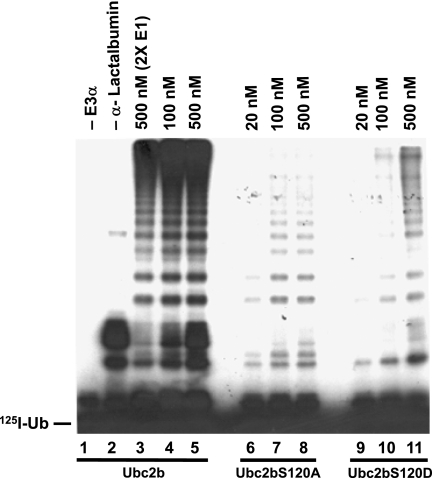
FIGURE 3.
Ser120 is critical for E3α-catalyzed polyubiquitination of human α-lactalbumin. Autoradiogram of a reducing SDS-PAGE gel illustrating the E3α-catalyzed polyubiquitination of human α-lactalbumin in 10-min incubations, as described under “Materials and Methods.” Concentrations refer to the corresponding recombinant wild type or mutant HsUbc2b. The migration position for free 125I-ubiquitin is shown to the left.
When either HsUbc2bS120A or HsUbc2bS120D is substituted for wild type HsUbc2b, there is a significant decrease in the initial velocity of substrate conjugation, shown by the reduced autoradiographic intensities for the E3α-catalyzed α-lactalbumin-linked polyubiquitin chains (Fig. 3, lanes 6–11). Because the autoradiographic intensities are proportional to the rate of product formation, the data of Fig. 3 qualitatively reveal very different mechanistic consequences arising from the two mutations. There is no significant increase in the autoradiographic densities of the polyubiquitinated α-lactalbumin bands when [HsUbc2bS120A]o is increased from 100 to 500 nm (Fig. 3, lanes 7 and 8); therefore, the reaction remains near saturation at these concentrations of the E2 mutant, setting an upper limit for the Km of HsUbc2bS120A-125I-ubiquitin thiolester binding to E3α. The overall attenuation in conjugation must, thus, result from a kcat effect on the rate-limiting step of polyubiquitin chain formation, which is thought to involve attachment of the first ubiquitin of the intact chain (34). In contrast, Fig. 3 shows that α-lactalbumin polyubiquitination is acutely dependent on the concentration of HsUbc2bS120D over the range examined (lanes 9–11), demonstrating that the consequences of this point mutation must minimally result in an increased Km for binding of HsUbc2bS120D-125I-ubiquitin thiolester to E3α, although additional effects on kcat cannot be precluded from the data of Fig. 3.
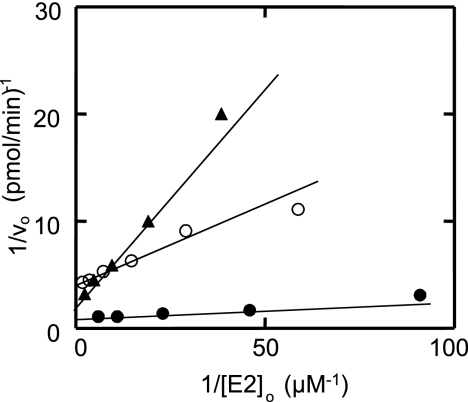
We quantitatively investigated the concentration dependence of the HsUbc2b mutants on the initial velocity of total 125I-ubiquitin α-lactalbumin conjugation under E3α-limiting conditions to confirm the conclusions of Fig. 3 (30, 34). Assays were conducted in parallel to preclude any variation in E3α activity with successive freeze-thaw cycles of the purified enzyme. The dependence of rate on wild type or mutant HsUbc2b concentration was hyperbolic, as indicated by the linearity of the corresponding double reciprocal plots (Fig. 4). Non-linear hyperbolic regression analysis of initial velocity versus [HsUbc2b]o yielded the corresponding Km and Vmax values, summarized in Table 1. The Km for HsUbc2b, representing binding of wild type HsUbc2b-125I-ubiquitin thiolester to E3α, was 22 ± 5 nm, in good agreement with the value of 54 ± 18 nm reported previously (30). As expected from the qualitative analysis of the autoradiogram of Fig. 3, HsUbc2bS120A exhibited a negligible change in Km (36 ± 4 nm) relative to wild type protein but an ∼5-fold decrease in Vmax (Table 1). In contrast, the HsUbc2bS120D mutant exhibited an ∼8-fold increase in Km (182 ± 30 nm), representing a 1.2 kcal/mol decrease in binding (ΔΔGbinding) of the corresponding 125I-ubiquitin thiolester, as expected from the autoradiogram of Fig. 3, lanes 9–11. Not evident in Fig. 3 but unambiguously demonstrated by the more detailed kinetic analysis of Fig. 4, HsUbc2bS120D also results in an 2.5-fold decrease in kcat, reflected by the lower Vmax (Table 1). Because there is no stoichiometric assay for E3α comparable with those for E1 and E2, we are unable to calculate kcat reliably; however, one can estimate the relative catalytic specificity of the enzyme, defined as Vmax/Km, and corresponding to the second order rate constant for the enzyme. A comparison of Vmax/Km values in Table 1 indicates that both mutations significantly affect E3α activity and that mutation of Ser120 to aspartate effectively inactivates the enzyme, as the residual activity is 22-fold lower than wild type HsUbc2b (Table 1).
