Abstract
Death receptor 5 (DR5) is a death domain-containing transmembrane receptor that triggers apoptosis upon binding to its ligand or when overexpressed. Its expression is induced by certain small molecule drugs, including celecoxib, through mechanisms that have not been fully elucidated. The current study has revealed a novel ERK/ribosomal S6 kinase (RSK)-dependent mechanism that regulates DR5 expression primarily using celecoxib as a DR5 inducer. Both C/EBP homologous protein (CHOP) and Elk1 are required for celecoxib-induced DR5 expression based on promoter deletion and mutation analysis and siRNA-mediated gene silencing results. Co-expression of both CHOP and Elk1 exhibited enhanced effects on increasing DR5 promoter activity and DR5 expression, indicating that CHOP and Elk1 co-operatively regulate DR5 expression. Because Elk1 is an ERK-regulated protein, we accordingly found that celecoxib increased the levels of phosphorylated ERK1/2, RSK2, and Elk1. Inhibition of either ERK signaling with a MEK inhibitor or ERK1/2 siRNA, or RSK2 signaling with an RSK2 inhibitor or RSK2 siRNA abrogated DR5 up-regulation by celecoxib as well as other agents. Moreover, these inhibitions suppressed celecoxib-induced CHOP up-regulation. Thus, ERK/RSK-dependent, CHOP and Elk1-mediated mechanisms are critical for DR5 induction. Additionally, celecoxib increased CHOP promoter activity in an ATF4-dependent manner, and siRNA-mediated blockade of ATF4 abrogated both CHOP induction and DR5 up-regulation, indicating that ATF4 is involved in celecoxib-induced CHOP and DR5 expression. Collectively, we conclude that small molecules such as celecoxib induce DR5 expression through activating ERK/RSK signaling and subsequent Elk1 activation and ATF4-dependent CHOP induction.
Keywords: Apoptosis, ER Stress, ERK, Gene Regulation, Signal Transduction, CHOP, Death Receptor 5, ElK1, RSK
Introduction
Death receptor 5 (DR54; also called TRAIL-R2 or killer/DR5) is a cell surface receptor for the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). It becomes oligomerized (trimerized) upon binding to its ligand TRAIL (or upon overexpression) and then transmits an apoptotic signal through rapid activation of caspase-8-dependent caspase cascades. This process involves trimerized DR5 interacting specifically with the adaptor protein Fas-associated death domain via death domain interaction and subsequent recruitment of caspase-8 through the death effector domain between Fas-associated death domain and caspase-8, leading to caspase-8 activation and ultimate apoptosis (1).
DR5 expression can be induced by certain stimuli, including small molecule anticancer drugs (2, 3). DR5 induction mediates enhancement of TRAIL-induced apoptosis or contributes to apoptosis induced by certain drugs (2, 3). The induction of DR5 expression generally occurs at the transcriptional level, involving activation of the transcriptional factors p53 (4, 5), NF-κB (6, 7), C/EBP homologous protein (CHOP; also known as growth arrest and DNA damage gene 153 (GADD153)) (8, 9) and YY1 (10). However, the signaling pathways that activate these transcriptional factors are largely unknown, except for the involvement of JNK activation in this process (11, 12).
In general, the Raf-MEK-ERK pathway represents a major survival signaling pathway in cancer cells to promote survival via inhibition of apoptosis, primarily through controlling the activity or abundance of Bcl-2 family members such as Bim, Bad, Bcl-1, and Mcl-1 (13, 14). However, a proapoptotic role of ERK signaling has also been documented in some studies (13, 15), but how ERK/RSK activates proapoptotic signaling remains largely unknown (13, 15).
The 90-kDa ribosomal S6 kinase 2 (RSK2) is a highly conserved Ser/Thr kinase and functions directly downstream of ERK1/2 to mediate ERK1/2 biological activities. RSK2 can be activated directly by ERK1/2 in response to growth factors, chemokines, and other stimuli and phosphorylates many cytosolic and nuclear proteins that are implicated in the regulation of diverse cellular process, including cell proliferation, cell survival, and cell motility (16). Although RSK2 inhibits apoptosis by phosphorylating proteins such as Bad (17), C/EBPβ (18), and death-associated protein kinase (19), it has also been shown that RSK can phosphorylate Nur77, leading to its mitochondrial translocation and induction of apoptosis (20). It is also known that RSK2 regulates transcription primarily through direct phosphorylation of transcriptional factors (16).
Celecoxib, a marketed antiinflammatory and antipain drug, is being tested in clinical trials for its chemopreventive and therapeutic effects against a broad spectrum of epithelial malignancies either as a single agent or in combination with other agents. The antitumor activity of celecoxib is thought to be associated with its ability to induce apoptosis in a variety of cancer cells (21). The molecular mechanism underlying celecoxib-mediated apoptosis has not been fully elucidated, although it appears to be associated with inactivation of PDK1/Akt, induction of endoplasmic reticulum (ER) stress involving up-regulation of CHOP and increase in Ca2+ levels, or down-regulation of the antiapoptotic protein survivin (22). We and others have shown that celecoxib induces apoptosis involving the activation of the extrinsic DR pathway through both DR5 induction and c-FLIP down-regulation independent of its cyclooxygenase-2-inhibitory activity (23–25). However, the mechanism underlying celecoxib-induced DR5 up-regulation has not been fully uncovered.
It has been shown that celecoxib and other agents induce DR5 expression through a CHOP-dependent mechanism (3, 8, 23, 26). In our effort to better define the mechanisms by which small molecules induce DR5 expression, we have revealed an additional mechanism involving Elk1, which cooperates with CHOP to induce DR5 expression primarily using celecoxib as a DR5 inducer. Importantly, we have shown that ERK/RSK signaling is involved in mediating DR5 expression induced by certain small molecules such as celecoxib through co-activation of CHOP and Elk1. Our findings thus highlight a novel mechanism accounting for DR5 induction.
EXPERIMENTAL PROCEDURES
Reagents
Celecoxib, human recombinant TRAIL, and antibodies against caspases, DR5, CHOP, Bip, activating transcription factor 4 (ATF4) and IRE1α were the same as described previously (12, 25). The MEK inhibitor U0126 was purchased from LC Laboratories (Woburn, MA). Tunicamycin and thapsigargin were purchased from Biomol (Plymouth Meeting, PA). The RSK inhibitor, fmk, was described previously (27). The pan-caspase inhibitor, Z-VAD-fmk, was purchased from Enzyme System Products (Livermore, CA). Rabbit polyclonal antibodies against Elk1, p-Elk1 (Ser383), ERK1/2, p-ERK1/2 (Thr202/Tyr204), and p-RSK (Ser380) were purchased from Cell Signaling Technology (Beverly, MA). Mouse monoclonal anti-RSK2 antibody (E-1) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-actin antibody was purchased from Sigma-Aldrich.
