Abstract
Glycogen synthase kinase-3 (Gsk-3) isoforms, Gsk-3α and Gsk-3β, are constitutively active, largely inhibitory kinases involved in signal transduction. Underscoring their biological significance, altered Gsk-3 activity has been implicated in diabetes, Alzheimer disease, schizophrenia, and bipolar disorder. Here, we demonstrate that deletion of both Gsk-3α and Gsk-3β in mouse embryonic stem cells results in reduced expression of the de novo DNA methyltransferase Dnmt3a2, causing misexpression of the imprinted genes Igf2, H19, and Igf2r and hypomethylation of their corresponding imprinted control regions. Treatment of wild-type embryonic stem cells and neural stem cells with the Gsk-3 inhibitor, lithium, phenocopies the DNA hypomethylation at these imprinted loci. We show that inhibition of Gsk-3 by phosphatidylinositol 3-kinase (PI3K)-mediated activation of Akt also results in reduced DNA methylation at these imprinted loci. Finally, we find that N-Myc is a potent Gsk-3-dependent regulator of Dnmt3a2 expression. In summary, we have identified a signal transduction pathway that is capable of altering the DNA methylation of imprinted loci.
Keywords: DNA Methylation, Embryonic Stem Cell, Epigenetics, Gene Expression, Signal Transduction, Imprinting
Introduction
Gsk-32 is functionally defined as the aggregate activity of both Gsk-3α and Gsk-3β isoforms that are encoded at distinct genetic loci. These highly redundant kinases are constitutively active and generally play an inhibitory role in the signal transduction pathways that they regulate (1), such as insulin signaling and canonical Wnt signaling (2). In addition, Gsk-3 has a major role in regulating differentiation of embryonic stem cells (3) and neural progenitors (4). Both insulin and Wnt signaling have been implicated in the regulation of stem cell pluripotency (5–13). Insulin signaling modulates Gsk-3 activity via the activity of upstream effectors; insulin or insulin-like growth factor binds to the insulin receptor, resulting in the activation of PI3K, which in turn phosphorylates and activates Akt (also called protein kinase B; PKB) (14). Akt subsequently phosphorylates several substrates, including Gsk-3 isoforms, on N-terminal serine residues (Gsk-3α Ser-21/Gsk-3β Ser-9), resulting in the inhibition of Gsk-3 activity (15).
Gsk-3 activity also represents a key regulatory step in the canonical Wnt signaling pathway. In the absence of ligand, a subset of cytoplasmic Gsk-3 is found in a protein complex that facilitates Gsk-3-mediated phosphorylation of β-catenin, targeting the protein for ubiquitination and degradation via the 26 S proteasome and keeping the Wnt pathway repressed (16). Upon ligand binding to the Wnt co-receptors, Gsk-3 redistributes to the cell surface (17), rendering the β-catenin destruction complex non-functional, causing the accumulation and subsequent nuclear translocation of β-catenin protein, which leads to the transcription of Wnt target genes. Although activated insulin and Wnt signaling both inhibit Gsk-3 activity, the mechanism of inhibition is distinct for each pathway; Wnt signaling does not affect insulin signaling, and insulin signaling does not activate Wnt target genes (18, 19).
Gsk-3 function in the insulin and Wnt signaling pathways has been thoroughly investigated, and numerous substrates of Gsk-3 have been described (2), yet the downstream effects of such a widely influential enzyme likely extend beyond our current knowledge. Here, we present evidence of a novel role for Gsk-3 isoforms in the regulation of DNA methylation at imprinted loci in mouse embryonic stem cells (ESCs). The de novo DNA methyltransferase, Dnmt3a2, is down-regulated in Gsk-3 double knock-out (DKO) ESCs, resulting in reduced DNA methylation and altered expression of imprinted genes. Inhibition of Gsk-3 activity with lithium mimics the effects of reducing DNA methylation in both wild-type ESCs and wild-type neural stem cells. Furthermore, inactivation of Gsk-3 via components of the insulin signaling pathway results in reduced DNA methylation at imprinted loci. Finally, microarray data reveal that N-myc mRNA is down-regulated in Gsk-3 DKO ESCs. We provide data that demonstrate that a highly conserved N-Myc binding site in the Dnmt3a2 promoter is required for normal expression, and we demonstrate that siRNA knockdown of N-Myc results in a decrease in Dnmt3a2 expression. Therefore, we have identified a novel function for Gsk-3 isoforms as key regulators of the epigenome, and our results add a new perspective on the consequences of altering Gsk-3 activity.
EXPERIMENTAL PROCEDURES
Cell Culture
Feeder-free wild-type, Gsk-3α−/−, Gsk-3β−/−, and Gsk-3 DKO ESCs (3) were grown on gelatin-coated plates in high glucose DMEM (Invitrogen) supplemented with 15% fetal bovine serum (HyClone), 1× non-essential amino acids, 1× sodium pyruvate, 2 mm l-glutamine, 1× penicillin/streptomycin (Invitrogen), 55 μm 2-mercaptoethanol, and 1000 units/ml ESGRO (Millipore). Media was replenished every other day. Neural stem cells were isolated from 12.5 days postcoitum embryos using NeuroCult neural stem cells (NSC) proliferation media (StemCell Technologies) following the manufacturer's protocol.
Microarray Analysis
Integrity of total RNA was evaluated using capillary electrophoresis (Bioanalyzer 2100, Agilent) and quantified using a Nanodrop 1000 (Nanodrop, Wilmington, DE). Following confirmation of RNA quality, OvationTM biotin RNA amplification and labeling system (NuGen Technologies, Inc., San Carlos, CA) was used to prepare amplified, biotin-labeled cDNA from total RNA following manufacturer's instructions. Briefly, first strand cDNA was synthesized from 25 ng of total RNA using a unique first strand DNA/RNA chimeric primer and reverse transcriptase. Following double strand cDNA generation, amplification of cDNA was achieved by utilizing an isothermal DNA amplification process that involves repeated SPIATM DNA/RNA primer binding, DNA duplication, strand displacement, and RNA cleavage. The amplified SPIATM cDNA was purified and subjected to a two-step fragmentation and labeling process. The fragmented/biotinylated cDNA content was measured in a ND-1000 spectrophotometer, and the quality was analyzed on an RNA 6000 Nano LabChip (Agilent) using an Agilent Bioanalyzer 2100.
For each array, 2.2 μg of cDNA was hybridized onto the GeneChips® mouse genome 430 2.0 array (Affymetrix Inc.), which contains ∼39,000 transcripts. The sequences from which these probe sets were derived were selected from GenBankTM, dbEST, and RefSeq. The sequence clusters were created from the UniGene data base (Build 107, June 2002) and then refined by analysis and comparison with the publicly available draft assembly of the mouse genome from the Whitehead Institute for Genome Research (Mouse Genome Sequencing Consortium (MGSC), April 2002). Hybridization was allowed to continue for 16 h at 45 °C followed by washing and staining of microarrays in a Fluidics Station 450 (Affymetrix Inc.). GeneChip arrays were scanned in a GeneChip Scanner 3000 (Affymetrix Inc.), and CEL files were generated from DAT files using the GeneChip® operating software (GCOS) software (Affymetrix Inc.). The probe set signals were generated using the RMA algorithm in ArrayAssist 3.4 (Stratagene) and were used to determine differential gene expression by pairwise comparisons. The genes that were altered by 2-fold either way and had a false discovery rate of <10% were sorted and used for further interpretation of the microarray data. Microarray data have been deposited in GEO (www.ncbi.nlm.nih.gov/geo) under accession number GSE20015.
