Abstract
The COG (conserved oligomeric Golgi complex) is a Golgi-associated tethering complex involved in retrograde trafficking of multiple Golgi enzymes. COG deficiencies lead to misorganization of the Golgi, defective trafficking of glycosylation enzymes, and abnormal N-, O- and ceramide-linked oligosaccharides. Here, we show that in Cog2 null mutant ldlC cells, the content of sphingomyelin (SM) is reduced to ∼25% of WT cells. Sphingomyelin synthase (SMS) activity is essentially normal in ldlC cells, but in contrast with the typical Golgi localization in WT cells, in ldlC cells, transfected SMS1 localizes to vesicular structures scattered throughout the cytoplasm, which show almost no signal of co-transfected ceramide transfer protein (CERT). Cog2 transfection restores SM formation and the typical SMS1 Golgi localization phenotype. Adding exogenous N-6-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)amino]hexanoyl-4-d-erythro-sphingosine (C6-NBD-ceramide) to ldlC cell cultures results in normal SM formation. Endogenous ceramide levels were 3-fold higher in ldlC cells than in WT cells, indicating that Golgi misorganization caused by Cog2 deficiency affects the delivery of ceramide to sites of SM synthesis by SMS1. Considering the importance of SM as a structural component of membranes, this finding is also worth of consideration in relation to a possible contribution to the clinical phenotype of patients suffering congenital disorders of glycosylation type II.
Keywords: Cell compartmentation, Confocal microscopy, Glycosylation, Golgi, Sphingolipid, CERT, COG, FAPP2, PIP4P, Sphingomyelin synthesis
Introduction
Protein and lipid oligosaccharide modifying enzymes are part of a complex machinery whose individual components are asymmetrically distributed along the cis/trans axis of the Golgi apparatus. Asymmetry is maintained despite a continuous anterograde membrane flow that moves secretory or membrane-bound cargoes along the secretory pathway, and a continuous COPI vesicle-mediated retrograde transport that moves residents from the late to early Golgi compartment (1). Fusion of COPI vesicles with target compartments occurs with participation of intra-Golgi soluble NSF attachment protein receptors; recent evidence indicates that this fusion step is preceded by one mediated by close-range acting oligomeric or monomeric tethering factors that may confer specificity to the fusion process. Among the oligomeric tethering factors, the COG (conserved oligomeric Golgi) complex is a soluble hetero-octamer associated to the cytoplasmic surface of the Golgi complex. It has been characterized in yeast (2) and mammalian cells (3) as a tethering complex necessary for Golgi retrograde trafficking of multiple Golgi processing proteins. COG complex is constituted by eight subunits organized in two lobules, lobule A (Cog1–4) and lobule B (Cog5–8) functionally interconnected via Cog1 and Cog8 (4, 5). Immunogold electron microscopy showed Cog1-HA localizing in the proximity of the tips and rims of the Golgi cisternae, in their associated vesicles and tubulovesicular structures, and in COPI-containing vesicles (6). Structural similarities between COG complex subunits and other multimeric tether subunits, such as exocysts and Dsl1p, support a common evolutionary origin and action mechanism for these complexes (7).
In yeast, a temperature-sensitive Cog3 mutant shows defective glycosylation but nearly normal secretion kinetics associated to mislocalization of two Golgi mannosyltransferases: Och1p and Mnn1p, which lead to the suggestion that COG is involved in the distribution of Och1p in retrograde vesicles (8). In mammals, two types of CHO mutant cells deficient in the LDL receptor, ldlB (Cog1 null mutant) and ldlC (Cog2 null mutant), have highly similar pleiotropic defects in processes associated to the Golgi complex and are characterized by the abnormal synthesis of N-linked, O-linked, and ceramide-linked oligosaccharides (9–13; for a review, see Ref. 14). These observations support the notion that the COG complex is a vesicle-tethering factor that normally tethers COPI retrograde transport vesicles originated in the late Golgi with cis Golgi membranes (15). COG also facilitates Golgi-ER traffic of some GEARs (COG-sensitive integral membrane proteins resident of the Golgi) that mislocalize and are rapidly degraded in COG mutant cells (16, 17). It has been proposed that COG and COPI participate together in the appropriate retention or recycling of GEARs and that COG prevents their accumulation in the ER and their degradation (16).