Cell Lines and Cell Culture
The human lung cancer cell lines used in this study were purchased from the American Type Culture Collection (Manassas, VA). 686LN head and neck cancer and HEK293T cells were provided by G. Chen and K. Ye (Emory University, Atlanta, GA), respectively. These cell lines were cultured in RPMI 1640 medium or DMEM containing 5% fetal bovine serum at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The immortalized normal human bronchial epithelial cell line, HBEC3KT, was kindly provided by Dr. J. D. Minna (University of Texas Southwestern Medical Center, Dallas, TX) and cultured as described previously (28).
Western Blot Analysis
Whole cell protein lysates were prepared and analyzed by Western blotting as described previously (25, 29).
Detection of Apoptosis
Apoptosis was evaluated by measuring cytoplasmic histone-associated DNA fragments using a Cell Death Detection ELISAPlus kit (Roche Applied Science) following the manufacturer's instructions. We also detected caspase activation by Western blotting as an additional indicator of apoptosis.
Expression Constructs and Transfection
CHOP expression construct was purchased from OriGene (Rockville, MD). Wild type Elk1 and constitutively activated Elk1-VP16 expression constructs were provided by Dr. A. D. Sharrocks (University of Manchester, Manchester, UK) (30) and R. Treisman (Imperial Cancer Research Fund, London, UK) (31), respectively. Cell transfection with the given plasmids was conducted using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol.
Gene Silencing using Small Interfering RNA (siRNA) or Short Hairpin RNA (shRNA)
Gene silencing was achieved by either transfecting siRNA using HiPerFect transfection reagent (Qiagen, Valencia, CA) following the manufacturer's instructions or infecting cells with lentiviruses harboring the given shRNA. Control (i.e. nonsilencing) and CHOP siRNAs were described previously (32). Elk1 and ATF4 siRNAs that target the sequences 5′-GGCAATGGCCACATCATCT-3′ and 5′-GCCTAGGTCTCTTAGATGA-3′ (33), respectively, were synthesized by Qiagen. ERK1/2 siRNA (no. 6560) was purchased from Cell Signaling Technology, Inc. siRNA SMARTpool® RSK2/MAPKAP kinase 1β (no. M-003026) was purchased from Upstate/Millipore. RSK2 shRNA (RHS3979-9607536) was purchased from Open Biosystems (Huntsville, AL). Gene silencing effects were evaluated by Western blot analysis as described above.
Reporter Plasmids, Transient Transfection, and Luciferase Activity Assay
DR5 reporter constructs used in this study were described previously (32, 34). The human CHOP promoter reporter constructs were kindly provided by Dr. P. Fafournoux (Unité Nutrition Humaine, INRA de Theix, Champanelle, France) (35). The pGL3 basic reporter plasmids with respective mutations in C/EBP-ATF, AP-1, and ER stress response element sites were kindly provided by Dr. A. B. Vaandrager (University of Utrecht, Utrecht, The Netherlands) (36). Plasmid transfection and luciferase assays were the same as described previously (32).
RESULTS
Celecoxib Increases CHOP Expression and ER Stress Accompanied by DR5 Up-regulation
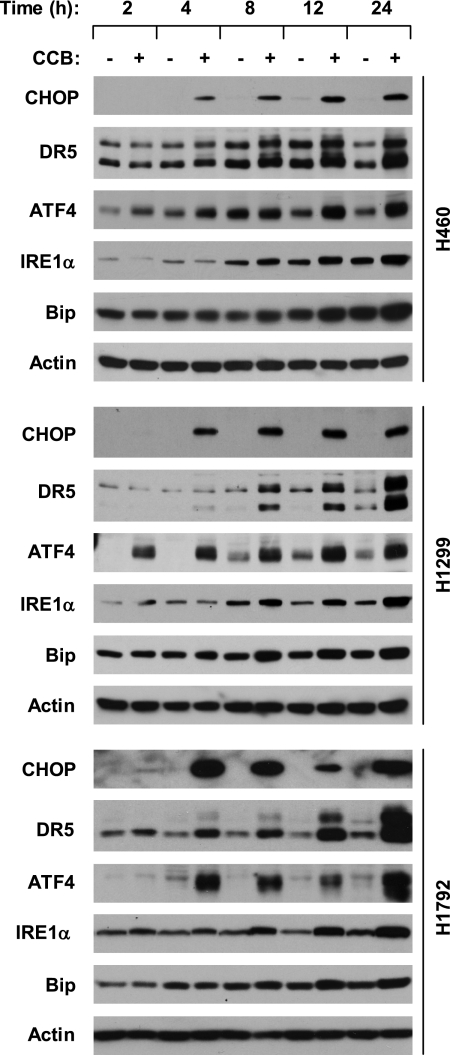
It has been shown that celecoxib induces ER stress, CHOP expression, and DR5 up-regulation (23, 25, 33, 37–40). Thus, we chose celecoxib as a tool agent to demonstrate the mechanisms underlying DR5 induction by small molecules in the current study. We first determined whether celecoxib at a concentration (e.g. 50 μm) that induces DR5 and apoptosis (25) induces CHOP expression in our cell systems. In three tested lung cancer cell lines, celecoxib increased CHOP levels accompanied by DR5 up-regulation, both of which occurred at 4 h and were sustained up to 24 h after treatment. Accordingly, we detected increased levels of ATF4, IRE1α, and Bip (Fig. 1), three well known ER stress marker proteins (41), indicating that celecoxib indeed induces ER stress in our cell systems.
FIGURE 1.
Celecoxib increases CHOP, ATF4, IRE1α, and DR5 in human lung cancer cells. The indicated cell lines were treated with and without 50 μm celecoxib (CCB) for the given times. The cells were then subjected to preparation of whole cell protein lysates and subsequent Western blot analysis for the indicated proteins.
CHOP Induction Contributes to Celecoxib-induced DR5 Expression
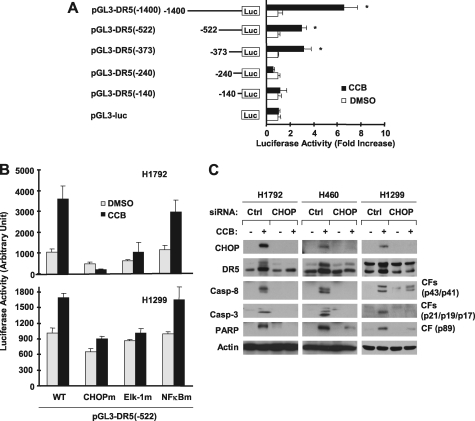
To explore the connection between CHOP induction and DR5 up-regulation, we analyzed the 5′-flanking region of the DR5 gene to determine whether CHOP is required for DR5 transactivation. Through deletion analysis, we found that the region between −373 bp and −240 bp was required for celecoxib to increase DR5 transcription because celecoxib could increase luciferase activity of reporter constructs carrying −373, −522, and −1400 bp DR5 5′-flanking regions, but failed to do so in cells transfected with reporter constructs harboring −240 and −140 bp DR5 promoter regions (Fig. 2A). Because the region between −373 bp and −240 bp of the DR5 promoter contains putative CHOP (−276/−264) and Elk1 (−323/−308) binding sites, we further analyzed the impact of mutations in these binding sites on celecoxib-induced DR5 transactivation. We included mutation of the NF-κB binding site (−236/−221) as a negative control in this analysis. Mutation of the NF-κB binding site did not affect celecoxib-induced DR5 transactivation; however, mutation of either the CHOP or Elk1 binding site abolished or reduced celecoxib-mediated DR5 transactivation (Fig. 2B). These data suggest that both CHOP and Elk1 binding sites are required for celecoxib to transactivate the DR5 gene. Moreover, we blocked CHOP induction by knocking down CHOP expression and determined its impact on celecoxib-induced DR5 up-regulation. As presented in Fig. 2C, celecoxib induced DR5 expression in control siRNA-transfected cells, but not in cells transfected with CHOP siRNA in every cell line tested. Thus, it is clear that celecoxib induces a CHOP-dependent DR5 expression. We noted that celecoxib-induced cleavage of caspase-8, caspase-3, and poly(ADP-ribose)polymerase was also abolished or attenuated in these cell lines transfected with CHOP siRNA compared with control siRNA-transfected cells (Fig. 2C), indicating that CHOP induction is also important for celecoxib-induced apoptosis.