Stable Expression of Dnmt3a2 and p110* in ESCs
Mouse Dnmt3a2 cDNA was subcloned into a modified pCAGEN plasmid (from Connie Cepko, Addgene plasmid 11160) with a preceding puromycin resistance gene and internal ribosomal entry site. The plasmid containing a constitutively active myristoylated mouse p110 subunit of PI3K (p110*) ((5); from Shinya Yamanaka, Addgene plasmid 15689) expressed a hygromycin resistance cassette. ES cells were transfected with either the Dnmt3a2 or the p110* expression constructs with Lipofectamine 2000 (Invitrogen) in serum-free Opti-MEM (Invitrogen). Cells were allowed to recover in non-selective ES cell media for 24 h, after which puromycin-resistant (Dnmt3a2) or hygromycin-resistant (p110*) cells were grown in ES cell media containing 1 μg/ml puromycin or 200 μg/ml hygromycin, respectively. Puromycin/hygromycin-resistant colonies were isolated after 14 days of selection and expanded in selective media.
Dnmt3a2 Reporter Assays
The Dnmt3a2 promoter was cloned by PCR amplification from bacterial artificial chromosome clone 330c18 RPCI-24 library using primers described in supplemental Table S1. Amplification products were TOPO TA-cloned into pCR8GW (Invitrogen) and then transferred by LR reaction to Gateway-modified pGL3-Basic, pGLF (from Glenn Maston, University of Massachusetts). Mutagenesis of the N-Myc binding site was accomplished by using the QuikChange II site-directed mutagenesis kit (Stratagene) (supplemental Table S1). Plasmids containing the Dnmt3a2 promoter constructs driving firefly luciferase were co-transfected into ESCs with pRL SV40 with Lipofectamine 2000 (Invitrogen) in serum-free Opti-MEM (Invitrogen). Cells were replated in a 24-well plate 24 h after transfection and then lysed 48 h after transfection, at which time firefly and Renilla Luciferase assays were performed according to the manufacturer's protocol (Biotium) in a Veritas microplate luminometer (Turner Biosystems). Three replicates were performed for each transfection.
Antibodies and Protein Expression Analysis
Protein expression was assayed by Western blotting. Cells were resuspended in lysis buffer (137 mm NaCl, 10 mm Tris, pH 7.4, 1% Nonidet P-40) containing protease inhibitor mixture (1:100 Sigma) and sonicated for 10 30-s pulses at 4 °C in a Bioruptor (Diagenode) on the highest setting. Lysates were electrophoresed (8–20 mg/lane) through 7.5% Tris/Tricine gels and transferred onto nitrocellulose membrane (Whatman BA85) at 100 V for 1 h. Blots were blocked for 1 h with 5% milk/TBST (150 mm NaCl, 50 mm Tris, pH 7.4, 0.1% Tween) and incubated in primary antibody diluted in 5% milk or 5% BSA (N-Myc and c-Myc antibodies)/TBST for 16–18 h at 4 °C. Antibodies were used under the following conditions. Anti-tubulin mouse mAb clone B-5-1-2 (Sigma) was diluted 1:10,000; anti-Dnmt3a mouse mAb clone 64B1446 (IMGENEX) was diluted 1:250; and anti-GSK-3α/β mouse mAb clone 1H8 (Calbiochem), anti-phospho-GSK-3α/β (Cell Signaling antibody 9331), anti-N-Myc (Cell Signaling antibody 9405), anti-c-Myc D84C12 XP (Cell Signaling antibody 5605), anti-PI3kinase p110α subunit C73F8 (Cell Signaling antibody 4249), anti-Akt1 2H10 (Cell Signaling antibody 2967), and anti-phospho-Akt Ser-473 193H12 (Cell Signaling antibody 4058) were all used at a 1:1000 dilution. Blots were incubated in anti-mouse or anti-rabbit IgG HRP secondary antibody (GE Healthcare) diluted 1:5000 in 5% milk in TBST for 45 min. Proteins were visualized using ECL detection reagent (GE Healthcare). Blots were stripped in a buffer consisting of 2% SDS, 62.5 mm Tris HCl, pH 6.7, and 100 mm β-mercaptoethanol at 50 °C for 30 min followed by repeated rinsing with TBST prior to reprobing with antibody.
DNA Isolation
High molecular weight genomic DNA was isolated after ESCs were washed in PBS and resuspended in lysis buffer (100 mm NaCl, 10 mm Tris, pH 8.0, 25 mm EDTA, pH 8.0, 0.5% SDS) and lysed 16–18 h at 55 °C. An equal volume of phenol:chloroform:isoamyl alcohol was added and gently rotated at ambient temperature for 2–3 h. DNA was precipitated with 2 volumes of ethanol and 0.1 volumes of sodium acetate, washed in 70% ethanol, and resuspended in sterile water.
Southern Blot
High molecular weight genomic DNA was digested with methylation-sensitive HpaII or methylation-insensitive MspI isoschizomers (New England Biolabs). Digested DNA was separated on 0.6% agarose in 0.5× Tris-acetate-EDTA and transferred to charged nylon membrane (Osmonics Inc.). The blot was hybridized with a [α-32P]dCTP random primed labeled (Prime-It II, Stratagene) intracisternal A particle (IAP) probe (20) in FBI buffer (21) at 65 °C for 16 h, washed twice with 2× SSC + 0.1% SDS at 65 °C for 30 min, and imaged with a Typhoon 9400 PhosphorImager (GE Healthcare).
RNA Isolation
Cells were resuspended in TRIzol (Invitrogen), and RNA was isolated following the manufacturer's protocol and then further purified with the RNeasy RNA cleanup procedure (Qiagen).
cDNA Synthesis and Quantitative RT-PCR
Complementary DNA was synthesized with the High Capacity cDNA reverse transcription kit (Applied Biosystems) following the manufacturer's protocol. Quantitative RT-PCR was done on an Applied Biosystems 7500 using TaqMan® master mix and one of the following TaqMan assays (Applied Biosystems): Dnmt3a2 (Mm00463987_m1), Igf2 (Mm00439564_m1), H19 (Mm01156721_g1), Igf2r (Mm01313554_m1), Airn (Mm03943369_s1), Snrpn (Mm02391920_g1), N-Myc (Mm00476449_m1), or c-Myc (Mm00487803_m1). Three biological replicates and three technical replicates were used for each target analyzed. All threshold cycle (Ct) values were normalized to a mouse GAPDH endogenous control (Applied Biosystems), and relative quantitation was calculated from the median Ct value.