In humans, mutations in genes encoding Cog7 (18), Cog1 (19), Cog4 (20), Cog8 (21), and Cog5 (22) subunits result in congenital disorders of glycosylation type II, which characterize by defects in the processing of N-linked glycans. Cog7 and Cog1 deficient patients suffer from motor and mental retardation and die in early infancy (for a review, see Ref. 23. A Cog8-deficient patient also showed motor and mental retardation and showed chronic axonal neuropathy. Analysis of serum N-glycans from the patient evidenced normal addition of one sialic acid but severe deficiency in subsequent sialylation of mostly normal N-glycans (21).
In the present work, we studied the lipid composition of ldlC cells. Biochemical studies showed that in addition to the glycosylation defect that affect GM3 formation, a defective synthesis of sphingomyelin (SM)3 also occurs in these cells. Results from enzyme activity determinations, metabolic labeling, and fluorescence microscopy indicate that reduced synthesis of SM is associated to a mislocalization of sphingomyelin synthase1 (SMS1) and defective utilization of the substrate ceramide.
EXPERIMENTAL PROCEDURES
Molecular Constructs
The expression plasmid pMH2-Cog2-HA was a generous gift of Dr. Françoise Foulquier (Structural and Functional Glycobiology Unit UMR8576, University of Lille I). The expression plasmid for human SMS1 (SMS1-His6-V5/pcDNA3.1) (24) was a generous gift of Dr. Joost Holthuis (Utrecht University, Utrecht, The Netherlands). The expression plasmids pEGFP-C2-CERT and pEGFP-FAPP2 were a generous gift of Dr. María Antonietta De Matteis (Telethon Institute of Genetics and Medicine, Napoli, Italy). The expression plasmid pCFP-PH from FAPP1 was a generous gift of Dr. José Luis Daniotti Centro de Investigación en Química Biológica de Córdoba (CIQUIBIC) National University of Córdoba, Córdoba, Argentina).
Cell Culture and Transfection
CHO-K1 cells WT (ATCC, U.S.A.) and CHO mutant cells deficient in LDL receptor (Cog2 null mutant, ldlC cells), were kindly provided by Dr. Monty Krieger (Massachusetts Institute of Technology, Cambridge, MA) (9, 11–13, 25). Cells were grown in DMEM nutrient mixture F-12 (Ham) (Invitrogen) containing 2 mm glutamine, 15 mm HEPES, 50 μg/ml penicillin, and 50 μg/ml streptomycin, supplemented with 10% fetal calf serum. At ∼70% confluence, cells were transfected with 1 μg/ml of the indicated cDNA using cationic liposomes following the manufacturer's instructions (Lipofectamine, Life Technologies, Carlsbad, CA) or by electroporation of 107 cells/ml with 45 μg of total DNA using the ECM 600 electroporation system, (BTX, Genetronics, San Diego, CA) following the manufacturer's instructions.
Fluorescence Microscopy
Cells grown on coverslips were fixed and permeabilized for 7 min in methanol at −20 °C or fixed with paraformaldehide 3% in PBS for 20 min and permeabilized with Triton X-100 0.1% of glycine (200 mm) for 10 min at room temperature. Fixed cells were incubated with the following specific antibodies: monoclonal mouse anti GM130 (1:300, BD Biosciences), monoclonal mouse anti V5 (1:500), polyclonal rabbit anti-HA (1:1000), monoclonal mouse anti-HA (1:800), polyclonal rabbit anti SMS1 (1:50, Santa Cruz Biotechnology) and appropriate secondary antibodies. Coverslips were mounted with FluorSave (Calbiochem, EMD Biosciences, Inc., La Jolla, CA) and observed under the spectral confocal microscope Olympus FluoViewTM FV1000 equipped with argon multiline 458, 488, and 514 nm; helium/neon 543 nm lasers; and 100× numerical aperture 1.4 UPlanSApo oil immersion objective (Olympus). Images were compiled with Adobe Photoshop CS2. Pearson's coefficient of co-localization was calculated using ImageJ software with the JACoP plug-in (for details, see supplemental “Experimental Procedures”) (26).