FIGURE 4.
Wild type and Ser120 mutants of HsUbc2b display hyperbolic kinetics in E3α-catalyzed human α-lactalbumin conjugation. Initial rates of E3α-catalyzed human α-lactalbumin conjugation were determined at the indicated wild type or mutant HsUbc2b concentrations, as described under “Materials and Methods.” The linearity of the resulting Lineweaver-Burk plots for wild type HsUbc2b (closed circles), HsUbc2bS120A (open circles), and HsUbc2bS120D (closed triangles) indicates hyperbolic kinetics for the three recombinant proteins.
TABLE 1.
Summary of kinetic constants for E3α-catalyzed conjugation of α-lactalbumin
| K1a | Vmax | Vmax/Km | |
|---|---|---|---|
| nm | nm·s−1 | s−1 × 10−2 | |
| HsUbc2b | 22 ± 5 | 0.70 ± 0.12 | 3.3 ± 0.3 |
| HsUbc2bS120A | 36 ± 4 | 0.16 ± 0.01 | 0.44 ± 0.10 |
| HsUbc2bS120D | 182 ± 30 | 0.28 ± 0.02 | 0.15 ± 0.06 |
HsUbc2b Ser120 Mutations Affect E3α -dependent Polyubiquitin Chain Initiation
The reduced kcat for E3α-dependent conjugation that results from mutating Ser120 (Table 1) could reflect effects on the step of chain initiation, chain elongation, or both, as product formation is monitored as total 125I-ubiquitin conjugated. To distinguish among these three alternatives, we examined the kinetics for conjugation of 125I-UbK48R, which is unable to support polyubiquitin chain formation (3, 44) but allows one kinetically to isolate the step of chain initiation represented by ligation of the first ubiquitin (34).
