Abstract
Endotoxin is a potent inducer of systemic inflammatory responses in human and rodents. Here, we show that in vivo endotoxin triggers a rapid and transient decline in ATP concentration in human peripheral blood leukocytes and murine peripheral blood leukocytes and liver, which is associated with a brief increase in expression of the autophagy indicator LC3-II. In both of these tissues, the ATP concentration reaches a nadir, and autophagy is induced between 2 and 4 h post-endotoxin infusion, and homeostasis is restored within 12 h. Mouse liver SIRT1 and AMP-activated protein kinase (AMPK) protein expression levels decline precipitously within 10 min and remain below detection levels for up to 12 h post-endotoxin administration. In marked contrast, the expression of HIF-1α is induced within 90 min and remains elevated for up to 12 h. The ATP recovery is delayed, and the increases in both HIF-1α expression and autophagy are prolonged in endotoxin-challenged SIRT1 liver knock-out mice. Resveratrol prevents the decline in ATP concentration and SIRT1 expression, as well as the increase in HIF-1α expression and autophagy in liver of endotoxin-challenged wild type mice but not in SIRT1 liver knock-out mice. These results provide novel insight into the state of both cellular bioenergetics and metabolic networks during the acute phase of systemic inflammation and suggest a role for SIRT1 in acute metabolic decline, as well as the restoration of metabolic homeostasis during an inflammatory challenge.
Keywords: Enzyme Kinetics, Enzyme Mutation, Enzymes, Heme, Metals, Porphyrin, Protein Metal Ion Interaction, Ferrochelatase, Tetrapyrrole
Introduction
Cells utilize multiple pathways to respond to metabolic needs that arise as a result of rapid changes in nutritional and environmental conditions. Several proteins that include silent information regulator T1 (SIRT1), AMP-activated protein kinase (AMPK),2 and hypoxia-inducible transcription factor-1 (HIF-1) play key roles in the regulation of metabolic pathways. SIRT1 is a NAD+-dependent sirtuin family member that catalyzes the deacetylation of proteins implicated in the regulation of multiple cellular processes, which include metabolism (1, 2). Among the abundant data that support its metabolic function (3, 4), the observations that SIRT1-null mice are metabolically inefficient as compared with normal mice, and that their liver mitochondria have a lower capacity to produce ATP, are particularly significant (5). SIRT1 regulates metabolic pathways through interactions with multiple transcription factors, which include peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), peroxisome proliferator-activated receptor γ, forkhead box, subgroup O (FOXO) family members, and p53 (6–9). Of these, PGC-1α is a predominant regulator of mitochondrial biogenesis as well as mitochondrial substrate usage (10–12). In fasting skeletal muscle, SIRT1-dependent deacetylation of PGC-1α promotes the shift in mitochondrial substrate utilization from glucose to fatty acid oxidation (13), whereas in fasting liver, it triggers transcription of gluconeogenic genes and an increase in hepatic glucose output (10). By acting through PGC-1α, SIRT1 also increases oxygen consumption and induces mitochondrial oxidative phosphorylation genes, as well as mitochondrial biogenesis in muscle (14). SIRT1 is one of the best characterized targets of resveratrol, a small polyphenolic compound found in red wine that has multiple physiologic effects, including the capacity to enhance mitochondrial activity and cellular metabolism (14, 15).
The Ser/Thr kinase AMPK, which is activated when the cellular ATP concentration is low, acts in parallel to SIRT1. Like SIRT1, AMPK also up-regulates catabolic pathways that include glycolysis, mitochondrial respiration, and mitochondrial biogenesis, while switching off energy-consuming anabolic processes such as lipid and protein synthesis (16). Studies revealed that SIRT1 and AMPK act on several common target genes that include PGC-1α and FOXO (3, 17, 18). In addition, there is evidence that AMPK modulates the activity of SIRT1 through the regulation of NAD+ availability (19–21) and that SIRT1 activates AMPK in several cell types that include HepG2 hepatocytes (22). Although the hierarchical functional relationship between SIRT1 and AMPK remains uncertain, these collective observations explain at least in part the apparent overlap between SIRT1- and AMPK-dependent metabolic outputs.
Under normoxic conditions, mitochondria are the primary site of cellular ATP production in cells that convert nutrients into energy through oxidative phosphorylation. Therefore, when shifted to conditions of low oxygen tension, or hypoxia, such cells might experience a decrease in mitochondrial function leading to a decline in ATP synthesis. However, hypoxic cells are able to maintain metabolic homeostasis through an increase in glycolysis, which is mediated by HIF-1 (23, 24). HIF-1 up-regulates the expression of glycolytic enzymes such as 6-phosphofructo-2-kinase, glucose transporters, as well as the expression of pyruvate dehydrogenase kinase, which inhibits pyruvate dehydrogenase and, consequently, the entry of pyruvate into the TCA cycle (24–26). As a result of these combined activities, HIF-1 enhances glycolysis while limiting mitochondrial function and the generation of reactive oxygen species (25, 26).
HIF-1 is a heterodimeric complex composed of an oxygen-regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit (27, 28). HIF-1α is degraded rapidly by the ubiquitin proteasome pathway under normoxic conditions but is stabilized in cells under hypoxia (29–33). Studies have demonstrated that endotoxin (lipopolysaccharide (LPS)), a well characterized inducer of systemic inflammatory responses and a toll-like receptor 4 (TLR4) agonist, triggers an increase in HIF-1α expression in myeloid cells and hepatocytes under both hypoxic and normoxic conditions (34–36). How the expressions of HIF-1α, AMPK, and SIRT1 are all balanced in cells during periods of systemic inflammation is currently undetermined.
SIRT1 (37), AMPK (38), and more recently HIF-1 (39, 40) are all implicated in the regulation of autophagy, a ubiquitous cell survival mechanism used by cells to compartmentalize and digest long lived and/or damaged proteins and organelles (41, 42). A family of autophagy-related (Atg) proteins, which include Atg 8/LC3, regulates the formation of specialized vesicles, known as autophagosomes. The autophagosomes shuttle their engulfed protein and organelle cargoes to lysosomes for degradation (43). During periods of nutrient deficiency, many cell types degrade cellular macromolecules and produce amino acids through autophagy to support their metabolic needs (44). Although it is known that endotoxin, as well as other toll-like receptors agonists, can trigger autophagy in immune cells and that this process is central to immune cell function (45, 46), the relationship between autophagy and cellular metabolism in cells that are exposed to an inflammatory challenge is currently undefined.