FIGURE 2.
Celecoxib induces CHOP-dependent transcription of DR5 (A and B) and DR5 expression (C). A, the given reporter constructs with different lengths of the 5′-flanking region of the DR5 gene were co-transfected with pCH110 plasmid into H1792 cells for 24 h. B, the given reporter constructs with and without different mutated binding sites were co-transfected with pCH110 plasmids into the given cell lines for 24 h. Afterward, the cells were treated with dimethyl sulfoxide (DMSO) or 50 μm celecoxib (CCB) for 12 h and then subjected to luciferase assay. Each column represents a mean ± S.D. (error bars) of triplicate determinations. C, the indicated cell lines were transfected with control (Ctrl) or CHOP siRNA. After 48 h, the cells were treated with 50 μm celecoxib for 24 h and then subjected to preparation of whole cell protein lysates and subsequent Western blot analysis. CF, cleaved form.
Elk1 Contributes to Celecoxib-induced DR5 Expression and Cooperates with CHOP to Regulate DR5 Expression
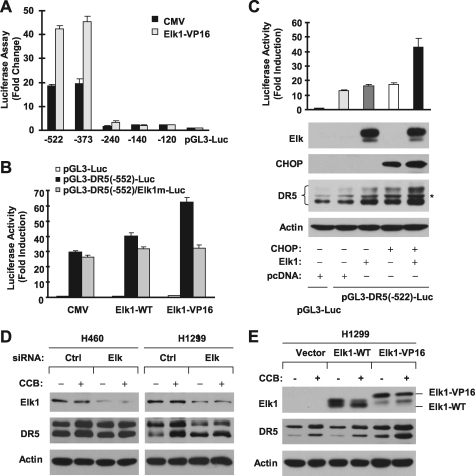
In the above experiments, we noted that mutation of the Elk1 binding site in the DR5 promoter also attenuated celecoxib-induced DR5 transactivation, suggesting that Elk1 may also be involved in celecoxib-induced DR5 expression. Because there was no previous study linking Elk1 to DR5 expression, we then determined whether Elk1 is indeed involved in regulating DR5 expression. To this end, we examined whether enforced expression of Elk1 alters DR5 transactivation and expression. We found that overexpression of a constitutively active Elk1 (i.e. Elk1-VP16) in HEK293T cells increased transcriptional activity of the DR5 promoter (Fig. 3A). Deletion analysis showed that Elk1-VP16 increased luciferase activity of reported constructs carrying −373 DR5 promoter region but did not do so in cells transfected with reporter constructs harboring −240, −140, and −120 DR5 promoter regions (Fig. 3A). Thus, the region between −373 and −240 bp is responsible for Elk1-mediated DR5 transactivation. Moreover, enforced expression of wild-type Elk1 or Elk1-VP16 increased luciferase activity of a DR5 promoter with wild type Elk1 binding site but did not increase transcriptional activity of a DR5 promoter in which the Elk1 binding site was mutated (Fig. 3B). Identical results were also generated from H1299 cells (supplemental Fig. S1). These results indicate that Elk1 indeed regulates DR5 transcription. Because the locations of the CHOP and Elk1 binding sites are close to each other, we tested whether CHOP and Elk1 cooperate to transactivate the DR5 gene. We co-expressed CHOP and Elk1 in HEK293T cells and then looked at their effects on DR5 promoter transcription and expression. As shown in Fig. 3C, transfection of CHOP or Elk1 alone weakly increased DR5 promoter activity and expression. In contrast, co-transfection of both CHOP and Elk1 genes had much more potent effects than either single gene alone in both increasing DR5 promoter transactivation and inducing DR5 expression. These data suggest that CHOP and Elk1 indeed cooperate to regulate DR5 expression. Taking the above data together, it is clear that Elk1 participates in regulation of DR5 expression, likely through cooperating with CHOP.
FIGURE 3.
Elk1 increases DR5 transactivation (A and B), cooperates with CHOP to increase DR5 transcription and DR5 expression (C), mediates the effect of celecoxib on DR5 induction (D), and enhances celecoxib-induced DR5 expression (E). A and B, HEK293T cells were co-transfected with the given DR5 promoter reporter and Elk1 expression plasmids for 36 h. C, HEK293T cells were co-transfected with the given DR5 promoter reporter plasmid with empty pcDNA, Elk1, CHOP, or CHOP plus Elk1 expression constructs for 40 h. After the aforementioned transfections, the cells were harvested for luciferase assay and Western blot analysis. D, the given cell lines were transfected with control (Ctrl) or Elk1 siRNA for 40 h and then treated with and without 50 μm celecoxib (CCB) for an additional 10 h. E, H1299 cells were transfected with the indicated plasmids for 36 h and then treated with and without 50 μm celecoxib for an additional 10 h. After treatment, the cells were harvested for preparation of whole cell protein lysates and subsequent Western blot analysis.
To demonstrate solidly that Elk1 contributes to DR5 up-regulation by celecoxib, we knocked down Elk1 expression and then examined its impact on celecoxib-induced DR5 expression. As presented in Fig. 3D, silencing of Elk1 abrogated the ability of celecoxib to induce DR5 expression, demonstrated by the ability of celecoxib to increase DR5 expression in control siRNA-transfected cells, but not in Elk1 siRNA-transfected cells. We also performed simultaneous knockdown of CHOP and Elk1 and found that co-silencing of both CHOP and Elk1 achieved results similar to knockdown of CHOP or Elk1 alone on blockage of DR5 induction by celecoxib (supplemental Fig. S2). Moreover, enforced expression of Elk-VP16 not only increased basal levels of DR5 expression, but also enhanced the effect of celecoxib on increasing DR5 expression because treatment of Elk1-VP16-transfected cells with celecoxib induced the highest DR5 expression compared with celecoxib treatment alone or Elk1-VP16 expression alone (Fig. 3E). Collectively, we conclude that celecoxib induces DR5 expression through a mechanism involving Elk1, which may cooperate with CHOP in regulation of DR5 expression.