Bisulfite Sequencing
High molecular weight genomic DNA was fragmented by rapid freezing on dry ice and thawing at 42 °C for five repetitions. 500 ng of fragmented DNA was bisulfite-converted with the methyl code bisulfite conversion kit (Invitrogen) per the manufacturer's instructions. Igf2/H19 differentially methylated domain (DMD) and Igf2r differentially methylated region 2 (DMR2) were amplified from bisulfite-converted DNA with Platinum Taq (Invitrogen) using primers shown in supplemental Table S1 (22) and the following conditions: 94 °C for 10 min, 35 cycles of 94 °C for 45 s, 54 °C for 45 s, and 72 °C for 60 s followed by 72 °C for 7 min. PCR products were then TOPO TA-cloned into pCR8GW (Invitrogen). Plasmid clones were sequenced with M13 reverse primer (supplemental Table S1) using Big Dye version 3.1 chemistry with the following conditions: 96 °C for 10 s, then 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min, and reaction products were resolved on an Applied Biosystems 3130XL genetic analyzer.
siRNA Transfection
125 pmol of Nmyc siRNA (Thermo SMARTpool L-058793-01-0005) or GFP siRNA (Dharmacon D-1300-20) was combined with 0.25 ml of Opti-MEM (Invitrogen). In parallel, 7.5 μl of Lipofectamine RNAiMAX (Invitrogen) was combined with 0.25 ml of Opti-MEM per transfection by gentle mixing. siRNA/Opti-MEM and RNAiMAX/Opti-MEM were then combined for a total volume of ∼0.5 ml and incubated for 20 min at room temperature. This mixture was then used to resuspend a pellet of 1 million wild-type ESCs. The cell suspension was then plated into 2 ml of ESC media in an individual well of a gelatin-coated 6-well plate. Media was replaced with fresh ESC media the next day. At 72 h, cells were harvested, and protein and RNA were isolated as described above.
RESULTS
Expression of the de Novo DNA Methyltransferase Dnmt3a2 Is Reduced in Gsk-3 DKO ESCs
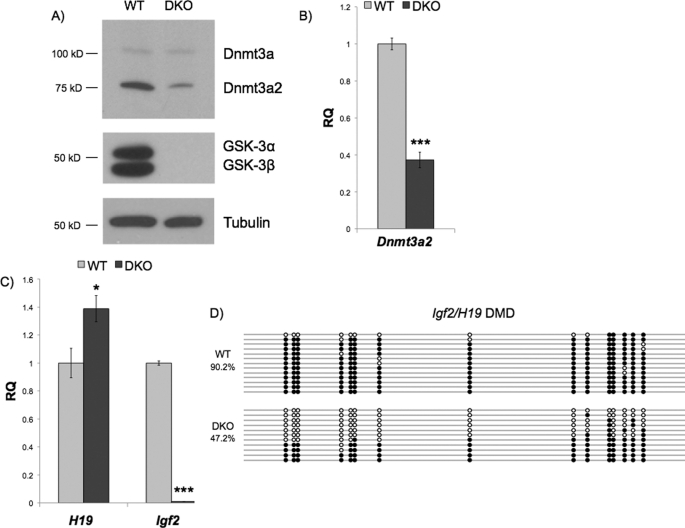
In an effort to better understand the downstream targets of Gsk-3, we performed microarray-based global gene expression analysis comparing wild-type (WT) mouse ESCs with those in which Gsk-3α, Gsk-3β, or both isoforms have been genetically deleted (Gsk-3α−/−; Gsk-3β−/− DKO) (3). Inspection of our array data revealed a 6.2-fold down-regulation of the de novo DNA methyltransferase Dnmt3a in Gsk-3 DKO ESCs but no effect in Gsk-3α−/− or Gsk-3β−/− ESCs (increased 1.1-fold in each). In addition to full-length Dnmt3a, there is a distinct smaller isoform, Dnmt3a2, which is transcribed from an alternative promoter within the Dnmt3a locus. Dnmt3a2 lacks the amino-terminal 219 amino acids found in Dnmt3a, but the remainder of the protein, including the domain containing methyltransferase activity, is identical between the isoforms (23). Importantly for this study, Dnmt3a2 is the isoform that is predominantly expressed in ESCs (24). The microarray probes are unable to differentiate between Dnmt3a isoforms, and the microarray expression data represent a composite of the expression of both Dnmt3a and Dnmt3a2. Therefore, we evaluated the protein expression of each individual isoform as the individual isoforms can be resolved by immunoblotting. We found that Dnmt3a2 protein expression is substantially reduced in Gsk-3 DKO cells, whereas Dnmt3a protein levels were essentially unchanged (Fig. 1A). Real-time quantitative PCR (qPCR) revealed that mRNA expression of Dnmt3a2 is reduced 2.7-fold in Gsk-3 DKO ESCs when compared with WT ESCs (Fig. 1B).
FIGURE 1.
Dnmt3a2 expression and DNA methylation are reduced in Gsk-3 DKO ESCs. A, Western blot of Dnmt3a isoforms in WT and Gsk-3 DKO ESCs. Protein expression of Gsk-3 isoforms in WT and Gsk-3 DKO ESCs is shown in the middle panel, whereas a Western blot for tubulin, which serves as a loading control, is shown in the bottom panel. B, real-time quantitative PCR of Dnmt3a2 showing mRNA expression of Dnmt3a2 in Gsk-3 DKO ESCs relative to WT ESCs (RQ = relative quantitation). Error bars represent S.D. between biological replicates, n = 3 (***, p < 0.001, two-tailed t test). C, real-time quantitative PCR showing H19 and Igf2 mRNA expression in Gsk-3 DKO ESCs relative to WT ESCs. Error bars represent S.D. between biological replicates, n = 3 (*, p < 0.05; ***, p < 0.001, two-tailed t test). D, bisulfite sequencing analysis of the Igf2/H19 DMD. Circles represent 15 CpG dinucleotides analyzed within the Igf2/H19 DMD. Open circles represent unmethylated CpG dinucleotides, whereas filled circles represent methylated CpG dinucleotides (p < 0.01, two-tailed t test).
Imprinted Loci Display Altered Gene Expression and DNA Methylation in Gsk-3 DKO ESCs
Dnmt3a/Dnmt3a2 activity is required for the establishment of DNA methylation at imprinted loci in germ cells (25). Mouse ESCs null for both Dnmt3a and Dnmt3b lose DNA methylation at imprinted loci after extended passaging, demonstrating a role for the de novo methyltransferases in the maintenance of DNA methylation at imprinted loci. Notably, re-expression of Dnmt3a2 alone in Dnmt3a−/−; Dnmt3b−/− ESCs is sufficient to fully restore DNA methylation at paternally imprinted loci, whereas re-expression of Dnmt3a or Dnmt3b1 is only able to minimally rescue DNA methylation at these loci (26). Based on these published observations, we hypothesized that reduction of Dnmt3a2 expression in ESCs would result in the loss of DNA methylation and disruption of gene expression at imprinted loci. The imprinted locus containing the genes insulin-like growth factor II (Igf2) and H19 has been well characterized. The reciprocal expression of Igf2 and H19 is regulated by a shared imprinting control region designated the DMD (27–29). In this imprinting control region, the paternal allele is methylated, resulting in expression of Igf2 and silencing of H19, whereas conversely, the maternal allele is unmethylated, promoting H19 expression while silencing Igf2 (30). Indeed, the imprinted genes H19 and Igf2 display altered expression in Gsk-3 DKO ESCs when compared with WT ESCs. Igf2 and H19 expression in Gsk-3 DKO cells was assayed by qPCR, and H19 expression is increased 1.4-fold, whereas Igf2 expression is decreased 100-fold (Fig. 1C). This pattern of gene expression is consistent with a scenario for the loss of DNA methylation at the Igf2/H19 DMD (29, 31, 32). We directly measured DNA methylation of the Igf2/H19 DMD by bisulfite sequencing and found a 47% reduction of DNA methylation in Gsk-3 DKO ESCs when compared with WT ESCs (Fig. 1D). These results strongly support the hypothesis that the alterations in Igf2 and H19 expression are due to a loss of DNA methylation at the Igf2/H19 DMD.