Severe inflammatory conditions resulting from sepsis, injury, or an endotoxin challenge trigger a decline in mitochondrial bioenergetics in both human and animal tissues (47–49). A decline in mitochondrial oxidative phosphorylation gene expression is similarly observed in human PBL from mice challenged with endotoxin in vivo (50). In this study, we begin to explore the mechanism through which in vivo endotoxin alters the cellular bioenergetics in two types of TLR4-expressing cell populations, PBL and liver. We show that the cellular ATP levels, autophagy, as well as HIF-1α, SIRT1, and AMPK protein expression levels are all coordinately dysregulated in human and mice leukocytes and in mice liver as early as 1–2 h after an in vivo endotoxin challenge. We also report that resveratrol suppresses the metabolic perturbations observed in endotoxin-challenged murine PBL and liver through a SIRT1-dependent mechanism. Finally, we show that the metabolic perturbations triggered by endotoxin are prolonged in the liver of liver-specific SIRT1-knock-out (L-KO) mice as compared with littermate controls. These findings shed new light on the state of SIRT1, AMPK, and HIF-1α expression and their potential roles during a critical period of systemic inflammation.
EXPERIMENTAL PROCEDURES
Human Subjects
Male and female subjects (age 18–29 years) were recruited by public advertisement to participate in a study approved by the Institutional Review Board of the Robert Wood Johnson Medical School after obtaining informed written consent. Subjects were administered a standard dose of endotoxin (2 ng/kg, clinical center reference endotoxin (CC-RE), lot 2, National Institutes of Health, Bethesda) (50). At the indicated time points of pre- and post-endotoxin infusion, blood was drawn into EDTA-containing tubes, and lysis buffer (bicarbonate-buffered ammonium chloride solution, 0.826% NH4Cl, 0.1% KHCO3, 0.0037% Na4EDTA in H2O) was added at a ratio of 20:1 (lysis buffer/blood). Once the erythrocytes lysed (∼5–7 min of incubation), the samples were centrifuged for 10 min at 400 × g. The leukocyte pellet was washed once with phosphate-buffered saline (PBS). The pellet was resuspended in PBS and divided into two fractions. One fraction (∼1/10 of the sample) was used for ATP analysis. The remaining sample was used for Western blot analysis.
Animals Studies
The animal studies were approved by the Institutional Animals Care and Use Committee of Robert Wood Johnson Medical School. Normal C57/BL6 mice were obtained from The Jackson Laboratory. The liver-specific SIRT1-null transgenic mice were described previously (51, 52). All transgenic mice studies were conducted using littermate animals homozygous for a floxed SIRT1 allele (SIRT1Δex4/Δex4), which either expressed Cre (liver-specific SIRT1 knock-out) or did not express Cre (designated wild type (WT)) (51, 52). Animals were challenged with a bolus dose of endotoxin (i.p. injection, 3 mg/kg in 300 μl of saline) or saline (300 μl). Where indicated, animals were given a bolus injection of resveratrol (Sigma) (i.p., 20 mg/kg in 5% ethanol (53)) or vehicle (5% ethanol) for 7 days. On day 8, the animals were challenged with endotoxin or saline as described above. The mice were sacrificed by CO2 inhalation. Blood was obtained by heart puncture, and leukocytes were isolated using the protocol outlined above. Liver segments were used for quantitative real time PCR, ATP, and Western blot analysis.
ATP Assays
Protein concentration of cells and tissues was determined using micro BCA reagents (Pierce). Cellular and tissue ATP levels were measured using the ATP bioluminescence assay kit (Roche Applied Science) as per the manufacturer's protocol. Briefly, cells and tissues were collected and resuspended in 500 μl of ATP assay dilution buffer (100 mm Tris, pH 7.75, and 4 mm EDTA). The samples were boiled for 2 min and spun for 5 min at 1000 × g. Supernatants were collected, and 50-μl samples were analyzed using a luminometer (PerkinElmer Life Sciences).
Western Blot Analysis
Leukocytes and liver tissue were lysed in RIPA buffer (1% Triton X-100, 1% deoxycholic acid, 10 mm Tris-HCl, pH 7.2, 158 mm NaCl, 0.1% SDS, and 1 mm PMSF and complete protease inhibitor mixture (Roche Applied Science)). Lysates containing equal protein amounts were subjected to Western blot analysis and probing with antibodies to LC3 (Sigma, L7543-200UL), SIRT1 (Santa Cruz Biotechnology, sc-15404), α-actinin (54), AMPK (Cell Signaling, 2532), HIF-1α (Santa Cruz Biotechnology, SC-10790), p62/SQSTM1 (Abgent, AP2183B), tubulin (Sigma, T6557), and actin (Sigma, A2066).
RNA Extraction and Gene Expression Analysis
Human or mouse blood was transferred to and stored in PAXgene tubes (Qiagen). Total RNA was extracted as per PAXgene blood RNA kit protocol (Qiagen). Mouse liver tissue was collected and stored at −70 °C in RNAlater (Qiagen). The tissue was homogenized using mortar and pestle, and total RNA was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Total RNA was quantified using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and reversed-transcribed to cDNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Gene expression was analyzed in duplicate by quantitative real time PCR as described previously (55). Data are expressed as fold change relative to time 0. SIRT1 (Mm00437764_m1) and B2M (Mm01168521_m1) gene expression assays were purchased from Applied Biosystems.
Statistics
Analysis was by one-way ANOVA with Newman-Keuls post test. The operations were carried out using Prism 4 software version 4.0b (GraphPad Software, Inc., La Jolla, CA). p values less than 0.05 were considered to be statistically significant.