Subcellular Fractionation
WT and ldlC CERT-GFP transfected cells were washed three times with cold PBS and harvested by scraping in 400 μl of 5 mm Tris-HCl (pH 7.4) in the presence of protease inhibitor mixture (1:1800). After homogenization, nuclear fractions and unbroken cells were removed by centrifuging twice at 4 °C for 5 min at 600 × g. Supernatants were then ultracentrifuged at 4 °C for 1 h at 400,000 × g in an OPTIMATM Ultracentrifuge (Beckman Coulter, Inc., Fullerton, CA). The supernatant was collected, and the pellet was resuspended in 400 μl of Tris-HCl 5 mm in the presence of protease inhibitor mixture. Protein in each fraction was precipitated with trichloroacetic acid to a final concentration of 10%, collected by centrifugation, and analyzed by SDS-PAGE and Western blot.
Lipid Analysis
Cells in culture (3 × 105 cells per 35-mm dish) were metabolically labeled overnight with 10 μCi/ml of d-[U-14C]galactose (329.5 mCi/mmol; DuPont NEN, Boston, MA) to label glycolipids (27) or with 10 μCi/ml of [9,10-3H]palmitic acid (53 Ci/mmol; Amersham Biosciences, Buckinghamshire, UK) to label phospholipids and ceramide. After washing with cold PBS, cells were scrapped from the plate, and lipids were extracted with chloroform:methanol (2:1 by volume) at room temperature for 30 min. Glycolipid composition was examined by HPTLC of the total lipid extract using as solvent chloroform:methanol (4:1 by volume) in a first run up to two-thirds of the plate and chloroform:methanol:0.2% CaCl2 (60:36:8 by volume) in a second run up to the front of the plate. Phospholipid and ceramide composition were examined in the lower phase after a Folch partition of the lipid extract. TLC of phospholipids was carried out using chloroform:methanol:acetic acid:water (40:10:10:1 by volume) as solvent in a first run up to the front of the plate and chloroform:methanol:acetic acid:water (120:46:19:3 by volume) in a second run up to half of the plate. Unlabeled phospholipids were visualized after dipping the chromatograms in 3% cupric acetate in 8% phosphoric acid and heating at 150 °C until development of the bands. For the analysis of lower phase sphingolipids, the lower phase was saponified as follows; it was evaporated to dryness under N2 in glass tubes, taken in 0.1 m KOH in methanol and either neutralized immediately or after keeping for 2 h at 37 °C with the same volume of 0.1 m HCl. Chloroform, methanol, and water were added to achieve a Folch partition, and lipids in the lower phase were run on HPTLC. For sphingophospholipids, the solvent system was as indicated above for total phospholipids. For ceramide separation, the solvent system used was chloroform:acetic acid (9:1 (by volume)).
Lipid radioactivity in chromatograms was recorded either by phosphorimaging or by exposure to radiographic films. Densitometric quantification of chromatograms and x-ray plates were done using ImageJ (image processing and Analysis in Java) software (National Institute of Health). Liquid chromatography mass spectrometry of unlabeled lower phase lipids was carried out at the Mass Spectrometry Facility at the University of California (Riverside, CA).
Lysenin Binding Studies
All manipulations were done at room temperature unless noted otherwise. Cells were grown on glass coverslips in DMEM medium at 37 °C for 24 h to ∼70% confluence. After washing with PBS, the cells were fixed with 3% formaldehyde in PBS for 20 min, washed with PBS, incubated with 0.1 m NH4Cl in PBS for 20 min, and washed again with PBS. After blocking with 2% BSA in PBS for 1 h, the cells were incubated with lysenin (400 ng/ml in 0.2% BSA/PBS, PeptaNova GmbH, Sandhausen, Germany) for 1 h and washed three times with PBS for 5 min with gentle shaking. Next, they were incubated with rabbit anti-lysenin antiserum (1:500 dilution in 0.2% BSA/PBS, PeptaNova GmbH) for 1 h and washed with PBS, followed by incubation with an appropriate secondary antibody. After washing with PBS, the cells were mounted and observed under a Olympus FluoViewTM FV1000 confocal microscope. Confocal images of lysenin-treated WT and ldlC cells were processed with ImageJ software (National Institute of Health). First, images were converted from 12 bits to 8 bits in order to use Counting Particles ImageJ plug-in, and then the same global threshold was defined for WT and ldlC cells. 30 cells were delimited with the free region of interest (ROI) tool of the program, and particles with sizes between 0.1 and 1 μm2 were counted.