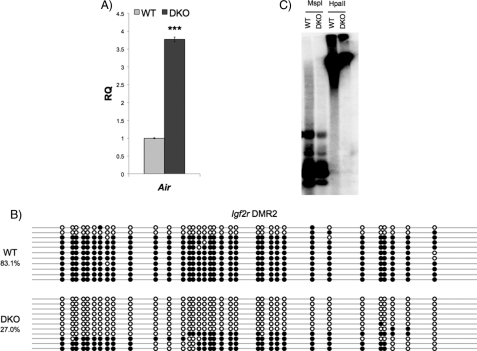
Expression of Air, another imprinted gene that encodes for a long non-coding RNA (34), is increased in Gsk-3 DKO ESCs. Air mRNA expression in Gsk-3 DKO ESCs was assayed by qPCR and is increased by 3.8-fold (Fig. 2A). We then determined whether the change in expression of Air is due to a loss of DNA methylation. We performed bisulfite sequencing on DMR2 of Igf2r, from which Air is transcribed and known to be methylated on the maternal allele (22, 35). DNA methylation at DMR2 is reduced by 67% in Gsk-3 DKO ESCs (Fig. 2B). These data suggest that the increase in Air expression is likely due to DNA hypomethylation within the Igf2r DMR2 and demonstrate that the loss of Gsk-3 activity on DNA methylation extends beyond the Igf2/H19 DMD, supporting a possible broader role for Gsk-3 in the regulation of imprinted genes in the mouse.
FIGURE 2.
Effect on DNA methylation in Gsk-3 DKO ESCs extends to other imprinted genes but is not global. A, real-time quantitative PCR analysis of Air mRNA expression in Gsk-3 DKO ESCs relative to WT ESCs (RQ = relative quantitation). Error bars represent S.D. between biological replicates, n = 3. B, bisulfite sequencing analysis of the Igf2r DMR2 in WT and Gsk-3 DKO ESCs. Circles represent 35 CpG dinucleotides analyzed within the Igf2r DMR2. Open circles represent unmethylated CpG dinucleotides, whereas filled circles represent methylated CpG dinucleotides (p < 0.01, two-tailed t test). C, Southern blot analysis showing methylation of IAP in WT and Gsk-3 DKO ESCs. Genomic DNA from WT ESCs or Gsk-3 DKO ESCs was digested with MspI or HpaII. MspI is not sensitive to DNA methylation and cuts DNA regardless of methylation status. HpaII is an isoschizomer of MspI that does not cut methylated DNA.
Global DNA Methylation Is Unchanged in Gsk-3 DKO ESCs
We next determined whether the DNA hypomethylation we observed at imprinted loci is due to a defect in global DNA methylation. Because Dnmt1 is the primary enzyme responsible for maintaining genome-wide DNA methylation (36), we examined whether the loss of Gsk-3 activity affects Dnmt1 function. Repetitive DNA sequences, such as IAP repeats, are highly methylated in WT mouse ESCs (37) and largely unmethylated in Dnmt1−/− and Dnmt1 hypomorph ESCs (38). IAP repeat methylation, although reduced, is largely retained in ESCs deficient in de novo DNA methyltransferase activity (Dnmt3a−/−; Dnmt3b−/−) (38, 39). Therefore, analyzing DNA methylation of IAP repeats provides an assay for Dnmt1 activity. Southern blotting using methylation-sensitive restriction enzymes revealed that Gsk-3 DKO ESCs do not exhibit a significant difference in DNA methylation of IAP repeats when compared with wild-type ESCs (Fig. 2C), suggesting that there is not a general defect in DNA maintenance methylation in Gsk-3 DKO cells. These data strengthen our hypothesis that the loss of DNA methylation observed at imprinted loci in Gsk-3 DKO ESCs is likely the result of specific down-regulation of Dnmt3a2 expression.
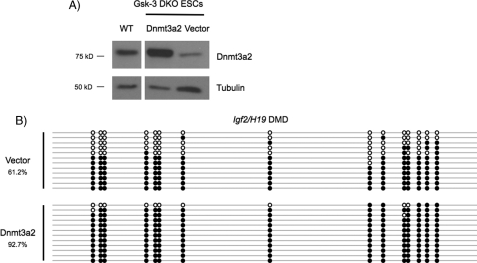
Ectopic Expression of Dnmt3a2 in Gsk-3 DKO ESCs Rescues the Deficits in DNA Methylation
To evaluate whether exogenous expression of Dnmt3a2 could rescue the loss of DNA methylation at imprinted loci in Gsk-3 DKO ESCs, we isolated puromycin-resistant Gsk-3 DKO ESCs stably expressing Dnmt3a2 under the control of the chicken β-actin (CAG) promoter and evaluated DNA methylation at the Igf2/H19 DMD by bisulfite sequencing. A puromycin-resistant clone in which Dnmt3a2 is stably overexpressed in DKO ESCs (Dnmt3a2) was selected for bisulfite sequencing analysis, and a puromycin-resistant clone that does not overexpress Dnmt3a2 (Vector) served as a negative control (Fig. 3A). Bisulfite sequencing of the Igf2/H19 DMD revealed that DNA methylation is restored to levels previously observed in WT ESCs in the overexpressing clone (92.7%; Fig. 3B). Rescue of the Igf2/H19 DMD methylation defect by stably expressing Dnmt3a2 in Gsk-3 DKO ESCs strongly supports our hypothesis that reduced Dnmt3a2 expression in Gsk-3 DKO ESCs is the cause of decreased DNA methylation at imprinted loci.
FIGURE 3.
Reintroduction of Dnmt3a2 into Gsk-3 DKO ESCs rescues DNA methylation. A, Western blot of Dnmt3a2 expression in Gsk-3 DKO ESCs stably expressing Dnmt3a2 from the chicken β-actin (CAG) promoter (Dnmt3a2), DKO ESCs stably expressing the puromycin-resistant empty vector (Vector), and WT ESCs. The WT, Dnmt3a2, and Vector lanes are from the same blot, with intervening lanes removed. B, bisulfite sequencing of the Igf2/H19 DMD in Dnmt3a2 rescued ESCs. Open circles represent unmethylated CpG dinucleotides, whereas filled circles represent methylated CpG dinucleotides.