RESULTS
In Vivo Endotoxin Triggers a Transient Decrease in Cellular ATP Concentration and a Parallel Increase in Autophagy in Peripheral Blood Leukocytes and Liver Tissue
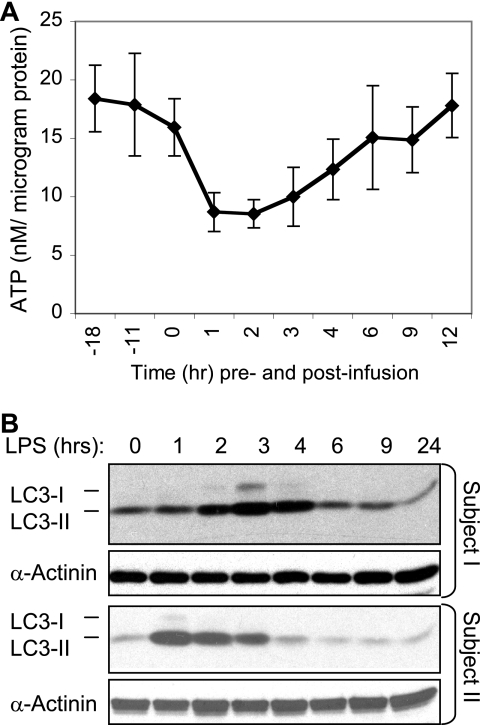
Endotoxin triggers transient changes in expression of genes associated with the mitochondrial oxidative phosphorylation activity in human PBL (50). To determine whether these changes in mitochondrial gene expression reflect changes in cellular bioenergetics, we examined the ATP concentration in PBL obtained from human subjects at multiple time points before and after an in vivo endotoxin challenge. The in vivo endotoxin challenge triggered an abrupt decrease in ATP concentration within 1 h (Fig. 1A). By 1–2 h post-endotoxin infusion, the ATP concentration decreased by ∼50% relative to base line. The ATP concentration began to recover after 4–6 h, with a full recovery by 12 h post-endotoxin administration.
FIGURE 1.
Endotoxin triggers a transient decline in ATP levels and a parallel increase in LC3-II expression in human PBL. A and B, blood was drawn from healthy volunteer donors (n = 4) at the indicated time before and after endotoxin (2 ng/kg body weight) infusion. Leukocytes were isolated and lysed, and samples containing equal protein amounts were used for ATP assays (A) (data are expressed as mean ± S.E.) and Western blotting and probing (B) with the indicated antibodies. Blotting for α-actinin was used as a loading control. Data are shown for two out of four subjects studied.
During periods of nutrient deficiency, cells can generate energy through autophagic degradation of cellular organelles and proteins (44). Hence, we sought to determine whether the transient decline in ATP concentration triggered by endotoxin was sufficiently robust so as to trigger an increase in expression of the autophagy indicator LC3-II (56, 57). LC3 (Atg8) is a 18-kDa cytosolic protein that is involved with autophagosome membrane formation. In the course of its recruitment to these membranes, LC3 is subjected to two post-translational modification steps, which yield the 16-kDa form, referred to as LC3-II (56). As shown (Fig. 1B), endotoxin triggered an increase in LC3-II expression within 2 h with a return to base-line expression levels between 4 and 6 h (Fig. 1B).
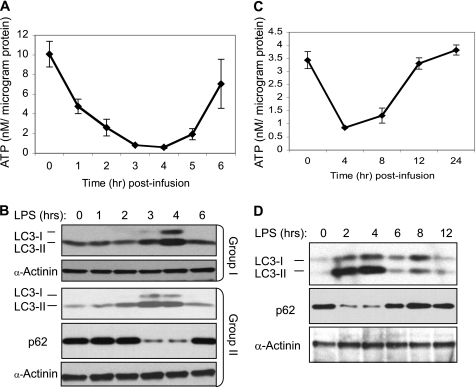
We then examined whether the metabolic changes observed in human PBL challenged with endotoxin in vivo are reproduced in mice. Indeed, a decrease in ATP was detected in murine PBL within 1 h, with a nadir between 3 and 4 h post-endotoxin infusion (Fig. 2A). Furthermore, consistent with the results observed in human PBL, LC3-II expression was increased (Fig. 2B) while the ATP concentration was at its lowest point (Fig. 2A). An indistinguishable transient decline in ATP concentration, accompanied by a parallel increase in LC3-II expression, was also observed in mice liver (Fig. 2, C and D). To further confirm that the transient decline in ATP was associated with an increase in autophagic flux, we examined the expression of p62/SQTM1, an LC3-binding protein that is degraded through autophagy (57, 58). Indeed, as shown in Fig. 2, B and D, the expression of p62/SQTM1 was significantly reduced in PBL and liver while the expression of LC3-II was at its peak. The parallel and inversely related changes in ATP levels and autophagy suggest that the two events are closely coordinated in multiple tissues during periods of systemic inflammation.
FIGURE 2.
Endotoxin triggers a transient decline in ATP levels and a parallel increase in LC3-II expression in murine PBL and liver. Blood (A and B) and liver tissue (C and D) were obtained from endotoxin (3 mg/kg body weight)-challenged mice (n = 4–6 per time point) at the indicated times post-infusion. Peripheral blood leukocytes and liver tissue samples containing equal protein amounts were used for ATP assays (A and C) (data are expressed as mean ± S.E.) and Western blotting and probing (B–D) with the indicated antibodies. Each lane represents a sample obtained from a single mouse. Blotting for α-actinin was used as a loading control. The samples marked as Group I and Group II in B are from two independent experiments, demonstrating reproducibility.
In Vivo Endotoxin Down-regulates SIRT1 and AMPK Expression and Up-regulates HIF-1α Expression in Peripheral Blood Leukocytes and Liver Tissue
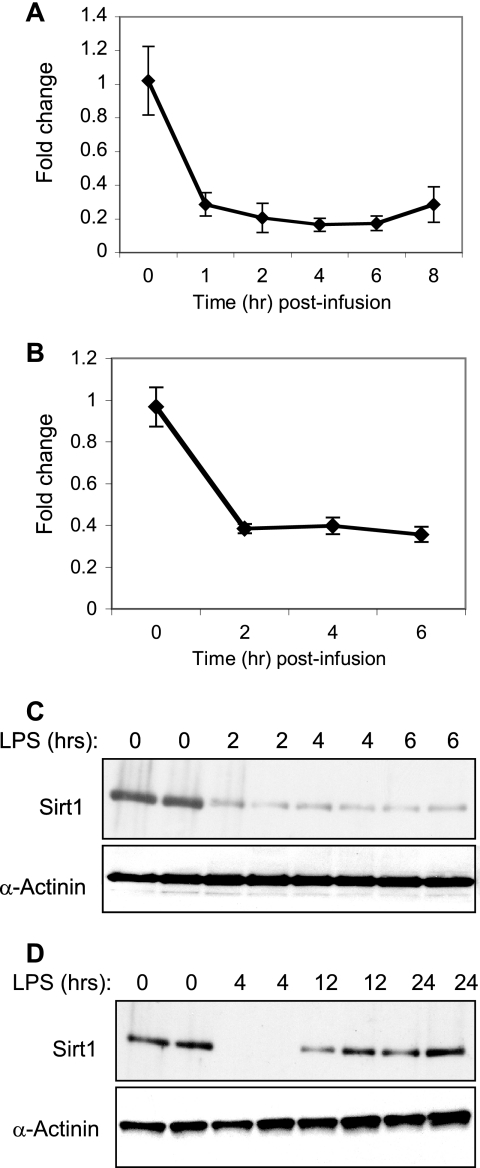
We next sought to determine whether the in vivo endotoxin challenge altered SIRT1, AMPK, or HIF-1α expression in mice PBL and liver. Using quantitative real time PCR, we found that endotoxin triggered a significant decline in SIRT1 gene expression levels in mice PBL (Fig. 3A) and liver (Fig. 3B). Furthermore, the in vivo endotoxin challenge also triggered an abrupt decline in liver SIRT1 protein expression level within 2 h (Fig. 3C). SIRT1 expression remained low for at least 6 h and recovered by 12 h post-endotoxin infusion (Fig. 3D).