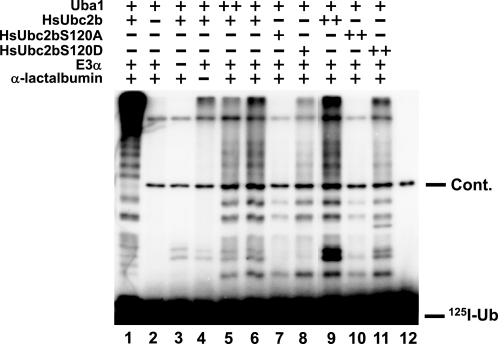
Lane 1 of Fig. 5 shows the pattern of robust polyubiquitination of α-lactalbumin observed with wild type HsUbc2b, E3α, and saturating wild type 125I-ubiquitin. In contrast, substitution of 125I-UbK48R for radioiodinated wild type ubiquitin results in significant attenuation of overall conjugation, particularly in the high molecular weight hyperconjugated α-lactalbumin that otherwise fails to enter the gel (Fig. 5, lane 6). As before, the kinetic assays of Fig. 5 were performed under conditions chosen to render overall ubiquitination rate-limiting with respect to [E3α]o, shown by the absence of a proportional increase in conjugation when HsUba1 levels were doubled (lanes 5 and 6). In addition, the incubation time was empirically shown to be within the initial velocity region of the reactions so that the autoradiographic intensities were proportional to the initial rates of monoubiquitination of human α-lactalbumin as the rate-limiting step of chain formation (30). No ubiquitination of α-lactalbumin was observed in the absence of HsUbc2b (lane 2), whereas omission of E3α resulted only in minor autoubiquitination of the E2 in the presence of HsUbc2b and HsUba1 (lane 3). The addition of E3α to the reaction mixture increased the multi-monoubiquitination of endogenous proteins in the incubation, which was significantly enhanced when the substrate α-lactalbumin was added (lanes 4 and 6). At equimolar HsUbc2b, the S120A mutant exhibits significantly less 125I-UbK48R monoubiquitination relative to wild type E2 than does the S120D mutant (lanes 6 and 7). If the assays are repeated at saturating wild type or mutant HsUbc2b concentrations to adjust for the effect of the S120D mutation on Km (Table 1), the effect on monoubiquitination is more pronounced for the S120A mutant (lanes 9–11). Indeed, the relative ranking in monoubiquitination found for HsUbc2bS120A compared with HsUbc2bS120D agrees well with their relative Vmax values found in Table 1. The results of Fig. 5, thus, indicate that the principal kinetic effect of mutating Ser120 is on the rate-limiting step of chain initiation catalyzed by E3α.
FIGURE 5.
Ser120 mutation affects E3α-catalyzed polyubiquitin chain initiation. Incubations identical to those used in Fig. 4 were conducted in the absence (−) or presence of 30 (+) or 60 (++) nm HsUba1, 20 (+) or 100 (++) nm wild type HsUbc2b, 30 (+) or 150 (++) nm HsUbc2bS120A, and 150 (+) or 750 (++) nm HsUbc2bS120D, as indicated. Incubations contained either 5 μm 125I-wild type ubiquitin (lane 1) or 125I-UbK48R (lanes 2–12). Lane 12 contained only 125I-UbK48R as a negative control, revealing the presence of a higher relative molecular weight contaminating band (Cont.). The migration position for free 125I-ubiquitin is shown to the right.
HsUbc2b Ser120 Mutations Fail to Support N-end Rule-dependent Proteolysis
The inability of the HsUbc2b Ser120 mutants fully to support E3α-dependent conjugation (Fig. 5 and Table 1), particularly with respect to Lys48-linked polyubiquitin chain formation, predicts that N-end rule-dependent 26 S proteasomal degradation should also be defective. To test this, we ectopically co-expressed wild type or mutant HsUbc2b constructs with a UbRYFP model N-end rule substrate in T47D human breast cancer monolayer cultures, as described under “Materials and Methods.” In UbRYFP, ubiquitin is fused to the amino terminus of yellow fluorescent protein harboring an amino-terminal arginine (36, 37). Expression of the fusion protein results in the co-translational processing of ubiquitin from the nascent polypeptide to expose the synthetic amino-terminal arginine of YFP, a primary destabilizing residue within the N-end rule pathway (36, 45). Because of endogenous fluorescence within the cell cultures, we found it more convenient and accurate to monitor YFP stability by quantitative Western blot using rabbit anti-GFP antibody, which fully cross-reacts with YFP rather than by the ablation of YFP fluorescence (36, 37).