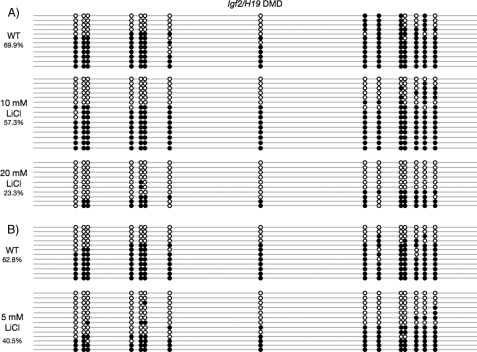
Lithium Reduces DNA Methylation at Imprinted Loci
Based on the data obtained from Gsk-3 DKO ESCs, we hypothesized that the Gsk-3 inhibitor lithium (40) should phenocopy the reduction in DNA methylation of imprinted genes. It was previously reported that Dnmt3a−/−; Dnmt3b−/− ESCs required a high number of passages before a loss of DNA methylation was detected at imprinted loci (26). In light of this observation, we grew WT ESCs in the presence of lithium (40) for 4 weeks and then examined DNA methylation at the Igf2/H19 DMD by bisulfite sequencing. Consistent with the reduction in DNA methylation in Gsk-3 DKO ESCs, treatment of wild-type ESCs with lithium reduces DNA methylation at the Igf2/H19 DMD in a dose-dependent manner. DNA methylation is reduced by 66% in wild-type ESCs treated with 20 mm lithium when compared with WT ESCs that had been cultured in parallel without lithium treatment (Fig. 4A).
FIGURE 4.
Lithium inhibition of Gsk-3 results in hypomethylation. A, WT ESCs were continuously grown in the presence of either 10 mm or 20 mm LiCl for 1 month. DNA methylation at CpG nucleotides within the Igf2/H19 DMD was assessed by bisulfite sequencing (WT versus 20 mm LiCl, p < 0.01, two-tailed t test). B, DNA methylation of CpG nucleotides within the Igf2/H19 DMD in WT NSCs that were continuously grown in the presence of 5 mm LiCl for 1 month (p < 0.01, two-tailed t test). Open circles represent unmethylated CpG dinucleotides, whereas filled circles represent methylated CpG dinucleotides.
Next we examined the effect of Gsk-3 inhibitors on DNA methylation in a cell type other than ESCs. Because the Gsk-3 inhibitor lithium is used clinically to treat bipolar disorder, we chose to use cells from the neuronal lineage, specifically mouse NSC. After 4 weeks of growth in the presence of 5 mm lithium, we observed a 35% reduction in DNA methylation at the Igf2/H19 DMD (Fig. 4B). These results demonstrate that the effect of lithium on DNA methylation extends to neural cells and expand the implications of our findings to other cell types.
Inactivation of Gsk-3 by PI3K results in Loss of DNA Methylation at Imprinted Loci
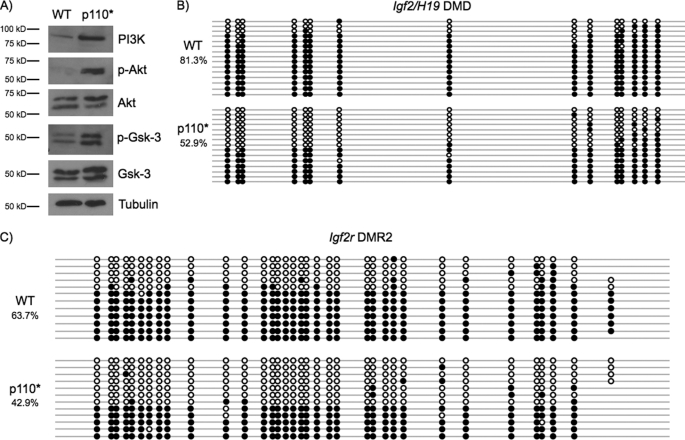
Activated insulin signaling negatively regulates Gsk-3 activity via inhibitory phosphorylation of an N-terminal serine by Akt (41). We investigated whether inactivation of Gsk-3 by constitutively active insulin signaling pathway could affect DNA methylation of imprinted genes. Expression of a myristoylated p110 subunit of PI3K (p110*) has previously been demonstrated to constitutively activate insulin signaling in ESCs (5). WT ESCs were stably transfected with p110*, and Western blots were performed to verify activation of the insulin pathway. Stable expression of p110* leads to robust phosphorylation of Akt on serine 473, which results in the activation of Akt. In addition, we found increased N-terminal phosphorylation of both Gsk-3α (serine 21) and Gsk-3β (serine 9), suggesting that the cellular pool of Gsk-3 regulated by Akt is inhibited (Fig. 5A). Bisulfite sequencing of the H19/Igf2 DMD and Igf2r DMR2 demonstrated a substantial reduction of DNA methylation levels in p110* stable cells, similar to that seen in Gsk-3 DKO ESCs (Fig. 5, B and C). These data demonstrate that activation of insulin signaling by constitutively active PI3K reduces DNA methylation at imprinted loci.
FIGURE 5.
Activation of components of the insulin signaling pathway reduces DNA methylation. A, Western blots showing that WT ESCs stably transfected with p110* constitutively activate insulin signaling. p-Akt, phospho-Akt; p-Gsk-3, phospho-glycogen synthase kinase-3. B, bisulfite sequencing analysis of the Igf2/H19 DMD in p110* ESCs when compared with WT ESCs (p < 0.01, two-tailed t test). C, bisulfite sequencing analysis of the Igf2r DMR2 in p110* ESCs when compared with WT ESCs. Open circles represent unmethylated CpG dinucleotides, whereas filled circles represent methylated CpG dinucleotides.
N-Myc Is a Potent Gsk-3-dependent Regulator of Dnmt3a2 Gene Expression
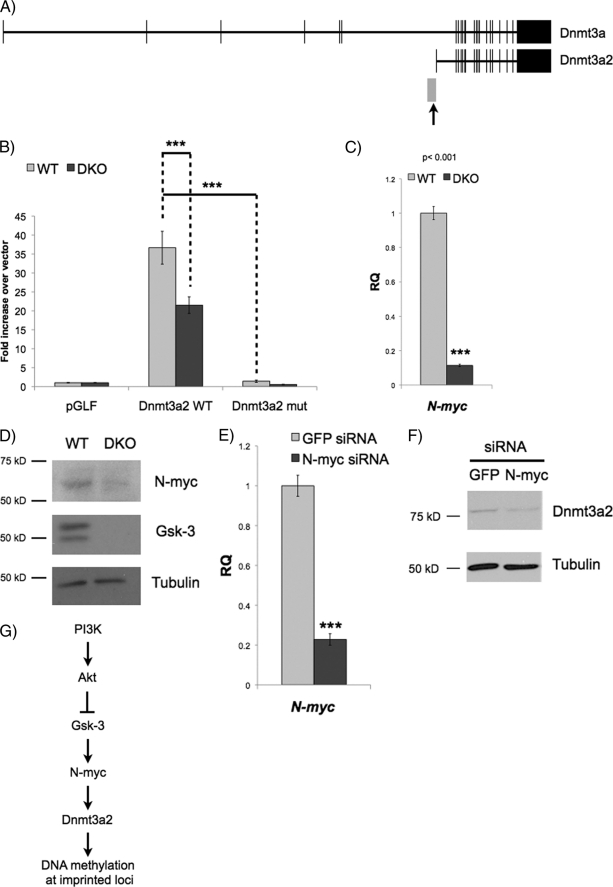
Our data show that the reduced levels of DNA methylation at imprinted loci in Gsk-3 DKO ESCs are likely due to a decrease in Dnmt3a2 levels. To establish the mechanism by which Gsk-3 activity regulates the expression of Dnmt3a2, we considered that both Dnmt3a2 mRNA and protein levels are decreased in Gsk-3 DKO ESCs, suggesting that the effect is likely due to altered transcriptional regulation. A role for miRNAs was excluded because both Dnmt3a and Dnmt3a2 share a common 3′-untranslated region (UTR) yet are not equally affected by the loss of Gsk-3 isoforms. Therefore, we focused on understanding the mechanism controlling Dnmt3a2 transcription. A 1.9-kb fragment of the Dnmt3a2 promoter was cloned (Fig. 6A), which is distinct from the regulatory regions of Dnmt3a and had previously been shown to be sufficient for driving high levels of reporter expression in ESCs (24). This 1.9-kb promoter fragment is capable of yielding robust (>35-fold) expression of a luciferase reporter in WT ESCs (Fig. 6B). The reporter also recapitulates the effect of Gsk-3 deletion on Dnmt3a2 mRNA, showing an ∼40% reduction in reporter activity when transfected into Gsk-3 DKO ESCs.