FIGURE 3.
Endotoxin alters SIRT1 transcript and protein expression levels in murine PBL and liver tissue. Peripheral blood leukocytes (A) and liver tissue (B–D) were obtained at the indicated time post-endotoxin infusion. A and B, SIRT1 transcript level was determined by quantitative PCR. Fold changes in mRNA levels are expressed as mean ± S.E. relative to the 0 time point (n = 4). C and D, liver tissue samples normalized for protein content were subjected to Western blot analysis. Blotting for α-actinin was used as a loading control. Each lane represents a sample obtained from a single mouse. The data shown in C and D are from two independent experiments.
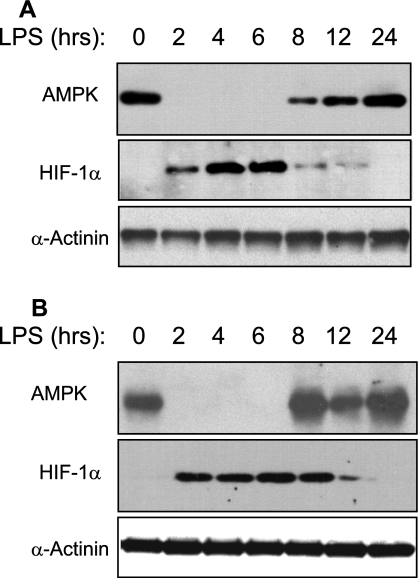
A decrease in ATP concentration is expected to trigger an increase in AMPK activity. Nonetheless, as observed for SIRT1 expression, AMPK expression was also transiently down-regulated in both PBL (Fig. 4A) and liver (Fig. 4B) of endotoxin-challenged mice. The abrupt decline in AMPK expression was seen within 2 h. AMPK expression remained significantly suppressed for at least 6 h but was fully recovery by 8–12 h post-endotoxin infusion (Fig. 4). In marked contrast with the fate of SIRT1 and AMPK expression, HIF-1α expression was up-regulated within 2 h in both mice PBL (Fig. 4A) and liver (Fig. 4B) and remained elevated for up to 8–12 h post-infusion. Together, these findings indicate that the expression of HIF-1α and SIRT1/AMPK is inversely regulated during periods of systemic inflammation.
FIGURE 4.
Endotoxin alters AMPK and HIF-1α expression in murine peripheral blood leukocytes and liver tissue. Peripheral blood leukocytes (A) and liver tissue (B) were obtained at the indicated time post-endotoxin infusion. Samples normalized for protein content were subjected to Western blot analysis. Blotting for α-actinin was used as a loading control. Each lane represents a sample obtained from a single mouse.
Resveratrol Prevents the Decrease in ATP Concentration and the Increase in LC3-II and HIF-1α Expression in Liver of Endotoxin-challenged Mice
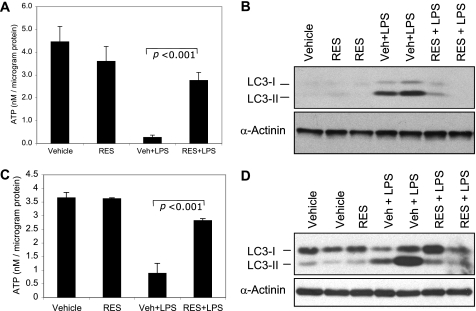
Studies have shown that resveratrol can improve mitochondrial function and cellular bioenergetics (14, 15). Hence, we sought to determine whether resveratrol could modify the bioenergetic consequences observed in endotoxin-challenged mice. To this end, mice were administered resveratrol or vehicle for 7 days as described previously (53). On day 8, the mice were challenged with endotoxin and were sacrificed 4 h later. The 4-h time point was chosen based on the results presented in Figs. 3 and 4. In contrast with the significant decrease in ATP concentration noted in PBL (Fig. 5A) and liver (Fig. 5C) of vehicle-pretreated mice, no significant changes in ATP concentration were observed in resveratrol-pretreated mice. Furthermore, the increases in LC3-II expression observed in PBL (Fig. 5B) and liver tissue (Fig. 5D) of vehicle-pretreated mice were not observed in tissues of resveratrol-pretreated mice.
FIGURE 5.
Resveratrol prevents the decline in ATP concentration and the increase in LC3-II expression in murine PBL and liver tissue when administered prior to the endotoxin challenge. Mice were given an intraperitoneal injection of vehicle (Veh) or resveratrol (RES) (20 mg/kg body weight) for 7 days. On day 8, the mice were challenged with saline or a bolus dose of endotoxin (LPS). Peripheral blood leukocytes (A and B) and liver tissue (C and D) were obtained at 4 h post-endotoxin infusion. The samples were used for ATP assays (A and C) and Western blot analysis (B and D) as described in the legend to Fig. 2. The data shown in A and C are expressed as means ± S.E. (n = 4). Statistical significance was determined by one-way ANOVA.
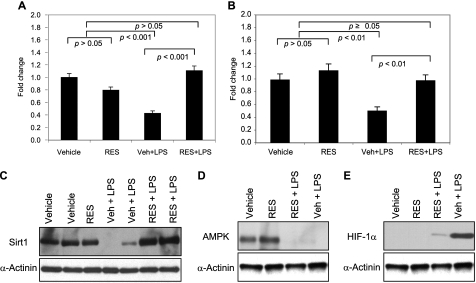
The decline in SIRT1 gene expression detected in vehicle-pretreated PBL (Fig. 6A) and liver (Fig. 6B) as well as the decline in SIRT1 protein expression in liver (Fig. 6C) were not observed in resveratrol-pretreated mice. Liver AMPK expression was suppressed in mice pretreated with either vehicle or resveratrol (Figs. 6D and 7C). In contrast, HIF-1α expression was detected in vehicle-pretreated but not in resveratrol-pretreated mouse liver (Figs. 6E and 7C).