Determination of Enzyme Activities
Homogenates of WT and ldlC cells were used as an enzyme source for determination of sialyltransferase 1 (27) and SMS (28) activities. Briefly, sialyltransferase 1 was determined in an incubation system that contained, in a final volume of 30 μl, 400 μm LacCer, 100 μm CMP[3H]NeuAc (250,000 cpm), 20 mm MnCl2, 1 mm MgCl2, 20 μg of Triton CF54/Tween 80 (2:1 w/w), 100 mm sodium cacodylate-HCl buffer (pH 6.5), and cell extract (40 μg of protein). Incubations were performed at 37 °C for 90 min. Samples without exogenous acceptor were used to discount the incorporation into endogenous acceptors. Reactions were stopped with 1 ml of 5% (w/v) trichloroacetic acid/0.5% phosphotungstic acid, and the radioactivity incorporated into lipids was determined as described (29). For determination of SMS and glucosylceramide synthase activities, the incubation system contained, in a final volume of 50 μl, 20 μm N-6-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)amino]hexanoyl-4-d-erythro-sphingosine (C6-NBD-ceramide, Molecular Probes) complexed with bovine serum albumin in a 1:1 molar ratio, 400 μm 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 400 μm UDP-glucose, 5 mm MgCl2, 5 mm MnCl2, 1 mm EDTA in 50 mm HEPES (pH 7.2), and cell homogenate (40 μg of protein). After 50 min of incubation at 37 °C, reactions were stopped by the addition of 250 μl of chloroform:methanol 2:1 (v/v) to extract C6-NBD lipids in the lower phase. After centrifugation at 1000 × g for 5 min, the lower phase was evaporated under nitrogen and subjected to TLC using chloroform:methanol:water (65:25:4, by volume) as solvent. C6-NBD lipids were visualized by UV illumination of the TLC.
Incubation of ldlC Cells with C6-NBD-ceramide
WT and ldlC cells were cultured 4 h in the presence of 1 μg/ml of C6-NBD-ceramide complexed with bovine serum albumin in a 1:1 molar ratio. After three washes with cold PBS, cells were harvested, and lipids were extracted with chloroform:methanol (2:1 by volume) at room temperature, and the extract was partitioned by adding 0.2 volumes of water. After centrifugation at 1000 × g for 5 min, the lower phase was evaporated under nitrogen and subjected to HPTLC by using chloroform:methanol:water (65:25:4 by volume) as solvent. C6-NBD lipids present in the chromatograms were visualized by UV illumination of the HPTLC.
Western Blot
For Western blot analysis, total protein from cells or cell fractions were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% (w/v) nonfat dried skim milk in PBS for 1 h at room temperature, followed by three 5-min washes with PBS, and reacted with the following primary antibodies: mouse monoclonal antibody anti-HA (1:1000; Sigma-Aldrich) or mouse monoclonal anti-GFP (1:2000, Roche Applied Science). Goat anti-mouse antibody coupled to IRDye 800 (1:20,000; LI-COR Biosciences, Lincoln, NE) was used as secondary antibody. Membranes were then scanned using the Odyssey IR Imager (LI-COR Biosciences, Lincoln, NE).
RESULTS
Lipid Composition of ldlC Cells
Cog2 null mutant ldlC cells belong to a group of CHO mutant cells deficient in LDL receptor activity. Deficiency is due to its decreased stability as a consequence of abnormal processing of its oligosaccharides in the late stage of Golgi. ldlC cells are able to add high mannose oligosaccharides in N-linked chains to glycoproteins (LDL receptor, vesicular stomatitis virus G protein) but are unable to add sialic acid residues to these oligosaccharide chains and to convert them to completely endo-β-N-acetylglucosaminidase H-resistant forms (11). The ldlC phenotype is corrected to the CHO WT phenotype by a cDNA coding for an 83-kDa Golgi-associated, brefeldin A-sensitive peripheral protein, later on referred to as Cog2. The endogenous LDLC mRNA was undetectable in ldlC cells (12). To investigate the lipid composition of ldlC cells, we carried out long term metabolic labeling of total lipids of CHO WT and ldlC cells in an overnight culture with appropriate radioactive precursors. Lipids were then extracted and analyzed by TLC.