Celecoxib Activates ERK1/2 Signaling Resulting in DR5 Expression
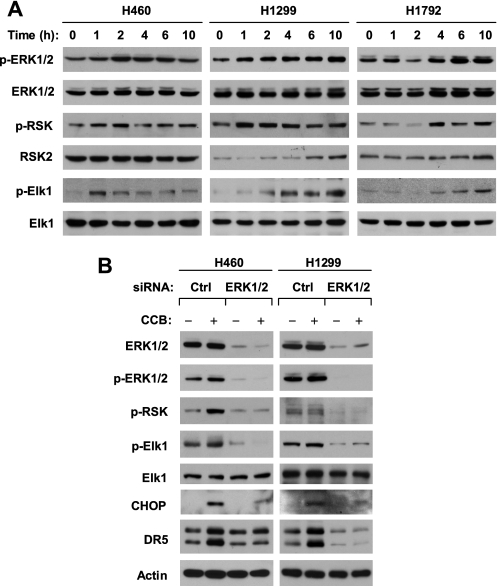
It is well known that Elk1 is a substrate of ERK1/2 and is directly phosphorylated by ERK1/2 (13, 42). We next determined whether celecoxib activates the ERK1/2 signaling pathway by examining phosphorylation of ERK1/2, RSK2, and Elk1 in cells exposed to celecoxib. As presented in Fig. 4A, p-ERK1/2 levels were increased in every celecoxib-treated lung cancer cell line, starting from 1–4 h after celecoxib treatment depending on the cell line. Similar increases were seen for p-RSK2 and p-Elk1. Thus, celecoxib activates the ERK1/2 signaling pathway. Following this study, we determined whether ERK activation is required for celecoxib to induce DR5 expression by blocking ERK activation with either MEK inhibitor or ERK1/2 siRNA. The presence of the MEK inhibitor, U0126, abolished ERK1/2 and RSK2 phosphorylation and in part inhibited Elk1 phosphorylation. Accordingly, DR5 induction by celecoxib was also abolished (e.g. H460 and H1792) or partly inhibited (e.g. H1299) (supplemental Fig. S3). We noted that two other ER stress-inducers, tunicamycin and thapsigargin, increased p-ERK/1/2, p-RSK2, and DR5 levels as well; these effects were also abolished by the presence of U0126 (supplemental Fig. S3B). These results suggest that the activation of ERK signaling is required for DR5 induction by celecoxib as well as other agents.
FIGURE 4.
Celecoxib activates ERK1/2 signaling (A), which contributes to celecoxib-induced DR5 up-regulation (B). A, the indicated cell lines were treated with 50 μm celecoxib for the given times. B, the indicated cell lines were transfected with control (Ctrl) or ERK1/2 siRNA for 48 h and then treated with and without 50 μm celecoxib (CCB) for an additional 10 h. After treatment, the cells were harvested for preparation of whole cell protein lysates and subsequent Western blot analysis for the given proteins.
To demonstrate robustly the involvement of ERK signaling in celecoxib-induced DR5 expression, we used ERK1/2 siRNA to silence ERK1/2 expression and inhibit its ERK1/2 signaling and then to investigate its impact on celecoxib-induced DR5 expression. Transfection of ERK1/2 siRNA reduced the basal levels of ERK1/2, p-ERK1/2, p-RSK2, and p-Elk1 and inhibited celecoxib-induced increases in p-ERK1/2, p-RSK2, and p-Elk1, indicating the successful inhibition of ERK1/2 signaling activated by celecoxib. Correspondingly, DR5 up-regulation by celecoxib was observed only in cells transfected with control siRNA, but not in ERK1/2 siRNA-transfected cells. We noted that CHOP induction by celecoxib was also inhibited in cells transfected with ERK1/2 siRNA (Fig. 4B). These results again indicate that ERK1/2 activation is required for DR5 induction by celecoxib.
RSK2 Mediates Celecoxib-induced CHOP and DR5 Up-regulation
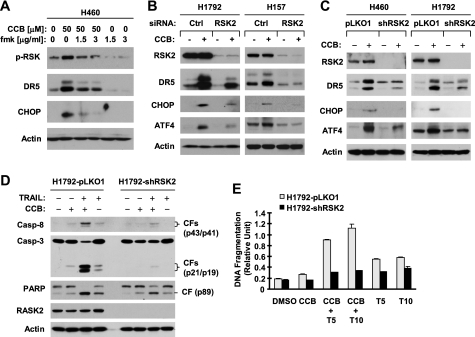
Given that RSK2 is a key downstream effector protein of ERK1/2 that mediates many biological functions (particularly apoptosis) of ERK1/2 signaling (13, 14), we questioned whether RSK2 is also a key player in ERK-mediated DR5 regulation. To this end, we first treated cells with celecoxib in the absence and presence of the RSK-specific inhibitor, fmk, and then detected p-RSK, DR5, and CHOP levels. As shown in Fig. 5A, the presence of 1.5 and 3 μg/ml fmk effectively blocked celecoxib-induced RSK phosphorylation and up-regulation of DR5 and CHOP. Moreover, we silenced RSK2 expression and then looked at its impact on celecoxib-induced DR5 and CHOP expression. Transient knockdown of RSK2 with RSK2 siRNA abrogated up-regulation of both DR5 and CHOP induced by celecoxib in both H1792 and H157 cells (Fig. 5B). In agreement, stable knockdown of RSK2 with lentiviral RSK2 shRNA generated similar results. Specifically, celecoxib increased DR5 and CHOP expression in both H460 and H1792 cells carrying empty vector, but not in cells expressing RSK2 shRNA (Fig. 5C). Similar results were also generated in 686LN cells (supplemental Fig. S4). These data collectively demonstrate that RSK2 activation is required for celecoxib to induce both CHOP and DR5 expression.
FIGURE 5.
RSK2 activation is required for celecoxib-induced DR5 induction (A–C) and enhancement of TRAIL-induced apoptosis (D–E). A, H460 cells were pretreated with the indicated concentrations of fmk for 30 min and then co-treated with 50 μm celecoxib (CCB) for an additional 10 h. B, the indicated cell lines were transfected with control (Ctrl) or RSK2 siRNA for 48 h and then treated with and without 50 μm celecoxib for an additional 10 h. C, the indicated pLKO1 and shRSK2 stable cell lines were treated with 50 μm celecoxib for 10 h. D, the indicated stable cell lines were treated with 50 μm celecoxib alone, 20 ng/ml TRAIL alone, and their combination for 24 h. After treatment (A--D), the cells were harvested for preparation of whole cell protein lysates and subsequent Western blot analysis for the given proteins. CF, cleaved from. E, the indicated cell lines were plated in 96-well plates and treated on the next day with dimethyl sulfoxide (DMSO), 50 μm celecoxib, 5 ng/ml (T5), or 10 ng/ml (T10) TRAIL, or celecoxib plus TRAIL. After 24 h, the cells were subjected to DNA fragmentation assay using the Cell Death Detection ELISAPlus kit. Columns represent means ± S.D. (error bars) of triplicate determinations.