Steady state levels of the processed RYFP substrate in T47D cells are considerably less than that of wild type YFP expressed in the absence of the amino-terminal arginine (Fig. 6A, lanes 1 and 2). Due to saturation of the x-ray film, the difference is more pronounced when fluorescence signal is directly quantitated (Fig. 6B). Co-expression of wild type HsUbc2b enhances proteasomal degradation, resulting in a still lower steady state level of the model N-end rule substrate (Fig. 6, A, lane 3, and B). Stimulation of RYFP degradation by ectopic expression of wild type HsUbc2b suggests endogenous E3α is not saturating with respect to its cognate E2 intracellularly and, therefore, is sensitive to changes in the steady state levels of its ubiquitin-charged cognate E2. In contrast, co-expression of HsUbc2bS120D results in a steady state level of RYFP comparable with that found in the absence of ectopically expressed wild type HsUbc2b, Fig. 6, A, lane 4, and B). Expression of the HsUbc2BS120A mutant is slightly more effective than the S120D mutant Fig. 6, A, lane 5, and B), presumably reflecting the residual activity of this point mutant in supporting Lys48-linked polyubiquitin chain formation (Figs. 3 and 5). The results of Figs. 6, A and B, are not due to differences in expression of wild type or mutant HsUbc2b as a parallel Western blot stained with anti-HsUbc2b antibody shows comparable levels of the slower migrating ectopic protein (supplemental Fig. S1). Densitometric quantitation of the higher relative molecular weight ectopic HsUbc2b bands averaged 59% of the endogenous wild type HsUbc2b bands. Separate counting of fluorescent cells yielded a transfection efficiency of 6.3% (not shown); therefore, ectopic expression of HsUbc2b increases the intracellular pool 9.4-fold. These results demonstrate that the enzymatic effect of Ser120 mutation on E3α-catalyzed polyubiquitin chain formation is recapitulated in the N-end rule targeting of RYFP catalyzed by E3α. The in vivo effects are unlikely to result from destabilization of the HsUbc2b mutants as the fractions of active protein for the bacterially expressed proteins were similar to that of wild type protein (“Materials and Methods”), and the activities of the point mutants in Uba1-catalyzed transthiolation were similar to that of wild type E2 (Figs. 1 and 2, supplemental Table 1). In addition, the steady state levels of ectopically expressed wild type and mutant HsUbc2b proteins were nearly identical (supplementary Fig. S1).
As a negative control, we ectopically expressed wild type and mutant forms of HsUbc2b with a second model substrate specific for an unrelated targeting pathway that does not require Ubc2 (37). In YFP-CL1, the fluorescent reporter harbors a carboxyl-terminal 16-residue hydrophobic peptide that is specifically targeted by the unfolded protein response pathway (38, 46). As noted by the lower steady state level of YFP-CL1 compared with RYFP, the unfolded protein response pathway is particularly active in the T47D cultures. Because this pathway is supported by the Ubc5 family of E2 carrier proteins, co-expression of wild type HsUbc2b is without effect (Fig. 6C). This important control obviates nonspecific effects of ectopic over expression of HsUbc2b or its point mutants on the steady state degradation of RYFP as might result by competition for charging of endogenous HsUbc2b by Uba1 within the cells.
DISCUSSION
The bulk of targeted protein degradation through the ubiquitin proteasome system is dependent on substrate availability and is controlled by changes in post-translational modification, ligand binding, or folding to trigger conjugation by the responsible ligase. Because each E2 family typically supports multiple E3 ligases that, in turn, target broader subsets of proteins for degradation, modulation of E2 availability offers a potential alternative mechanism for regulating system-wide changes of state in the proteome by coordinating protein targeting among otherwise-unrelated pathways. Evidence that Ubc2 is subject to Cdk1/2-dependent Ser120 phosphorylation and that this modification independently affects histone monoubiquitination and cell cycle progression provides a potential test of such orchestrated regulation (18). Phosphorylation of Ser120 has significant potential for influencing rates of Uba1-catalyzed transthiolation to form the requisite E2-ubiquitin thiolester intermediate and E3-catalyzed target protein conjugation as it is positioned in the active site groove adjacent to Cys88 to which the ubiquitin thiolester is formed (supplementary Fig. S2). Serine 120 also lies on the same face as the loop 1 and 2 residues Asn65 and Thr99, respectively, that are important for binding of E2-ubiquitin thiolesters to both Ring and Hect domains that serve as catalytic modules essential for E3 activity (47, 48) (supplementary Fig. S2). In the present work we have examined the potential consequence of Ser120 phosphorylation on another critical activity supported by Ubc2, the E3α/Ubr1-dependent targeting of proteins through the N-end rule pathway. To quantitatively address this question, we have used the HsUbc2bS120D point mutant as a phosphomimic of Cdk1/2 modification. The data predict that phosphorylation of HsUbc2b at Ser120 should markedly affect both E2 charging and E3α-dependent conjugation. These observations together with earlier work of Sarcevic et al. (18) suggest Ser120 phosphorylation can affect system-wide coordinated changes in ubiquitin-dependent signaling through the Ubc2-dependent pathways.