FIGURE 6.
N-Myc regulates Dnmt3a2 expression in a Gsk-3-dependent manner. A, schematic representation of the locus encoding Dnmt3a isoforms. Exons are shown as the vertical bars. The Dnmt3a2 promoter we used to drive the luciferase promoter is shown as the gray box. The arrow denotes the putative N-Myc binding site that we mutated in our reporter construct. B, Dnmt3a2 promoter activity was assessed with a luciferase reporter. Reporters containing WT 1.9-kb Dnmt3a2 promoter (WT) or promoter with two point mutations (bases underlined) in putative Myc binding site (CACGTG → CAGCTG; mutation (mut)) were transfected into WT and Gsk-3 DKO ESCs. Bars represent -fold change over promoterless luciferase vector (pGLF). Error bars represent S.D. between replicate transfections, n = 3 (***, p < 0.001, two-tailed t test). C, real-time quantitative PCR analysis of N-myc mRNA expression in Gsk-3 DKO ESCs relative to WT ESCs (RQ = relative quantitation). Error bars represent S.D. between biological replicates, n = 3 (***, p < 0.001, two-tailed t test). D, Western blots showing N-Myc, Gsk-3α, Gsk-3β, and tubulin in WT and Gsk-3 DKO ESCs. E, real-time quantitative PCR analysis of N-myc mRNA expression in WT ESCs transfected with GFP siRNA and N-myc siRNA. Error bars represent S.D. between biological replicates, n = 3 (***, p < 0.001, two-tailed t test). F, Western blot for Dnmt3a2 in WT cells transfected with GFP siRNA and N-myc siRNA. G, model depicting a pathway that leads from PI3K activation to changes in DNA methylation in the nucleus.
In silico analysis of the Dnmt3a2 promoter revealed a region of highly conserved sequence just proximal to the exon unique to Dnmt3a2, which encodes the 5′-UTR. Within this conserved region is a putative Myc binding site. Our microarray data showed a 2.8-fold decrease in N-Myc levels in Gsk-3 DKO ESCs. Real-time qPCR revealed an 8.8-fold decrease in N-myc mRNA (Fig. 6C), and Western blot analysis confirmed the corresponding decrease in N-Myc protein levels (Fig. 6D). To test the importance of the Myc binding site in the Dnmt3a2 promoter, we performed site-directed mutagenesis on the putative Myc binding site in the context of the 1.9-kb promoter fragment and transfected this reporter into ESCs. We observed a complete loss of reporter activity, confirming the functional importance of this binding site (Fig. 6B). Finally, transfection of WT ESCs with N-myc siRNA resulted in a 77% reduction in N-Myc levels (Fig. 6E) and a decrease in Dnmt3a2 protein (Fig. 6F), confirming the importance of N-Myc on the regulation of Dnmt3a2 in ESCs.
DISCUSSION
Mechanisms to explain how extracellular environmental signals are transduced to the nucleus to affect epigenetic modifications, such as DNA methylation, have been elusive (44). Here, we provide evidence for a novel role for Gsk-3 in the epigenetic regulation of imprinted genes. In mouse ESCs, deletion of Gsk-3 isoforms or expression of myristoylated PI3-kinase both lead to constitutive activation of downstream components of the insulin pathway, ultimately resulting in attenuated DNA methylation of imprinted loci. Although we provide evidence that several imprinted genes are hypomethylated as a consequence of the loss of Gsk-3 activity, it remains to be determined whether DNA methylation is reduced at all known imprinted loci in Gsk-3 DKO ESCs or whether the effect on DNA methylation extends to non-imprinted genes.
Interestingly, we observed that ∼90% of CpG dinucleotides analyzed in the Igf2/H19 DMD were methylated in WT ESCs. It is expected that the number of methylated CpG dinucleotides would be closer to 50% for this imprinted locus because we did not discriminate between alleles in our bisulfite sequencing assay. The high amount of DNA methylation observed in WT ESCs is not likely due to PCR bias in our bisulfite sequencing procedure as we observed 44% methylation of the Igf2/H19 DMD in NSC derived from WT mice using the same bisulfite sequencing assay (supplemental Fig. 1). It has been reported that cultured ESC lines may exhibit almost complete methylation of the Igf2/H19 DMD on both parental alleles (33). In light of these previously reported observations, it is likely that the high degree of DNA methylation we detect at the Igf2/H19 DMD in WT ESCs is due to biallelic methylation.
Our data show an increase in N-terminal phosphorylation of Ser-9/Ser-21 on Gsk-3 isoforms upon ectopic expression of PI3K, resulting in decreased DNA methylation at the Igf2/H19 imprinting control region. Although our experimental approach utilized constitutively active PI3-kinase as a means of inducing phosphorylation on Gsk-3α Ser-21/Gsk-3β Ser-9, signal transduction of several pathways, such as protein kinase A (PKA) (45), also result in Gsk-3α Ser-21/Gsk-3β Ser-9 phosphorylation. Therefore, our data predict that signaling pathways that lead to the phosphorylation and inhibition of Gsk-3 akin to insulin signaling would lead to decreased DNA methylation at imprinted loci. We have not ruled out the possibility that inactivation of Gsk-3 by Wnt signaling can lead to hypomethylation of Igf2/H19. Further investigation is needed to determine whether activation of the Wnt pathway has the same effect on Dnmt3a2 expression.