ATF4 is known to be an RSK2 substrate (43). Thus, we also looked at whether knockdown of RSK2 affects ATF4 induction by celecoxib. Indeed, both transient and stable knockdown of RSK2 abolished celecoxib-induced increase of ATF4 in all of the tested cell lines (Fig. 5, B and C, and supplemental Fig. S4). Thus, celecoxib-induced ATF4 increase is also RSK2-dependent.
We also looked at the importance of RSK2 activation in induction of DR5, CHOP, and ATF4 by tunicamycin and thapsigargin. Like celecoxib, tunicamycin and thapsigargin increased the levels of DR5, CHOP, and ATF4 in the control H1792 cells carrying an empty vector (H1792-pLKO1) but failed to do so in H1792 cells expressing RSK2 shRNA (H1792-shRNA) (supplemental Fig. S5), indicating that knockdown of RSK2 abolishes the ability of these agents to up-regulate DR5, CHOP, and ATF4 expression. Thus, our findings regarding RSK2-dependent DR5 and CHOP induction are also applicable to other agents.
We showed previously that celecoxib up-regulates DR5 expression, leading to enhancement of TRAIL-induced apoptosis (25). Here, we show further that celecoxib and TRAIL combination exerted enhanced effect on decreasing cell survival in H1299 lung cancer cells, but not in HBEC3KT normal human bronchial epithelial cells (supplemental Fig. S6). Moreover, the enhanced induction of apoptosis by celecoxib and TRAIL in lung cancer cell lines is caspase-mediated because this effect was abolished by the presence of the pan-caspase inhibitor, Z-VAD-fmk (supplemental Fig. S7). Thus, these results validate our previous finding on celecoxib-induced enhancement of TRAIL-induced apoptosis in human lung cancer cells (25). In this study, we further asked whether RSK2 activation is involved in mediating the cooperative induction of apoptosis by celecoxib and TRAIL. We compared induction of apoptosis by the combination of celecoxib and TRAIL between H1792-pLKO1 and H1792-shRSK2 cell lines. As reported previously (25), the combination of celecoxib and TRAIL was much more potent than each single agent alone in inducing the cleavage of caspase-8, caspase-3, and poly(ADP-ribose)polymerase and in increasing DNA fragmentation in H1792-pLKO1 cells; however, these effects were substantially attenuated in H1792-shRSK2 cells (Fig. 5, D and E). Thus, it is clear that RSK2 activation is critical for celecoxib to enhance TRAIL-induced apoptosis.
Celecoxib Induces an ATF4-dependent CHOP Up-regulation
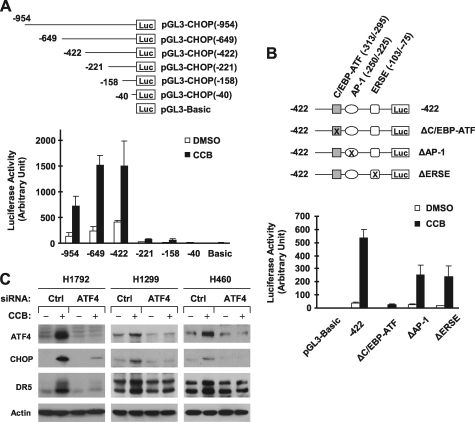
The aforementioned results indicate that both CHOP and ATF4 are increased upon treatment with celecoxib or other agents through an RSK2-dependent mechanism. Moreover, ATF4 can directly regulate CHOP transcription and expression through binding to the CHOP promoter region (44–46). Thus, we further addressed whether celecoxib increases CHOP expression involving ATF4. Through analyzing the effects of celecoxib on the transcriptional activity of different lengths of CHOP promoter regions, we found that celecoxib substantially increased luciferase activity in cells transfected with reporter constructs carrying −954/+91, −649/+91, and −422/+91 CHOP promoter regions, but only minimally in cells transfected with the reporter constructs harboring −221/+91, −158/+91, and −40/+91 promoter regions (Fig. 6A). Thus, the region between −422 and −221 contains elements that are likely responsible for celecoxib-induced CHOP expression.
FIGURE 6.
Promoter analysis (A and B) and siRNA-mediated gene silencing (C) demonstrate that ATF4 mediates celecoxib-induced CHOP up-regulation. A and B, the given reporter constructs with different lengths of the 5′-flanking region of CHOP gene (A) or with different mutations as indicated (B) were co-transfected with pCH110 plasmid into H1792 cells for 24 h. The cells were treated with dimethyl sulfoxide (DMSO) or 50 μm celecoxib (CCB) for 12 h and then subjected to luciferase assay. Each column represents a mean ± S.D. (error bars) of triplicate determinations. C, the indicated cell lines were transfected with control (Ctrl) or ATF4 siRNA for 48 and then treated with and without 50 μm celecoxib for an additional 10 h. The cells were harvested for preparation of whole cell protein lysates and subsequent Western blot analysis for the given proteins.
In the region between −422 and −221, two known stress-responsive elements are present, an AP-1 site (−250/−225) and a C/EBP-ATF site (−313/−295) within an amino acid response element (45). Thus, we further analyzed the effects of celecoxib on the transcriptional activity of the CHOP promoter (−422/+91) in which the AP-1 and C/EBP-ATF sites were respectively mutated. We also included another reporter construct in which the ER stress response element site (−103/−75) was mutated as a control. As presented in Fig. 6B, celecoxib potently increased luciferase activity in cells transfected with reporter constructs with wild type (−422/+91) CHOP promoter region, AP-1 site-mutated region or ER stress response element site-mutated region, but only minimally in cells transfected with reporter plasmid harboring CHOP promoter region with mutated C/EBP-ATF site. Collectively, these results indicate that the C/EBP-ATF site is critical for celecoxib-mediated CHOP up-regulation, implying that ATF4 is involved in CHOP induction by celecoxib because ATF4 binds to this site to regulate CHOP expression (44).
To provide direct evidence for ATF4 mediation of CHOP induction by celecoxib, we blocked ATF4 increase through siRNA-mediated ATF4 knockdown and then examined its impact on celecoxib-induced CHOP and DR5 up-regulation. As presented in Fig. 6C, celecoxib could increase CHOP and DR5 expression in cells transfected with control siRNA, but failed to do so in every tested cell line transfected with ATF4 siRNA. Thus, blockade of ATF4 increase abolishes not only CHOP induction, but also DR5 up-regulation in cells exposed to celecoxib, indicating that celecoxib induces CHOP and DR5 expression through an ATF4-dependent mechanism.
DISCUSSION
The effects of celecoxib on induction of ER stress, including CHOP and up-regulation of DR5, have been demonstrated in several previous studies (23, 25, 33, 37–40, 47). CHOP-dependent DR5 induction by celecoxib was also documented (23). In the present study, we showed that celecoxib at a DR5- and apoptosis-inducing concentration initiated ER stress, including CHOP, ATF4, Bip, and IRE1α up-regulation in a rapid fashion, which was accompanied by DR5 induction in different lung cancer cell lines (Fig. 1). DR5 promoter analyses and CHOP knockdown study have demonstrated that CHOP induction is required for celecoxib-mediated DR5 induction and subsequent apoptosis (Fig. 2). These findings are not novel and basically confirm the previous findings in this regard.