Our observation that ectopic expression of wild type HsUbc2b significantly increases degradation of the RYFP model N-end rule substrate but not an irrelevant YFP-CL1 unfolded protein response substrate in T47D human breast cancer cells is consistent with the rate of E3α-dependent conjugation being E2 limiting under the conditions of the experiment (Fig. 6). Similar results have been obtained in A549 human lung fibroblasts (not shown), indicating that this is not a cell line-specific effect. Previous estimates of intracellular E2 pools show that Ubc2 is the most abundant of E2 families and is present at ∼1 μm (30); therefore, E3α should be saturated with respect to endogenous Ubc2. That ectopic expression of HsUbc2b stimulates RYFP degradation suggests the intracellular pool of this E2 may be much lower or not stoichiometrically present as the corresponding ubiquitin thiolester in steady state. The inability of the ectopically expressed HsUbc2bS120D phosphomimic to similarly stimulate degradation of RYFP suggests that Ser120 phosphorylation can block the ability of HsUbc2b to support N-end rule-dependent targeting. This is the first demonstration of an inhibitory effect for the S120D mutation and, thus, of differential responses of ligases to the modification (18). Equally important, the inability of the HsUbc2bS120A mutant to support N-end rule degradation of RYFP in vivo agrees with the in vitro kinetic results obtained with E3α (Table 1). The in vitro data of Table 1 further illustrates that the principle effect on kcat (reflected in the Vmax) is due to loss of Ser120, as the largest effect is seen with the HsUbc2bS120A mutant. Additionally, imposition of a negative charge at this position results in a modest additional ablation of binding affinity for the HsUbc2bS120D-ubiquitin thiolester. Micromolar concentrations of uncharged E2 or the corresponding E2-ubiquitin thiolester have the potential to competitively inhibit Uba1-catalyzed transthiolation so that ectopic expression of a ubiquitin carrier protein could inhibit downstream targeting pathways dependent on the paralog (30). That expression of neither wild type HsUbc2b nor the S120D or S120A point mutants alter YFP-CL1 degradation precludes such off-pathway effects and validates the inability of the mutants to support the N-end rule-dependent degradation of RYFP (Fig. 6).
Initial rate studies of α-lactalbumin conjugation demonstrate that Ser120 is important for human E3α-catalyzed conjugation (Fig. 3). Mutation of Ser120 to alanine results in a significant ablation in chain formation on the α-lactalbumin N-end rule substrate (Fig. 3) that derives from a marked decrease in Vmax but with little change in the affinity for the binding of the HsUbc2bS120A-ubiquitin thiolester, as reflected in the Km (Fig. 4 and Table 1). This Vmax effect likely accounts for the modest stimulation of RYFP degradation in vivo observed in response to ectopic expression of HsUbc2bS120A (Fig. 6, A and B). Similar effects on in vitro monoubiquitination with 125I-reductively methylated ubiquitin show that the S120A mutation alters the rate for conjugation of the initial ubiquitin moiety, although similar effects on subsequent steps of chain elongation cannot be precluded (Fig. 5). Reductions in kcat at constant enzyme (i.e. Vmax) by definition reflect destabilization of the transition state formed during attack of the target protein primary amine nucleophile on the ubiquitin carbonyl of the E2-ubiquitin thiolester bond. The proximity of Ser120 to the Cys88-ubiquitin thiolester bond suggests that in the E3α-catalyzed reaction, the side chain of the Ser120 might partially stabilize the incipient transition state by hydrogen bonding. It is significant that the paralogous position among all E2 orthologs is occupied by amino acids with hydrogen bonding potential (Fig. 2A). Notably, a similar catalytic role has been shown for Asn79, which forms part of the conserved HPN consensus sequence of the E2 catalytic core domain (49). However, the net decrease in kcat for isopeptide bond formation on mutation of Asn79 to alanine was considerably larger (36-fold) than that found for the HsUbc2S120A mutant (Table 1), consistent with a corresponding smaller role for this putative stabilization.