FIGURE 6.
Resveratrol alters SIRT1 and HIF-1α expression in liver tissue when administered prior to the endotoxin challenge. Mice were treated as described in the legend to Fig. 5. Blood (A) and liver tissue (B–F) were obtained at 4 h post-infusion. SIRT1 transcripts levels in peripheral blood leukocytes (A) and liver tissue (B) were examined by quantitative PCR. Data are expressed as means ± S.E. (n = 4) relative to the untreated group. C–E, liver tissue samples normalized for protein content were used for Western blot analysis with the indicated antibodies. Statistical significance was determined by one-way ANOVA. Veh, vehicle; Res, resveratrol.
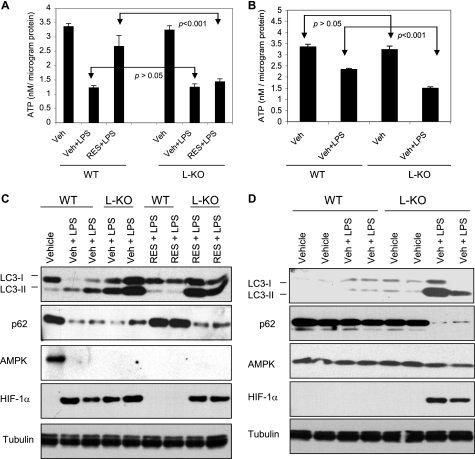
FIGURE 7.
ATP levels, autophagy, and HIF-1α and AMPK expression in liver of wild type and SIRT1 L-KO mice challenged with endotoxin in vivo. Wild type (WT) littermates and liver SIRT1 KO (SIRT1 L-KO) mice were given an intraperitoneal injection of vehicle (Veh) or resveratrol (RES) as described in the legend to Fig. 5. On day 8, the mice were challenged with saline or endotoxin (LPS). Liver was extracted at 4 h (A and C) or 24 h (B and E) post-endotoxin infusion. A and B, samples normalized for protein content were used for ATP analyses. Data are expressed as means ± S.E. (n = 4). C and D, liver tissue samples normalized for protein content were subjected to Western blot analysis with the indicated antibodies. Blotting for tubulin was used as a loading control. Each lane represents a sample obtained from a single mouse. Statistical significance was determined by one-way ANOVA.
SIRT1 Contributes to the Restoration of Bioenergetic Homeostasis in Liver Challenged with Endotoxin in Vivo
Next, using the resveratrol pretreatment protocol described earlier, we sought to determine whether the beneficial effects of resveratrol shown above (Figs. 5 and 6) are dependent on SIRT1 expression. Indeed, as shown in Fig. 7A, by 4 h post-endotoxin infusion, the ATP concentration in liver of resveratrol-pretreated SIRT1 L-KO mice was significantly lower than that of resveratrol-pretreated WT mice and was indistinguishable from the ATP concentration observed in vehicle-pretreated SIRT1 L-KO or WT mice. Consistent with these findings, resveratrol failed to prevent the increase in autophagy in SIRT1 L-KO mice liver (Fig. 7C). Furthermore, resveratrol also prevented the increase in HIF-1α expression in the liver of endotoxin-challenged WT mice but not in SIRT1 L-KO mice (Fig. 7C). Collectively, these data indicate that SIRT1 mediates the beneficial effects of resveratrol in endotoxin-challenged mice.
To further explore the role of bioenergetics function of SIRT1 in this model of systemic inflammation, we next examined the cellular ATP levels as well as the changes in LC3-I/II and p62/SQSTM1 expression in the liver of liver-specific SIRT1 knock-out (SIRT1 L-KO) mice and wild type (WT) littermate controls. No difference in the liver ATP concentration was observed between vehicle-pretreated SIRT1 L-KO and WT mice groups (Fig. 7, A and B), and the increase in autophagy, determined based on the increase in LC3-II expression and the decrease in p62/SQSTM1 expression, was noted in both groups at 4 h post-endotoxin infusion (Fig. 7C). In addition, AMPK expression was suppressed, and HIF-1α expression was up-regulated in the livers of either SIRT1 L-KO or WT mice (Fig. 7C). However, differences in ATP concentration as well as autophagy and HIF-1α expression were observed by 24 h post-endotoxin infusion (Fig. 7, B and D). At that time, the ATP concentration was significantly lower in the liver of SIRT1 L-KO mice as compared with WT controls (Fig. 7B), and autophagy remained robust in SIRT1 L-KO but not in WT mice liver (Fig. 7D). In addition, HIF-1α remained up-regulated in the liver of SIRT1 L-KO. These data suggested that SIRT1 facilitates the restoration of bioenergetic homeostasis during periods of systemic inflammation.
Kinetics of Acute Inflammatory Response
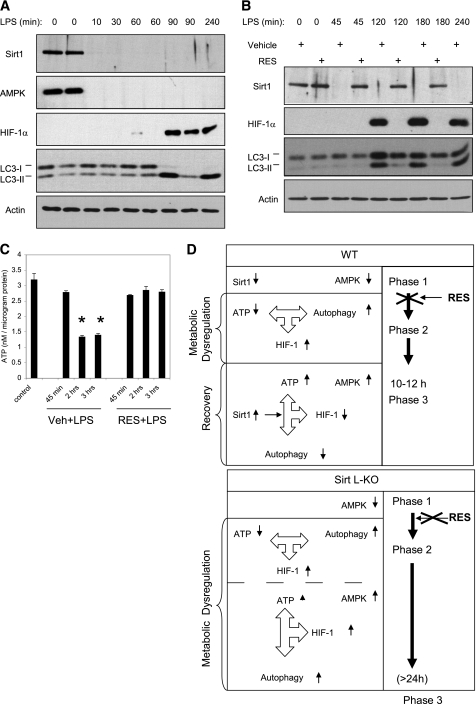
The data presented in Fig. 7 suggested that the decrease in SIRT1 expression sets the stage for endotoxin-induced changes in ATP concentration, autophagy, and HIF-1α expression. To address this possibility, we examined the status of SIRT1, AMPK, and HIF-1α expression as early as 10 min post-endotoxin challenge. Remarkably, SIRT1 and AMPK expression declined below detection levels within 10 min (Fig. 8A). In contrast, the increase in HIF-1α was not detected for at least 60 min (Fig. 8A). Similarly, the decline in ATP concentration and the increase in autophagy were not observed during the first 45 min post-challenge (Fig. 8B). Neither the decrease in ATP nor the increases in autophagy and HIF-1α expression were observed in resveratrol-pretreated mice (Fig. 8, B and C). These data established that the decline in SIRT1 and AMPK expression noted in liver of endotoxin-challenged mice precedes the decline in ATP, as well as the increases in autophagy and HIF-1α expression (Fig. 8D).