Moreover, we report the novel finding that the transcriptional factor Elk1 is involved in celecoxib-induced DR5 expression, based on the following evidence: 1) the presence of the putative Elk1 binding site is required for celecoxib to increase DR5 promoter or transcriptional activity (Fig. 2); 2) enforced expression of constitutively active Elk1 (Elk1-VP16) increased DR5 transcription and expression through an Elk1-dependent manner (Fig. 3B), implying that Elk1 indeed regulates DR5 expression; and 3) knockdown of Elk1 abolished the ability of celecoxib to induce DR5 expression (Fig. 3D and supplemental Fig. S2), indicating that Elk1 is involved in celecoxib-induced DR5 expression. Given the co-induction of CHOP and Elk1 by celecoxib and the close locations of CHOP and Elk1 binding sites in the DR5 promoter region, we speculated that CHOP and Elk1 might cooperate to transactivate the DR5 gene and induce DR5 expression. This hypothesis is supported by our finding that co-expression of ectopic CHOP and Elk1 is more potent than each single gene transfection in increasing DR5 promoter activity and expression (Fig. 3C). Moreover, silencing of either CHOP or Elk1 was sufficiently abrogated DR5 induction by celecoxib (supplemental Fig. S2). Therefore, it is very likely that both CHOP and Elk1 are essential for DR5 regulation, and they cooperate to induce DR5 expression upon celecoxib treatment.
It is known that the Elk1 protein is directly phosphorylated by ERK1/2 (13, 48). Thus, the finding of the involvement of Elk1 in regulation of celecoxib-induced DR5 expression resulted in our subsequent novel finding that celecoxib and other agents activate the ERK1/2 signaling pathway, which contributes to DR5 induction by these agents. In addition to Elk1, RSK2 is another well known protein kinase that is directly phosphorylated and activated by ERK1/2 (16, 49). In our study, celecoxib increased phosphorylation of not only ERK1/2, but also RSK2 and Elk1 in every cell line tested, indicating that celecoxib activates ERK1/2 signaling. Inhibition of either ERK1/2 or RSK2 activation with both small molecule inhibitors and siRNA- or shRNA-mediated gene silencing abolished celecoxib-induced DR5 expression (Figs. 4 and 5). These compelling lines of evidence clearly indicate that the activation of ERK/RSK signaling is required for celecoxib-induced DR5 expression. Similarly, we found that other well known ER stress inducers, tunicamycin and thapsigargin, also increased ERK1/2 and RSK2 phosphorylation and induced DR5 expression. Inhibition of ERK1/2 and RSK activation with the MEK inhibitor U0126 or knockdown of RSK2 abrogated DR5 induction by these agents (supplemental Figs. S3 and S5), indicating that the ERK/RSK signaling is also involved in DR5 up-regulation induced by these agents. Thus, ERK/RSK-mediated DR5 regulation is not only a mechanism of action of celecoxib, but also of other drugs. To the best of our knowledge, this constitutes the first demonstration that the ERK/RSK signaling participates in the regulation of DR5 expression.
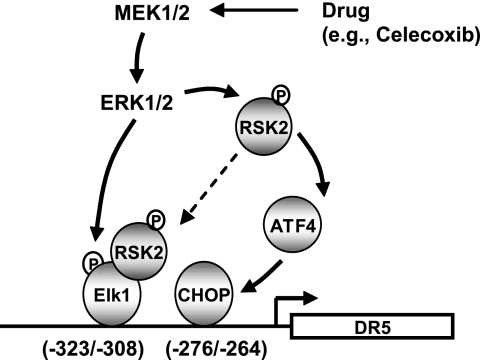
It has been proposed that RSK2 can enhance Elk1-mediated transcription through direct interaction with Elk1 (50). Therefore, it is possible that RSK2 is involved in Elk1-mediated DR5 transactivation (e.g. by celecoxib) via direct interaction with Elk1 (Fig. 7). Given the essential role of RSK2 activation in DR5 up-regulation by celecoxib as demonstrated in this study, further investigation in this regard is warranted to understand fully how Elk1 regulates DR5 expression.
FIGURE 7.
Schematic working model for ERK/RSK-mediated signaling of drug (e.g. celecoxib)-induced DR5 expression through co-activation of both CHOP and Elk1. The data in the present study suggest that ERK1/2 activation can result in CHOP expression through RSK2-mediated ATF4 activation, which concurrently activates Elk1 through direct phosphorylation. It is also possible that activated RSK2 may enhance Elk1-mediated DR5 transactivation through direct interaction with Elk1. CHOP and Elk1 then co-operate to transactivate the DR5 gene and expression.
It has been shown that ERK1/2 regulates CHOP expression either positively or negatively depending on environmental stimuli (51). Moreover, ERK1/2-mediated CHOP up-regulation induced by small molecules, including butyrate, green tea plus celecoxib, and resveratrol, has also been reported (52–54). In agreement with these findings, we observed that inhibition of both ERK1/2 and RSK2 attenuated the ability of celecoxib to induce CHOP and DR5 expression (Figs. 4 and 5), indicating that ERK/RSK signaling regulates celecoxib-induced CHOP expression. Similarly, RSK2 knockdown abolished CHOP induction by tunicamycin and thapsigargin (supplemental Fig. S5). However, how ERK/RSK signaling regulates drug-induced CHOP expression has not been demonstrated. In an effort to understand the mechanism by which celecoxib increases CHOP expression, we have shown that ATF4 is important for celecoxib-induced CHOP expression based on the following findings: 1) the C/EBP-ATF site in the CHOP promoter region to which ATF4 can bind (45, 46) is critical for CHOP transcription induced by celecoxib (Fig. 6); 2) celecoxib increases ATF4 levels, which occurs even ahead of CHOP induction upon celecoxib treatment (Fig. 1); and 3) silencing of ATF4 with ATF4 siRNA abolished celecoxib-induced CHOP expression. Therefore, our current study also highlights a mechanism underlying CHOP up-regulation induced by small molecule drugs such as celecoxib.
It has been documented that RSK2 directly phosphorylates and activates ATF4 (43). In our study, we found that inhibition of RSK2 by knocking down RSK2 expression attenuated up-regulation of ATF4, in addition to CHOP and DR5, induced by celecoxib as well as tunicamycin and thapsigargin (Fig. 5 and supplemental Figs. S4 and S5), indicating that RSK2 functions upstream of ATF4 to mediate ATF4 up-regulation by these agents. Therefore, it is plausible to propose that agents like celecoxib activate ERK1/2 and RSK2 signaling, leading to ATF4 activation, which in turn promotes CHOP induction and subsequent DR5 expression (Fig. 7).