A markedly different catalytic phenotype was observed when Ser120 is mutated to aspartate, which is also present in the paralogous position of several other wild type E2 isoforms (Fig. 2A). Kinetic studies of E3α-catalyzed conjugation of α-lactalbumin in the presence of HsUbc2bS120D showed a marked increase in Km and a more modest decrease in Vmax than observed for the S120A mutant (Table 1), consistent with the qualitative results of Fig. 3. Mutation of Ser120 to aspartate has a negligible effect on the in vitro conjugation of the initial ubiquitin to the N-end rule model substrate human α-lactalbumin, indicating that the defect is principally in chain elongation (Fig. 5). The attenuated effect of substituting aspartate for serine is consistent with a role for this position in transition state stabilization by hydrogen bonding. Significantly, the HsUbc2bS120D phosphomimic shows an overall 20-fold decrease in catalytic specificity, as measured by Vmax/Km (Table 1). Therefore, phosphorylation at Ser120 is predicted effectively to inactivate E3α-catalyzed N-end rule targeting. Because phosphate modification at Ser120 would also provide additional bulk within the sterically restrained active site groove, one could anticipate a more marked effect on E3α activity than shown by Asp120 substitution.
The kinetic results of Fig. 2B and supplemental Table 1 rule out effects on Uba1-catalyzed transthiolation of HsUbc2b as accounting for the marked ablation in α-lactalbumin conjugation in vitro or RYFP degradation in vivo. Mutation of Ser120 has only a modest effect on the Km for binding of the uncharged E2 to the activating enzyme and no effect on kcat (Fig. 2); therefore, position 120 functions exclusively in binding of the uncharged E2 to the Uba1 ternary complex. Notably, the HsUbc2bS120D mutation shows a significant enhancement in affinity for HsUba1on the order of that observed for wild type HsUbc5B (Fig. 2B). This observation together with the results on the converse mutation in the paralogous position of HsUbc5B demonstrate that the difference in affinity of HsUba1 for the E2 paralogs can be accounted for quantitatively by the identity of position 120 (HsUbc2b numbering). The results of Fig. 2B suggest that phosphorylation of Ser120 may equally enhance the ability of HsUbc2 to compete with other E2 paralogs for charging by HsUba1, further favoring downstream Ubc2-dependent pathways.
Regulation of E2 activity by phosphorylation has not been systematically examined; however, Ser120 and its paralogous positions among other E2 isoforms are predicted to be potential substrates for a number of protein kinases (Table 2). Although not all sites predicted to possess phosphorylation motifs are actual substrates for such post-translational modification, Ser120 and its paralogous positions reside within an exposed loop region sterically accessible to potential kinases (supplemental Fig. S2). The SP motif (the underline denotes the site of phosphorylation) common to Ubc2, Ubc3, Ubc7/E2G1, and Ubc10 is a predicted site for phosphorylation by ERK1/2 kinases that integrate various extracellular signals within regulatory cascades (50) and the cyclin-dependent CDK5 kinase that is localized to post-mitotic neurons and whose gain-of-function dysregulation is strongly implicated in several neurodegenerative diseases including Alzheimer (51). Additionally, Ubc2 and Ubc10 are each potentially subject to regulation by casein kinase I, glycogen synthase kinase 3, and MAPK-activated protein kinase 2 (MAPKAPK2), although in each case activity would be dependent on prior phosphorylation of Ser/Thr at the +4 position by either ataxia telangiectasia mutated kinase (ATM) or protein kinase. In addition to a role in DNA repair, MAPKAPK2 is associated with stress and inflammatory responses, presumably mediated through Ubc10 and its cognate Pex4 ubiquitin ligase, which are required for peroxisome maturation (52–54).
TABLE 2.