FIGURE 8.
Kinetics of the acute inflammatory response in liver of mice challenged with endotoxin. Liver tissue was obtained at the indicated time post-endotoxin infusion. Samples normalized for protein content were subjected to Western blot analysis (A). Blotting for actin was used as a loading control. B and C, mice were given an intraperitoneal injection of vehicle (Veh) or resveratrol (RES) as described in the legend to Fig. 5. On day 8, the mice were challenged with saline or a bolus dose of endotoxin (LPS). Liver tissue was obtained at the indicated time post-endotoxin infusion. Samples were normalized for protein content and subjected to Western blot analysis (B) and ATP assays (C). Data are expressed as means ± S.E. (n = 4). Statistical significance was determined by one-way ANOVA. *, p < 0.001. D, model describing the temporal changes in SIRT1, AMPK, and HIF-1α expression post-endotoxin infusion and their relationship to the metabolic dysregulation and recovery in WT and SIRT1 L-KO mice.
DISCUSSION
Although it is known that severe systemic inflammatory responses can alter the cellular bioenergetics in various tissues, the molecular mechanism that underlies this outcome is currently largely undetermined (59–62). This study provides direct evidence that in vivo endotoxin triggers profound metabolic perturbations that include parallel and opposite changes in ATP concentration and autophagy, a robust decrease in SIRT1 and AMPK expression, and an increase in HIF-1α expression in mice liver. Most of these outcomes were also observed in both human and mouse leukocytes. Given that systemic inflammatory conditions are associated with numerous human diseases, it is possible that bioenergetic perturbations similar to those that are described in this study exist and/or contribute to the etiology of many diseases.
Decreases in ATP concentration and mitochondrial activity were previously observed in the liver of endotoxin-challenged animals and in the skeletal muscle of septic patients or burn-injured mice (47–49, 63, 64). Our data extend these results by demonstrating that similar metabolic perturbations occur simultaneously in multiple tissues during a period of systemic inflammation. These findings are significant because they suggest that the PBL bioenergetics is representative of those in less accessible tissues and organs. Although the decline in ATP concentration reported here is consistent with the trend observed by others, our study is the first to reveal that the changes in ATP concentration occur within a well defined time window. Furthermore, we show that during the same time period the expressions of SIRT1, AMPK, and HIF-1α are all also profoundly altered with distinct, protein-specific outcomes.
A decrease in SIRT1 expression was observed in MonoMac6 cells and in inflammatory cells of rat lungs following exposure to cigarette smoke extract (65). A decrease in SIRT1 expression was also observed in Raw264.7 cells that were serum-starved and then challenged with endotoxin overnight (66). In another study, Yang et al. (67) reported that endotoxin, as well as free fatty acid-induced inflammatory conditions, suppresses both AMPK and SIRT1 expression in murine macrophages in vitro and in mice epididymal adipose tissue in vivo. Building on these observations, our data demonstrate not only that both AMPK and SIRT1 expressions are transiently suppressed in the liver during the acute phase of systemic inflammation but also that HIF-1α expression is elevated in both PBL and liver within the very same time period. Lim et al. (68) recently reported that SIRT1 and HIF-1α interact in vitro and in vivo and that SIRT1 represses HIF-1α signaling by deacetylating HIF-1α at Lys-674. That study also revealed that the expression of SIRT1 is suppressed under hypoxic conditions due to a decrease in the NAD+/NADH ratio and that the expression of HIF-1α is consequently enhanced (68). It is plausible that the expression of SIRT1 and HIF-1α is regulated by a similar mechanism during the acute phase of systemic inflammation.
SIRT1 and AMPK can be viewed as positive regulators of mitochondrial function, in that they trigger increases in mitochondrial biogenesis as well as mitochondrial respiration and oxygen consumption. In contrast, HIF-1 induces the expression of genes associated with glycolysis and actively represses mitochondrial function and oxygen consumption (24–26). Based on these seemingly opposite metabolic functions, the activity of HIF-1 should be optimal while the activities of both SIRT1 and AMPK are suppressed. In further support of this possibility, Lim et al. (68) recently proposed that by promoting a low NAD+/NADH ratio, glycolysis potentiates HIF-1α signaling through the suppression of SIRT1 expression. The shifts that we see in HIF-1, SIRT1, and AMPK expression may therefore provide the means for the cells to optimize HIF-1-regulatory outputs.
HIF-1 is a central regulator of myeloid cell metabolism, bacterial killing potency, migration, and pro-inflammatory cytokine production (35, 69). Mice lacking HIF-1α in myeloid cells were protected against endotoxin-induced mortality. This positive outcome was attributed to a decrease in pro-inflammatory cytokines that are produced by myeloid cells (69). In contrast, targeted deletion of HIF-1α in T cells was associated with an increase in pro-inflammatory cytokine levels and improved septic mouse survival (70). These observations indicate that the impact of HIF-1α expression during inflammation is cell type-specific. Our data extend these previous observations and show that the increase in HIF-1α expression during inflammation unfolds not only in immune cells that may transmigrate and accumulate at sites of inflammation but also in a solid organ such as liver, which is predominantly populated by TLR4-expressing parenchymal cells/hepatocytes (71). This raises the possibility that when challenged with a TLR4 agonist, hepatocytes might mount an HIF-1α adaptive response similar to the one activated in immune cells. Once activated, HIF-1α may trigger a metabolic switch leading to inhibition of mitochondrial respiration and enhanced glycolysis (72). This would imply that rather than absorbing lactate via the Cori cycle, hepatocytes, like other cells that express HIF-1α (25), might also secrete lactate during periods of systemic inflammation. This could provide the basis for the decrease in lactate clearance that is observed in critically ill patients (73).