We previously reported that celecoxib induces apoptosis and enhances TRAIL-induced apoptosis in part through up-regulation of DR5 (25). In this study, we further show that celecoxib-induced DR5 expression is dependent on the activation of ERK/RSK signaling. Therefore, we speculated that blockade of celecoxib-induced ERK/RSK activation will impair the ability of celecoxib to enhance TRAIL-induced apoptosis. Indeed, the combination of celecoxib and TRAIL exhibited augmented effects on inducing caspase cleavage and increasing DNA fragmentation compared with the effect of each single agent in the control cells. These effects were substantially attenuated in cells in which RSK2 was silenced (Fig. 5), indicating that RSK2 activation is critical for celecoxib-mediated enhancement of TRAIL-induced apoptosis. This result reinforces our notion that DR5 induction is important for celecoxib-mediated enhancement of TRAIL-induced apoptosis.
In this study, we have not addressed how celecoxib as well as other agents activate the ERK/RSK signaling. It has been demonstrated previously that induction of ER stress activates MEK/ERK signaling in various cell types, although it is largely a survival mechanism (55–59). In our cell systems, tunicamycin and thapsigargin also increased ERK1/2 and RSK phosphorylation (supplemental Fig. S2), confirming that induction of ER stress activates the ERK/RSK signaling pathway. Thus, it is likely that celecoxib-induced ERK/RSK activation is associated with ER stress. The question is what the relationship between ER stress and the ERK/RSK signaling pathway is. A recent study has shown that local K-Ras (i.e. ER surface K-Ras) activation turns on ERK1/2, leading to regulation of ER stress signaling (60). Given that Ras activation induces MEK-dependent DR5 up-regulation (61, 62), it is plausible to ask whether celecoxib and other ER stress inducers activate the ERK/RSK signaling through a Ras-dependent mechanism. The ongoing work in this direction may provide answer to this question.
In summary, the present study has revealed a novel mechanism by which celecoxib and other agents increase DR5 expression. The activation of ERK1/2 and RSK2 appears central in this mechanism. The activated ERK1/2 may directly phosphorylate and activate Elk1-mediated transcription of DR5 and concurrently promote CHOP-dependent transcription of DR5 through RSK2-dependent, ATF4-mediated CHOP up-regulation. Moreover, phosphorylated RSK2 may also enhance Elk1-mediated DR5 transcription through direct interaction with Elk1. It is likely that CHOP and Elk1 cooperate to mediate DR5 up-regulation induced by celecoxib as well other agents (Fig. 7). Our findings thus highlight a novel mechanism underlying drug-induced DR5 expression and apoptosis.
Supplementary Material
Acknowledgments
We thank Drs. H. G. Wang, A. D. Sharrocks, R. Treisman, P. Fafournoux, and A. B. Vaandrager for providing important constructs and Drs. G. Chen, K. Ye, and J. D. Minna for cell lines. We also thank Dr. A. Hammond for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health/NCI SPORE P50 Grant CA128613 (to S.-Y. S. for Project 2). This work was also supported by a Georgia Cancer Coalition Distinguished Cancer Scholar award (to S.-Y. S.) and Department of Defense VITAL Grant W81XWH-04-1-0142 (to S.-Y. S. for Project 4).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- DR
- death receptor
- ATF
- activating transcription factor
- CHOP
- C/EBP homologous protein
- ER
- endoplasmic reticulum
- fmk
- fluoromethyl ketone
- RSK
- ribosomal S6 kinase
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- Z
- benzyloxycarbonyl.
REFERENCES
- 1.Ashkenazi A. (2008) Cytokine Growth Factor Rev. 19, 325–331 [DOI] [PubMed] [Google Scholar]
- 2.Sun S. Y. (2005) Apoptosis 10, 1203–1210 [DOI] [PubMed] [Google Scholar]
- 3.Elrod H. A., Sun S. Y. (2008) Cancer Biol. Ther. 7, 163–173 [DOI] [PubMed] [Google Scholar]
- 4.Wu G. S., Burns T. F., McDonald E. R., 3rd, Jiang W., Meng R., Krantz I. D., Kao G., Gan D. D., Zhou J. Y., Muschel R., Hamilton S. R., Spinner N. B., Markowitz S., Wu G., el-Deiry W. S. (1997) Nat. Genet. 17, 141–143 [DOI] [PubMed] [Google Scholar]
- 5.Takimoto R., El-Deiry W. S. (2000) Oncogene 19, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 6.Ravi R., Bedi G. C., Engstrom L. W., Zeng Q., Mookerjee B., Gélinas C., Fuchs E. J., Bedi A. (2001) Nat. Cell Biol. 3, 409–416 [DOI] [PubMed] [Google Scholar]
- 7.Shetty S., Graham B. A., Brown J. G., Hu X., Vegh-Yarema N., Harding G., Paul J. T., Gibson S. B. (2005) Mol. Cell. Biol. 25, 5404–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi H., Wang H. G. (2004) J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 9.Yoshida T., Shiraishi T., Nakata S., Horinaka M., Wakada M., Mizutani Y., Miki T., Sakai T. (2005) Cancer Res. 65, 5662–5667 [DOI] [PubMed] [Google Scholar]
- 10.Baritaki S., Katsman A., Chatterjee D., Yeung K. C., Spandidos D. A., Bonavida B. (2007) J. Immunol. 179, 5441–5453 [DOI] [PubMed] [Google Scholar]
- 11.Zou W., Liu X., Yue P., Zhou Z., Sporn M. B., Lotan R., Khuri F. R., Sun S. Y. (2004) Cancer Res. 64, 7570–7578 [DOI] [PubMed] [Google Scholar]
- 12.Zou W., Yue P., Khuri F. R., Sun S. Y. (2008) Cancer Res. 68, 7484–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z., Xu S. (2006) IUBMB Life 58, 621–631 [DOI] [PubMed] [Google Scholar]
- 14.Balmanno K., Cook S. J. (2009) Cell Death Differ. 16, 368–377 [DOI] [PubMed] [Google Scholar]
- 15.Cagnol S., Chambard J. C. (2010) FEBS J. 277, 2–21 [DOI] [PubMed] [Google Scholar]
- 16.Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 17.Tan Y., Ruan H., Demeter M. R., Comb M. J. (1999) J. Biol. Chem. 274, 34859–34867 [DOI] [PubMed] [Google Scholar]
- 18.Buck M., Poli V., Hunter T., Chojkier M. (2001) Mol. Cell 8, 807–816 [DOI] [PubMed] [Google Scholar]
- 19.Anjum R., Roux P. P., Ballif B. A., Gygi S. P., Blenis J. (2005) Curr. Biol. 15, 1762–1767 [DOI] [PubMed] [Google Scholar]