Predicted protein kinase sites involving Ser120 paralogous sites
Phosphorylation sites predicted by NetPhosK (Center for Biological Sequence Analysis, Technical University of Denmark) and PhosphoMotif Finder (Institute of Bioinformatics, Johns Hopkins University).
| Ubc2/Rad6 | PNPNSPANS | CDK2 |
| CKI (ATM, PKA) | ||
| GSK3 (ATM, PKA) | ||
| MAPKAPK2 (ATM, PKA) | ||
| ERK1/2, CDK5 | ||
| Ubc3/Cdc34 | PNPDSSPANAE | ERK1/2, CDK5 |
| Ubc7/E2G1 | PNDESSPANVE | ERK1/2, CDK5 |
| Ubc10 | PNIDSSPLNTH | CKI (ATM, PKA) |
| GSK3 (ATM, PKA) | ||
| MAPKAPK2 (ATM, PKA) | ||
| ERK1/2, CDK5 |
The present work suggests the basis for a novel regulatory mechanism whereby E2 phosphorylation/dephosphorylation coordinates activity among different ligase-dependent processes, enhancing some pathways (18) and inhibiting others (this study). Although roles for protein kinases in regulating ubiquitin-dependent regulatory pathways remain speculative, these potential functional associations provide a testable framework for future study.
Supplementary Material
Acknowledgment
We gratefully acknowledge Dr. Nico Dantuma of the Karolinska Institute for providing the pUBRYFP and pYFPCL1 ubiquitin-proteasome reporter constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM034009 (NIGMS; to A. L. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
We use E1 and E2 generically to refer to the paralogous activating enzymes and carrier proteins/conjugating enzymes, respectively, for Class 1 ubiquitin-like proteins.
We chose to mutate Asp116 of HsUbc5B to asparagine to correspond to the paralogous position 119 of HsUbc2b (Fig. 2B).
- Uba1
- ubiquitin (Ub)-activating enzyme (gene name, UBE1)
- E1
- generic term for activating enzymes of Class 1 ubiquitin-like proteins
- E3α/Ubr1
- N-end rule ubiquitin-protein isopeptide ligase (gene name, UBR1)
- HsUba1
- human ubiquitin activating enzyme (gene name: UBE1)
- HsUbc2b
- “b” isoform of the human/rabbit ortholog of S. cerevisiae Rad6/Ubc2 (also termed E214kb
- gene name
- UBE2B)
- HsUbc5B
- “B” isoform of the human ortholog of the S. cerevisiae Ubc5 (also termed UbcH5B
- gene name
- UBE2D2).
REFERENCES
- 1.Welchman R. L., Gordon C., Mayer R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 2.Haglund K., Dikic I. (2005) EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 4.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 5.Streich F. C., Haas A. L. (2010) in Conjugation and Deconjugation of Ubiquitin Family Modifiers (Groettrup M. ed.) pp. 1–16, Landes Bioscience and Springer, Austin, TX [Google Scholar]
- 6.Haas A. L., Warms J. V., Hershko A., Rose I. A. (1982) J. Biol. Chem. 257, 2543–2548 [PubMed] [Google Scholar]
- 7.Haas A. L., Rose I. A. (1982) J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 8.Pickart C. M., Rose I. A. (1985) J. Biol. Chem. 260, 1573–1581 [PubMed] [Google Scholar]
- 9.Haas A. L., Bright P. M. (1988) J. Biol. Chem. 263, 13258–13267 [PubMed] [Google Scholar]
- 10.Haas A. L. (1991) in Red Blood Cell Aging (Magnani M., DeFlora A. eds.) pp. 191–205, Plenum Press, New York [Google Scholar]
- 11.Schwartz L. M., Myer A., Kosz L., Engelstein M., Maier C. (1990) Neuron 5, 411–419 [DOI] [PubMed] [Google Scholar]
- 12.Haas A. L., Baboshina O., Williams B., Schwartz L. M. (1995) J. Biol. Chem. 270, 9407–9412 [DOI] [PubMed] [Google Scholar]
- 13.Wing S. S., Haas A. L., Goldberg A. L. (1995) Biochem. J. 307, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Gene Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 15.Froese N., Schwarzer M., Niedick I., Frischmann U., Köster M., Kröger A., Mueller P. P., Nourbakhsh M., Pasche B., Reimann J., Staeheli P., Hauser H. (2006) Mol. Cell. Biol. 26, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J. M. (2003) EMBO J. 22, 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block K., Boyer T. G., Yew P. R. (2001) J. Biol. Chem. 276, 41049–41058 [DOI] [PubMed] [Google Scholar]