The beneficial effects of resveratrol have been documented in numerous studies. The findings presented here show that when administered prior to the endotoxin challenge, resveratrol restores the expression of both SIRT1 and HIF-1α to base-line levels and prevents the decline in ATP levels and the induction of autophagy. The ability of resveratrol to prevent these endotoxin-mediated changes did not occur in the SIRT1 L-KO. It is interesting to note that resveratrol did not prevent the decline in AMPK expression, indicating that SIRT1 and AMPK are differentially regulated when resveratrol and endotoxin are combined. Studies have shown that resveratrol can enhance the enzymatic activities of SIRT1 (74), AMPK (75, 76), as well as other enzymes (15). Resveratrol could prevent the decrease in SIRT1 expression if, as shown for PGC-1 (77), SIRT1 can also regulate its own expression by acting within a positive auto-regulatory feedback loop independent of or through the diminution of NF-κB-dependent pro-inflammatory cytokine production (65, 78). In addition, it is known that resveratrol can exert anti-oxidative effects by modulating the expression of enzymes such as quinone reductase 2 (79) and cyclooxygenases (15, 80, 81). A recent study suggested that under oxidative stress conditions SIRT1 is subject to post-translational modification and that this modification marks SIRT1 for proteasomal degradation (82). This raises the possibility that the anti-oxidative effects of resveratrol, rather than a direct interaction with SIRT1, protect SIRT1 from post-translational modification and degradation. Although a decrease in intracellular oxygen species could in principle also explain how resveratrol prevents the increase in HIF-1α expression (83), the inability of resveratrol to prevent HIF-1α expression in liver of SIRT1 L-KO clearly indicates that SIRT1 is a critical component in the resveratrol-mediated protection against HIF-1α induction and ATP decline during inflammation.
When taken as a whole, our data suggest that the time period during which the cellular bioenergetics are perturbed post-endotoxin infusion can be separated into at least three phases. In the first phase, SIRT1 and AMPK expression are significantly reduced (Fig. 8D). This phase is completed within 10 min. We speculate that in parallel to these events endotoxin also alters the mitochondrial function, setting the stage for the second phase. During this phase the ATP concentration declines, and HIF-1α expression and autophagy are induced (Fig. 8D). This phase begins 1–1.5 h post-endotoxin infusion and lasts ∼10–12 h. Our comparative analyses of WT and SIRT1 L-KO mice (Fig. 7) indicate that the second phase is SIRT1-independent. In the third phase, the expression of HIF-1α and SIRT1 returns to normal. In liver of WT mice, the metabolic homeostasis is re-established within 24 h (Fig. 7). However, in SIRT1 L-KO mice, this recovery does not occur. These observations provide a strong indication that SIRT1 facilitates the restoration of bioenergetic homeostasis after the acute phase of systemic inflammation.
In conclusion, we have found that endotoxin triggers profound, transient, and parallel changes in expression of three key metabolic regulators: SIRT1, AMPK, and HIF-1α in liver and in PBL. We hypothesize that these changes reflect a shift from AMPK/SIRT1-dependent oxidative metabolism to a HIF-1-regulated glycolytic pathway and that HIF-1 may be the chief regulator of cellular metabolism as long as the inflammatory conditions remain unresolved. In support of this possibility, our analyses of gene expression patterns in PBL obtained from trauma patients revealed a significant increase in genes that include PFKFB3, SLC2A3, and PDK3, which are HIF-1-regulated targets. These genes remained elevated in PBL for at least 12 days after trauma (84). Our data raise the possibility that SIRT1 could represent a potential target for regulating HIF-1α expression during the acute phase of ATP loss and also for facilitating the return to metabolic homeostasis after an inflammatory challenge.
Acknowledgments
We thank Susette M. Coyle and Marie A. Macor for clinical assistance and Steve Calvano for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R21GM079255 (to B. H.) and RO1 GM 034695 (to S. F. L.) from NIGMS and Grant ES005022 from NIEHS Center for Excellence (to B. H.).
- AMPK
- AMP-activated protein kinase
- PBL
- peripheral blood leukocytes
- L-KO
- liver knockout
- LPS
- lipopolysaccharide/endotoxin
- ANOVA
- analysis of variance.
REFERENCES
- 1.Blander G., Guarente L. (2004) Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 2.Michan S., Sinclair D. (2007) Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantó C., Auwerx J. (2009) Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis M. C., Sinclair D. A. (2010) Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boily G., Seifert E. L., Bevilacqua L., He X. H., Sabourin G., Estey C., Moffat C., Crawford S., Saliba S., Jardine K., Xuan J., Evans M., Harper M. E., McBurney M. W. (2008) PLoS One 3, e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 8.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 9.Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 10.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 11.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 12.Lin J. D. (2009) Mol. Endocrinol. 23, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 15.Baur J. A. (2010) Biochim. Biophys. Acta 1804, 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 17.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. (2007) J. Biol. Chem. 282, 30107–30119 [DOI] [PubMed] [Google Scholar]
- 19.Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., Bass J. (2009) Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semenza G. L., Wang G. L. (1992) Mol. Cell. Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semenza G. L., Roth P. H., Fang H. M., Wang G. L. (1994) J. Biol. Chem. 269, 23757–23763 [PubMed] [Google Scholar]
- 25.Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 26.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 27.Wang G. L., Semenza G. L. (1995) J. Biol. Chem. 270, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 28.Wang G. L., Jiang B. H., Semenza G. L. (1995) Biochem. Biophys. Res. Commun. 212, 550–556 [DOI] [PubMed] [Google Scholar]
- 29.Jiang B. H., Semenza G. L., Bauer C., Marti H. H. (1996) Am. J. Physiol. 271, C1172–C1180 [DOI] [PubMed] [Google Scholar]
- 30.Huang L. E., Arany Z., Livingston D. M., Bunn H. F. (1996) J. Biol. Chem. 271, 32253–32259 [DOI] [PubMed] [Google Scholar]
- 31.Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallio P. J., Wilson W. J., O'Brien S., Makino Y., Poellinger L. (1999) J. Biol. Chem. 274, 6519–6525 [DOI] [PubMed] [Google Scholar]