- 20.Wang A., Rud J., Olson C. M., Jr., Anguita J., Osborne B. A. (2009) J. Immunol. 183, 3268–3277 [DOI] [PubMed] [Google Scholar]
- 21.Thun M. J., Henley S. J., Patrono C. (2002) J. Natl. Cancer Inst. 94, 252–266 [DOI] [PubMed] [Google Scholar]
- 22.Schönthal A. H. (2006) Neurosurg. Focus 20, E21. [DOI] [PubMed] [Google Scholar]
- 23.He Q., Luo X., Jin W., Huang Y., Reddy M. V., Reddy E. P., Sheikh M. S. (2008) Oncogene 27, 2656–2660 [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Yue P., Schönthal A. H., Khuri F. R., Sun S. Y. (2006) Cancer Res. 66, 11115–11119 [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Yue P., Zhou Z., Khuri F. R., Sun S. Y. (2004) J. Natl. Cancer Inst. 96, 1769–1780 [DOI] [PubMed] [Google Scholar]
- 26.Chen S., Liu X., Yue P., Schönthal A. H., Khuri F. R., Sun S. Y. (2007) Mol. Pharmacol. 72, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 27.Cohen M. S., Zhang C., Shokat K. M., Taunton J. (2005) Science 308, 1318–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez R. D., Sheridan S., Girard L., Sato M., Kim Y., Pollack J., Peyton M., Zou Y., Kurie J. M., Dimaio J. M., Milchgrub S., Smith A. L., Souza R. F., Gilbey L., Zhang X., Gandia K., Vaughan M. B., Wright W. E., Gazdar A. F., Shay J. W., Minna J. D. (2004) Cancer Res. 64, 9027–9034 [DOI] [PubMed] [Google Scholar]
- 29.Sun S. Y., Yue P., Wu G. S., El-Deiry W. S., Shroot B., Hong W. K., Lotan R. (1999) Oncogene 18, 2357–2365 [DOI] [PubMed] [Google Scholar]
- 30.Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D. (2003) Mol. Cell 12, 63–74 [DOI] [PubMed] [Google Scholar]
- 31.Price M. A., Rogers A. E., Treisman R. (1995) EMBO J. 14, 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S. Y., Liu X., Zou W., Yue P., Marcus A. I., Khuri F. R. (2007) J. Biol. Chem. 282, 18800–18809 [DOI] [PubMed] [Google Scholar]
- 33.Tsutsumi S., Namba T., Tanaka K. I., Arai Y., Ishihara T., Aburaya M., Mima S., Hoshino T., Mizushima T. (2006) Oncogene 25, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 34.Lin Y. D., Chen S., Yue P., Zou W., Benbrook D. M., Liu S., Le T. C., Berlin K. D., Khuri F. R., Sun S. Y. (2008) Cancer Res. 68, 5335–5344 [DOI] [PubMed] [Google Scholar]
- 35.Bruhat A., Jousse C., Carraro V., Reimold A. M., Ferrara M., Fafournoux P. (2000) Mol. Cell. Biol. 20, 7192–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Sanden M. H., Meems H., Houweling M., Helms J. B., Vaandrager A. B. (2004) J. Biol. Chem. 279, 52007–52015 [DOI] [PubMed] [Google Scholar]
- 37.Pyrko P., Kardosh A., Schönthal A. H. (2008) Biochem. Pharmacol. 75, 395–404 [DOI] [PubMed] [Google Scholar]
- 38.Tsutsumi S., Gotoh T., Tomisato W., Mima S., Hoshino T., Hwang H. J., Takenaka H., Tsuchiya T., Mori M., Mizushima T. (2004) Cell Death Differ. 11, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 39.Chen S. T., Thomas S., Gaffney K. J., Louie S. G., Petasis N. A., Schönthal A. H. (2010) Leukemia Res. 34, 250–253 [DOI] [PubMed] [Google Scholar]
- 40.Kim S. H., Hwang C. I., Juhnn Y. S., Lee J. H., Park W. Y., Song Y. S. (2007) Carcinogenesis 28, 223–231 [DOI] [PubMed] [Google Scholar]
- 41.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 42.Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H. C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T. M., Hanauer A., Karsenty G. (2004) Cell 117, 387–398 [DOI] [PubMed] [Google Scholar]
- 44.Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. (1999) Biochem. J. 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 45.Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. (2004) J. Biol. Chem. 279, 5288–5297 [DOI] [PubMed] [Google Scholar]
- 46.Chérasse Y., Maurin A. C., Chaveroux C., Jousse C., Carraro V., Parry L., Deval C., Chambon C., Fafournoux P., Bruhat A. (2007) Nucleic Acids Res. 35, 5954–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K., Tomisato W., Hoshino T., Ishihara T., Namba T., Aburaya M., Katsu T., Suzuki K., Tsutsumi S., Mizushima T. (2005) J. Biol. Chem. 280, 31059–31067 [DOI] [PubMed] [Google Scholar]
- 48.Yordy J. S., Muise-Helmericks R. C. (2000) Oncogene 19, 6503–6513 [DOI] [PubMed] [Google Scholar]
- 49.Hauge C., Frödin M. (2006) J. Cell Sci. 119, 3021–3023 [DOI] [PubMed] [Google Scholar]
- 50.Aksan Kurnaz I. (2004) Biotechnol. Bioeng. 88, 890–900 [DOI] [PubMed] [Google Scholar]
- 51.Lawrence M. C., McGlynn K., Naziruddin B., Levy M. F., Cobb M. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11518–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo K. J., Lee T. J., Lee S. H., Lee J. M., Seo J. H., Jeong Y. J., Park J. W., Kwon T. K. (2007) Biochem. Pharmacol. 73, 68–76 [DOI] [PubMed] [Google Scholar]
- 53.Suganuma M., Kurusu M., Suzuki K., Tasaki E., Fujiki H. (2006) Int. J. Cancer 119, 33–40 [DOI] [PubMed] [Google Scholar]
- 54.Scott D. W., Longpre J. M., Loo G. (2008) DNA Cell Biol. 27, 607–614 [DOI] [PubMed] [Google Scholar]
- 55.Zhang L. J., Chen S., Wu P., Hu C. S., Thorne R. F., Luo C. M., Hersey P., Zhang X. D. (2009) Cancer Lett. 274, 40–46 [DOI] [PubMed] [Google Scholar]
- 56.Jiang C. C., Chen L. H., Gillespie S., Wang Y. F., Kiejda K. A., Zhang X. D., Hersey P. (2007) Cancer Res. 67, 9750–9761 [DOI] [PubMed] [Google Scholar]
- 57.Hu P., Han Z., Couvillon A. D., Exton J. H. (2004) J. Biol. Chem. 279, 49420–49429 [DOI] [PubMed] [Google Scholar]
- 58.Hung C. C., Ichimura T., Stevens J. L., Bonventre J. V. (2003) J. Biol. Chem. 278, 29317–29326 [DOI] [PubMed] [Google Scholar]
- 59.Arai K., Lee S. R., van Leyen K., Kurose H., Lo E. H. (2004) J. Neurochem. 89, 232–239 [DOI] [PubMed] [Google Scholar]
- 60.Wu R. F., Ma Z., Liu Z., Terada L. S. (2010) Mol. Cell. Biol. 30, 3553–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drosopoulos K. G., Roberts M. L., Cermak L., Sasazuki T., Shirasawa S., Andera L., Pintzas A. (2005) J. Biol. Chem. 280, 22856–22867 [DOI] [PubMed] [Google Scholar]
- 62.Nesterov A., Nikrad M., Johnson T., Kraft A. S. (2004) Cancer Res. 64, 3922–3927 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.