The deficits we observe in DNA methylation are due to reduced levels of the de novo DNA methyltransferase Dnmt3a2. Dnmt3a2 co-purifies with Dnmt3L in ES cells, whereas Dnmt3a does not (46, 47). Although Dnmt3L is catalytically inactive, it is required for establishment of DNA methylation at imprinted loci (48, 49) and recruits Dnmt3a2 to DNA sequences associated with unmethylated lysine 4 of histone H3 (47). Dnmt3L levels were not significantly changed in Gsk-3 DKO ESCs (decreased 1.3-fold). These findings suggest that Dnmt3a2 plays a prominent role in the maintenance of DNA methylation at imprinted genes in early development. Heretofore, understanding the specific function of Dnmt3a2 in vivo has been difficult because targeted disruptions of the Dnmt3a locus in the mouse result in the simultaneous deletion of both Dnmt3a and Dnmt3a2 as the isoforms share common 3′ exons. Our identification of a signal transduction pathway capable of specifically reducing levels of Dnmt3a2 provides a new approach for the study of this isoform in vivo.
To better understand the mechanism by which Dnmt3a2 mRNA is regulated, we cloned and analyzed a 1.9-kb fragment of mouse genomic DNA that contained a previously characterized promoter region. Computational analysis of the promoter revealed very little conservation of sequence across different species, with the exception of an ∼200-bp region just upstream of the novel Dnmt3a2 exon. Within this region, we found a putative Myc binding site that is highly conserved, and a point mutation in this site completely impaired reporter activation. Recent chromatin immunoprecipitation-sequencing (ChIP-Seq) data showed that both c-Myc and N-Myc share an identical DNA binding motif (50); however, we did not observe a reduction in c-Myc protein in Gsk-3 DKO ESCs. Furthermore, ChIP-Seq data in mouse ESCs show that N-Myc, but not c-Myc, binds specifically to the Dnmt3a2 promoter (50). We have experimentally verified the importance of N-Myc on regulating Dnmt3a2 expression by knocking down N-myc via siRNA. Taken together with the decreased levels of N-Myc and unchanged levels of c-Myc (supplemental Fig. 2, A and B), we conclude that N-Myc regulates Dnmt3a2 transcription. Gsk-3-mediated phosphorylation and degradation of c-Myc and N-Myc proteins by Gsk-3 have been reported in COS-7 cells (c-Myc) and cerebellar granule neuron precursors (N-Myc) (42, 43, 51). We did not observe this regulation of Myc proteins in our ESCs. In fact, we see an opposite effect of Gsk-3 on N-Myc expression and no effect on c-Myc expression. Our data likely reflect cell type-specific differences in the regulation of Myc proteins by Gsk-3. Furthermore, our real-time qPCR data indicate that N-Myc down-regulation in Gsk-3 DKO ESCs is occurring at the level of transcriptional regulation. The mechanism by which Gsk-3 regulates N-myc transcription in ESCs remains to be determined.
The ability of the Gsk-3 inhibitor lithium to phenocopy the reduction in DNA methylation of imprinted genes seen in Gsk-3 DKO ESCs is particularly intriguing. We have shown that DNA methylation is reduced at the Igf2/H19 imprinting control region in both ESCs and NSCs after prolonged exposure to lithium. The effectiveness of lithium in the treatment of bipolar disorder was initially reported more than 60 years ago (52). Lithium has been widely prescribed ever since, although the therapeutic mechanism of action has not been fully defined. Growing evidence suggests that alterations in DNA methylation have a role in the development of bipolar disorder (53, 54). Our data raise the possibility that reduced DNA methylation of imprinted genes may be related to the mechanism of lithium action in bipolar disorder. It is noteworthy that another drug widely used for the treatment of bipolar disorder, valproic acid, also alters the epigenome by inhibiting histone deacetylase activity (55, 56). In the context of our findings, it is tempting to speculate that lithium and valproic acid are effective in treating bipolar disorder because they modify the expression of a common subset of genes via changes in DNA methylation and histone acetylation, respectively.
Our findings may also provide a new focus for the study of neurological diseases. Gsk-3 activity has been linked to schizophrenia (57), whereas alterations in the epigenome have been proposed to be a major factor in the development of schizophrenia (53). Of the postnatal tissues that have been examined to date, imprinting is clearly predominant in the brain, and alterations in these imprinting events have been shown to affect behavior (58). One example in humans is Prader-Willi syndrome, which is caused by a deletion on chromosome 15q11–13, an area that contains the imprinted genes SNRPN and Necdin (NDN) (59). This raises the possibility that altered Gsk-3 activity, resulting in aberrant DNA methylation of imprinted genes, could contribute to the development of schizophrenia or other behavioral disorders.
In summary, we have discovered a novel role for Gsk-3 in maintaining DNA methylation of imprinted loci in mouse ESCs. Because Gsk-3 is a nexus for numerous signal transduction pathways, the potential for Gsk-3 to regulate epigenetic changes could have profound consequences for our understanding of diverse human diseases.
Supplementary Material
Acknowledgments
We thank Drs. Jeff Kuret, Jing Yang, and Scott Harper for reading the manuscript and helpful discussions. We are also grateful to Dr. Glenn Maston for the pGLF construct, Dr. Carlos Miranda for assistance in isolating neural stem cells, and Dr. Yulei Wang for assistance with c-Myc qPCR.
This work was supported, in whole or in part, by National Institutes of Health Grants P30 DK078392 and U54RR025216 (to B. A.), R01CA63517 (to J. G.), and R01AG031833 (to C. J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- Gsk-3
- glycogen synthase kinase-3
- Dnmt1
- DNA methyltransferase 1
- Dnmt3a
- DNA methyltransferase 3a
- Dnmt3a2
- DNA methyltransferase 3a, isoform 2
- ESC
- embryonic stem cell
- DMD
- differentially methylated domain
- DMR2
- differentially methylated region 2
- IAP
- intracisternal A-particle
- Igf2
- insulin-like growth factor II
- Igf2r
- insulin-like growth factor II receptor
- NSC
- neural stem cells
- qPCR
- quantitative polymerase chain reaction
- DKO
- double knock-out
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Force T., Woodgett J. R. (2009) J. Biol. Chem. 284, 9643–9647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kockeritz L., Doble B., Patel S., Woodgett J. R. (2006) Curr. Drug. Targets 7, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 3.Doble B. W., Patel S., Wood G. A., Kockeritz L. K., Woodgett J. R. (2007) Dev. Cell 12, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim W. Y., Wang X., Wu Y., Doble B. W., Patel S., Woodgett J. R., Snider W. D. (2009) Nat. Neurosci. 12, 1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K., Mitsui K., Yamanaka S. (2003) Nature 423, 541–545 [DOI] [PubMed] [Google Scholar]