- 33.Salceda S., Caro J. (1997) J. Biol. Chem. 272, 22642–22647 [DOI] [PubMed] [Google Scholar]
- 34.Kim H. Y., Kim Y. H., Nam B. H., Kong H. J., Kim H. H., Kim Y. J., An W. G., Cheong J. (2007) Exp. Cell Res. 313, 1866–1876 [DOI] [PubMed] [Google Scholar]
- 35.Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S. (2003) Cell 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., Karin M. (2008) Nature 453, 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., Tsokos M., Alt F. W., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3374–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meley D., Bauvy C., Houben-Weerts J. H., Dubbelhuis P. F., Helmond M. T., Codogno P., Meijer A. J. (2006) J. Biol. Chem. 281, 34870–34879 [DOI] [PubMed] [Google Scholar]
- 39.Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., Semenza G. L. (2008) J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., Mazure N. M. (2009) Mol. Cell. Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 45.Xu Y., Jagannath C., Liu X. D., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. (2007) Immunity 27, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgado M. A., Elmaoued R. A., Davis A. S., Kyei G., Deretic V. (2008) EMBO J. 27, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crouser E. D., Julian M. W., Blaho D. V., Pfeiffer D. R. (2002) Crit. Care Med. 30, 276–284 [DOI] [PubMed] [Google Scholar]
- 48.Padfield K. E., Astrakas L. G., Zhang Q., Gopalan S., Dai G., Mindrinos M. N., Tompkins R. G., Rahme L. G., Tzika A. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5368–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brealey D., Brand M., Hargreaves I., Heales S., Land J., Smolenski R., Davies N. A., Cooper C. E., Singer M. (2002) Lancet 360, 219–223 [DOI] [PubMed] [Google Scholar]
- 50.Calvano S. E., Xiao W., Richards D. R., Felciano R. M., Baker H. V., Cho R. J., Chen R. O., Brownstein B. H., Cobb J. P., Tschoeke S. K., Miller-Graziano C., Moldawer L. L., Mindrinos M. N., Davis R. W., Tompkins R. G., Lowry S. F. and Inflamm. and Host Response to Injury Large Scale Collaborative Research Program (2005) Nature 437, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 51.Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen D., Bruno J., Easlon E., Lin S. J., Cheng H. L., Alt F. W., Guarente L. (2008) Genes Dev. 22, 1753–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebai H., Ben-Attia M., Sani M., Aouani E., Ghanem-Boughanmi N. (2009) Arch. Toxicol. 83, 335–340 [DOI] [PubMed] [Google Scholar]
- 54.Izaguirre G., Aguirre L., Ji P., Aneskievich B., Haimovich B. (1999) J. Biol. Chem. 274, 37012–37020 [DOI] [PubMed] [Google Scholar]
- 55.Haimovich B., Calvano J., Haimovich A. D., Calvano S. E., Coyle S. M., Lowry S. F. (2010) Crit. Care Med. 38, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 58.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowry S. F. (2005) Shock 24, Suppl. 1, 94–100 [DOI] [PubMed] [Google Scholar]
- 60.Talwar S., Munson P. J., Barb J., Fiuza C., Cintron A. P., Logun C., Tropea M., Khan S., Reda D., Shelhamer J. H., Danner R. L., Suffredini A. F. (2006) Physiol. Genomics 25, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreasen A. S., Krabbe K. S., Krogh-Madsen R., Taudorf S., Pedersen B. K., Møller K. (2008) Curr. Med. Chem. 15, 1697–1705 [DOI] [PubMed] [Google Scholar]
- 62.Copeland S., Warren H. S., Lowry S. F., Calvano S. E., Remick D. (2005) Clin. Diagn. Lab. Immunol. 12, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crouser E. D., Julian M. W., Huff J. E., Struck J., Cook C. H. (2006) Crit. Care Med. 34, 2439–2446 [DOI] [PubMed] [Google Scholar]
- 64.Singer M. (2007) Crit. Care Med. 35, S441–S448 [DOI] [PubMed] [Google Scholar]
- 65.Yang S. R., Wright J., Bauter M., Seweryniak K., Kode A., Rahman I. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L567–L576 [DOI] [PubMed] [Google Scholar]
- 66.Shen Z., Ajmo J. M., Rogers C. Q., Liang X., Le L., Murr M. M., Peng Y., You M. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1047–G1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z., Kahn B. B., Shi H., Xue B. Z. (2010) J. Biol. Chem. 285, 19051–19059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim J. H., Lee Y. M., Chun Y. S., Chen J., Kim J. E., Park J. W. (2010) Mol. Cell 38, 864–878 [DOI] [PubMed] [Google Scholar]
- 69.Peyssonnaux C., Cejudo-Martin P., Doedens A., Zinkernagel A. S., Johnson R. S., Nizet V. (2007) J. Immunol. 178, 7516–7519 [DOI] [PubMed] [Google Scholar]
- 70.Thiel M., Caldwell C. C., Kreth S., Kuboki S., Chen P., Smith P., Ohta A., Lentsch A. B., Lukashev D., Sitkovsky M. V. (2007) PLoS One 2, e853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu S., Gallo D. J., Green A. M., Williams D. L., Gong X., Shapiro R. A., Gambotto A. A., Humphris E. L., Vodovotz Y., Billiar T. R. (2002) Infect. Immun. 70, 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu C. W., Lin S. C., Chen K. F., Lai Y. Y., Tsai S. J. (2008) J. Biol. Chem. 283, 28106–28114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen H. B., Rivers E. P., Knoblich B. P., Jacobsen G., Muzzin A., Ressler J. A., Tomlanovich M. C. (2004) Crit. Care Med. 32, 1637–1642 [DOI] [PubMed] [Google Scholar]
- 74.Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 75.Zang M., Xu S., Maitland-Toolan K. A., Zuccollo A., Hou X., Jiang B., Wierzbicki M., Verbeuren T. J., Cohen R. A. (2006) Diabetes 55, 2180–2191 [DOI] [PubMed] [Google Scholar]
- 76.Dasgupta B., Milbrandt J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7217–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Handschin C., Rhee J., Lin J., Tarr P. T., Spiegelman B. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buryanovskyy L., Fu Y., Boyd M., Ma Y., Hsieh T. C., Wu J. M., Zhang Z. (2004) Biochemistry 43, 11417–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jang M., Cai L., Udeani G. O., Slowing K. V., Thomas C. F., Beecher C. W., Fong H. H., Farnsworth N. R., Kinghorn A. D., Mehta R. G., Moon R. C., Pezzuto J. M. (1997) Science 275, 218–220 [DOI] [PubMed] [Google Scholar]
- 81.Murias M., Handler N., Erker T., Pleban K., Ecker G., Saiko P., Szekeres T., Jäger W. (2004) Bioorg. Med. Chem. 12, 5571–5578 [DOI] [PubMed] [Google Scholar]
- 82.Caito S., Rajendrasozhan S., Cook S., Chung S., Yao H., Friedman A. E., Brookes P. S., Rahman I. (2010) FASEB J. 24, 3145–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida K., Kirito K., Yongzhen H., Ozawa K., Kaushansky K., Komatsu N. (2008) Int. J. Hematol. 88, 43–51 [DOI] [PubMed] [Google Scholar]
- 84.Haimovich B., Reddell M. T., Calvano J. E., Calvano S. E., Macor M. A., Coyle S. M., Lowry S. F. (2010) Crit. Care 14, R177. [DOI] [PMC free article] [PubMed] [Google Scholar]