- 18.Sarcevic B., Mawson A., Baker R. T., Sutherland R. L. (2002) EMBO J. 21, 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jentsch S., McGrath J. P., Varshavsky A. (1987) Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 20.Ulrich H. D., Jentsch S. (2000) EMBO J. 19, 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohmen R. J., Madura K., Bartel B., Varshavsky A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7351–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varshavsky A. (2003) Nat. Cell Biol. 5, 373–376 [DOI] [PubMed] [Google Scholar]
- 23.Eisele F., Wolf D. H. (2008) FEBS Lett. 582, 4143–4146 [DOI] [PubMed] [Google Scholar]
- 24.Nillegoda N. B., Theodoraki M. A., Mandal A. K., Mayo K. J., Ren H. Y., Sultana R., Wu K., Johnson J., Cyr D. M., Caplan A. J. (2010) Mol. Biol. Cell 21, 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 26.Yamashita K., Shinohara M., Shinohara A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11380–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osley M. A. (2004) Biochim. Biophys. Acta 1677, 74–78 [DOI] [PubMed] [Google Scholar]
- 28.Kume K., Iizumi Y., Shimada M., Ito Y., Kishi T., Yamaguchi Y., Handa H. (2010) Genes Cells 15, 339–349 [DOI] [PubMed] [Google Scholar]
- 29.Baboshina O. V., Haas A. L. (1996) J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 30.Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L. (2003) J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 31.Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas A. L., Rose I. A. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 6845–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas A. L. (2005) Methods Mol. Biol. 301, 23–35 [DOI] [PubMed] [Google Scholar]
- 34.Baboshina O. V., Crinelli R., Siepmann T. J., Haas A. L. (2001) J. Biol. Chem. 276, 39428–39437 [DOI] [PubMed] [Google Scholar]
- 35.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 36.Salomons F. A., Verhoef L. G., Dantuma N. P. (2005) Essays Biochem. 41, 113–128 [DOI] [PubMed] [Google Scholar]
- 37.Menéndez-Benito V., Heessen S., Dantuma N. P. (2005) Methods Enzymol. 399, 490–511 [DOI] [PubMed] [Google Scholar]
- 38.Menéndez-Benito V., Verhoef L. G., Masucci M. G., Dantuma N. P. (2005) Hum. Mol. Genet. 14, 2787–2799 [DOI] [PubMed] [Google Scholar]
- 39.Loeb K. R., Haas A. L. (1992) J. Biol. Chem. 267, 7806–7813 [PubMed] [Google Scholar]
- 40.Haas A. L., Bright P. M. (1985) J. Biol. Chem. 260, 12464–12473 [PubMed] [Google Scholar]
- 41.Bohnsack R. N., Haas A. L. (2003) J. Biol. Chem. 278, 26823–26830 [DOI] [PubMed] [Google Scholar]
- 42.Tokgöz Z., Bohnsack R. N., Haas A. L. (2006) J. Biol. Chem. 281, 14729–14737 [DOI] [PubMed] [Google Scholar]
- 43.Reiss Y., Kaim D., Hershko A. (1988) J. Biol. Chem. 263, 2693–2698 [PubMed] [Google Scholar]
- 44.Haas A. L., Reback P. B., Chau V. (1991) J. Biol. Chem. 266, 5104–5112 [PubMed] [Google Scholar]
- 45.Varshavsky A. (1997) Genes Cells 2, 13–28 [DOI] [PubMed] [Google Scholar]
- 46.Gilon T., Chomsky O., Kulka R. G. (1998) EMBO J. 17, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 48.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 49.Ogunjobi O., Ramage R. (1990) Biochem. Soc. Trans. 18, 1322–1323 [DOI] [PubMed] [Google Scholar]
- 50.Calvo F., Agudo-Ibáñez L., Crespo P. (2010) Bioessays 32, 412–421 [DOI] [PubMed] [Google Scholar]
- 51.Monaco E. A., 3rd (2004) Curr. Alzheimer Res. 1, 33–38 [DOI] [PubMed] [Google Scholar]
- 52.Reinhardt H. C., Yaffe M. B. (2009) Curr. Opin. Cell Biol. 21, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platta H. W., El Magraoui F., Bäumer B. E., Schlee D., Girzalsky W., Erdmann R. (2009) Mol. Cell. Biol. 29, 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiebel F. F., Kunau W. H. (1992) Nature 359, 73–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.