- 6.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004) Nat. Med. 10, 55–63 [DOI] [PubMed] [Google Scholar]
- 7.Paling N. R., Wheadon H., Bone H. K., Welham M. J. (2004) J. Biol. Chem. 279, 48063–48070 [DOI] [PubMed] [Google Scholar]
- 8.Watanabe S., Umehara H., Murayama K., Okabe M., Kimura T., Nakano T. (2006) Oncogene 25, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 9.Lu J., Hou R., Booth C. J., Yang S. H., Snyder M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5688–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storm M. P., Bone H. K., Beck C. G., Bourillot P. Y., Schreiber V., Damiano T., Nelson A., Savatier P., Welham M. J. (2007) J. Biol. Chem. 282, 6265–6273 [DOI] [PubMed] [Google Scholar]
- 11.Miyabayashi T., Teo J. L., Yamamoto M., McMillan M., Nguyen C., Kahn M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5668–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niwa H., Ogawa K., Shimosato D., Adachi K. (2009) Nature 460, 118–122 [DOI] [PubMed] [Google Scholar]
- 14.Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 15.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 16.MacDonald B. T., Tamai K., He X. (2009) Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X., Tamai K., Doble B., Li S., Huang H., Habas R., Okamura H., Woodgett J., He X. (2005) Nature 438, 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding V. W., Chen R. H., McCormick F. (2000) J. Biol. Chem. 275, 32475–32481 [DOI] [PubMed] [Google Scholar]
- 19.Ng S. S., Mahmoudi T., Danenberg E., Bejaoui I., de Lau W., Korswagen H. C., Schutte M., Clevers H. (2009) J. Biol. Chem. 284, 35308–35313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong K. B., Maksakova I. A., Mohn F., Leung D., Appanah R., Lee S., Yang H. W., Lam L. L., Mager D. L., Schübeler D., Tachibana M., Shinkai Y., Lorincz M. C. (2008) EMBO J. 27, 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budowle B., Baechtel F. S. (1990) Appl. Theor. Electrophor. 1, 181–187 [PubMed] [Google Scholar]
- 22.Yamasaki Y., Kayashima T., Soejima H., Kinoshita A., Yoshiura K., Matsumoto N., Ohta T., Urano T., Masuzaki H., Ishimaru T., Mukai T., Niikawa N., Kishino T. (2005) Hum. Mol. Genet. 14, 2511–2520 [DOI] [PubMed] [Google Scholar]
- 23.Okano M., Xie S., Li E. (1998) Nat. Genet. 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 24.Chen T., Ueda Y., Xie S., Li E. (2002) J. Biol. Chem. 277, 38746–38754 [DOI] [PubMed] [Google Scholar]
- 25.Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004) Nature 429, 900–903 [DOI] [PubMed] [Google Scholar]
- 26.Chen T., Ueda Y., Dodge J. E., Wang Z., Li E. (2003) Mol. Cell. Biol. 23, 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeChiara T. M., Robertson E. J., Efstratiadis A. (1991) Cell 64, 849–859 [DOI] [PubMed] [Google Scholar]
- 28.Bartolomei M. S., Webber A. L., Brunkow M. E., Tilghman S. M. (1993) Genes Dev. 7, 1663–1673 [DOI] [PubMed] [Google Scholar]
- 29.Tremblay K. D., Duran K. L., Bartolomei M. S. (1997) Mol. Cell. Biol. 17, 4322–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ideraabdullah F. Y., Vigneau S., Bartolomei M. S. (2008) Mutat. Res. 647, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinno Y., Sengoku K., Nakao M., Tamate K., Miyamoto T., Matsuzaka T., Sutcliffe J. S., Anan T., Takuma N., Nishiwaki K., Ikeda Y., Ishimaru T., Ishikawa M., Niikawa N. (1996) Hum. Mol. Genet. 5, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 32.Thorvaldsen J. L., Duran K. L., Bartolomei M. S. (1998) Genes Dev. 12, 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean W., Bowden L., Aitchison A., Klose J., Moore T., Meneses J. J., Reik W., Feil R. (1998) Development 125, 2273–2282 [DOI] [PubMed] [Google Scholar]
- 34.Lyle R., Watanabe D., te Vruchte D., Lerchner W., Smrzka O. W., Wutz A., Schageman J., Hahner L., Davies C., Barlow D. P. (2000) Nat. Genet. 25, 19–21 [DOI] [PubMed] [Google Scholar]
- 35.Stöger R., Kubicka P., Liu C. G., Kafri T., Razin A., Cedar H., Barlow D. P. (1993) Cell 73, 61–71 [DOI] [PubMed] [Google Scholar]
- 36.Li E., Bestor T. H., Jaenisch R. (1992) Cell 69, 915–926 [DOI] [PubMed] [Google Scholar]
- 37.Howlett S. K., Reik W. (1991) Development 113, 119–127 [DOI] [PubMed] [Google Scholar]
- 38.Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 39.Lei H., Oh S. P., Okano M., Jüttermann R., Goss K. A., Jaenisch R., Li E. (1996) Development 122, 3195–3205 [DOI] [PubMed] [Google Scholar]
- 40.Klein P. S., Melton D. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doble B. W., Woodgett J. R. (2003) J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory M. A., Qi Y., Hann S. R. (2003) J. Biol. Chem. 278, 51606–51612 [DOI] [PubMed] [Google Scholar]
- 43.Sjostrom S. K., Finn G., Hahn W. C., Rowitch D. H., Kenney A. M. (2005) Dev. Cell 9, 327–338 [DOI] [PubMed] [Google Scholar]
- 44.Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. (2009) Genes Dev. 23, 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M., Wang X., Meintzer M. K., Laessig T., Birnbaum M. J., Heidenreich K. A. (2000) Mol. Cell. Biol. 20, 9356–9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nimura K., Ishida C., Koriyama H., Hata K., Yamanaka S., Li E., Ura K., Kaneda Y. (2006) Genes Cells 11, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 47.Ooi S. K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S. P., Allis C. D., Cheng X., Bestor T. H. (2007) Nature 448, 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001) Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 49.Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. (2007) Hum. Mol. Genet. 16, 2272–2280 [DOI] [PubMed] [Google Scholar]
- 50.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 51.Kenney A. M., Cole M. D., Rowitch D. H. (2003) Development 130, 15–28 [DOI] [PubMed] [Google Scholar]
- 52.Cade J. F. (1949) Med. J. Aust 2, 349–352 [DOI] [PubMed] [Google Scholar]
- 53.Mill J., Tang T., Kaminsky Z., Khare T., Yazdanpanah S., Bouchard L., Jia P., Assadzadeh A., Flanagan J., Schumacher A., Wang S. C., Petronis A. (2008) Am. J. Hum. Genet. 82, 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuratomi G., Iwamoto K., Bundo M., Kusumi I., Kato N., Iwata N., Ozaki N., Kato T. (2008) Mol. Psychiatry 13, 429–441 [DOI] [PubMed] [Google Scholar]
- 55.Phiel C. J., Zhang F., Huang E. Y., Guenther M. G., Lazar M. A., Klein P. S. (2001) J. Biol. Chem. 276, 36734–36741 [DOI] [PubMed] [Google Scholar]
- 56.Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., Sleeman J. P., Lo Coco F., Nervi C., Pelicci P. G., Heinzel T. (2001) EMBO J. 20, 6969–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao Y., Ge X., Frank C. L., Madison J. M., Koehler A. N., Doud M. K., Tassa C., Berry E. M., Soda T., Singh K. K., Biechele T., Petryshen T. L., Moon R. T., Haggarty S. J., Tsai L. H. (2009) Cell 136, 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson L. S., Davies W., Isles A. R. (2007) Nat. Rev. Neurosci. 8, 832–843 [DOI] [PubMed] [Google Scholar]
- 59.Horsthemke B., Wagstaff J. (2008) Am. J. Med. Genet. A. 146A, 2041–52 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.