The glycolipid fraction was analyzed in the total lipid extract from cells cultured overnight in the presence of [14C]galactose. A relative accumulation of GlcCer (∼1.5-fold higher than in WT cells) and LacCer (∼10-fold higher than in WT cells) and a marked decrease of GM3 (∼5-fold lower than in WT cells) was evident in the chromatogram (Fig. 1A).
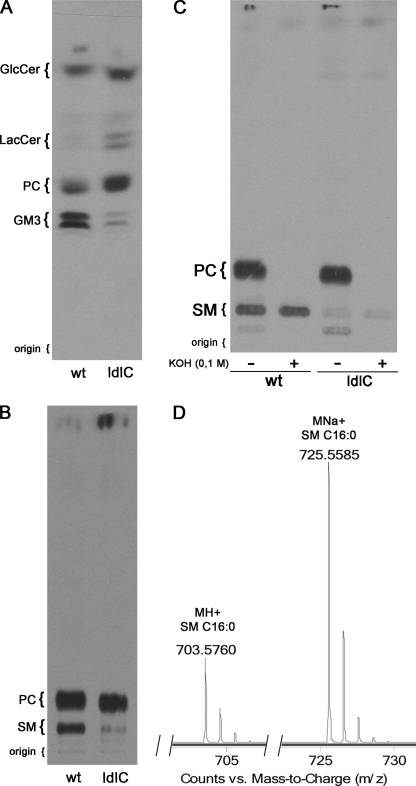
FIGURE 1.
ldlC cells show reduced sphingomyelin and GM3 content. A, lipids from WT and ldlC cells cultured overnight in the presence of [14C]galactose were extracted with chloroform:methanol (2:1, by volume) and total lipids separated by HPTLC as described under “Experimental Procedures.” B, lipid extracts of cells as in A, but cultured with [3H]palmitate were Folch-partitioned into a lower and an upper phase, and lipids from the lower phase were separated by TLC as indicated under “Experimental Procedures.” C, aliquots of [3H]palmitate-radiolabeled lipids from the lower phase of both WT and ldlC cells were subjected to mild alkaline hydrolysis as indicated under “Experimental Procedures” and then chromatographed as in B. The position of co-chromatographed standard lipids is shown at the left of each chromatogram. D, lower phase lipids from ldlC cells were separated by liquid chromatography, and the peak that eluted as the standard oleic acid containing Sphingomyelin (C18:1 SM) (elution time, 12.1 min) was subjected to mass spectrometry.
The phospholipid fraction was analyzed in the lower phase of the lipid extract from cells cultured in the presence of [3H]palmitate, after partition against water. Compared with WT cells, ldlC cells extracts showed essentially no difference in a compound migrating as the standard phosphatidylcholine (PC) but showed a clear decrease of ∼75% of a compound migrating in the chromatogram as the standard SM (Fig. 1B), showing that besides the defects in the glycosylation of glycolipids (11), the composition of other lipids is also affected in ldlC cells.
ldlC Cells Have Reduced Sphingomyelin Content
To identify the compound running as SM and partially missing from ldlC cells, the lower phase lipids of both WT and ldlC cells, were subjected to mild alkaline hydrolysis. If, as suspected, this compound is SM, it should be resistant to saponification, whereas the major glycerophospholipid PC should not. It is clear in Fig. 1C that after alkaline hydrolysis, the bands running as PC in both WT and ldlC cells completely disappeared, whereas the bands running as SM remained intact, indicating that the compound is not a glycerolipid and most probably is SM. A liquid chromatography mass spectrometric analysis of lower phase lipids from WT and ldlC cells confirmed that the compound eluted from the liquid chromatograph at the time at which a standard sample of oleic acid containing Sphingomyelin (C18:1 SM) elutes (12.1 min) shows ions at m/z 703.6 and 725.56, corresponding to the protonated and Na adduct of 16:0 SM, respectively (Fig. 1D). The quantification of the SM content in radioactive samples derived from long term metabolic labeling from [3H]palmitate was confirmed by direct measurement of SM mass in lipid extracts from both cell types. For this, lower phase lipids from WT and ldlC cells were separated by TLC, and stained lipid bands were quantified by densitometry. The SM content in ldlC cells lipid extract was ∼65% lower than in WT cells (supplemental Fig. 1), consistent with ∼75% lower content determined from metabolic labeling experiments.
ldlC Cells Show Reduced Lysenin Aggregates at Plasma Membrane
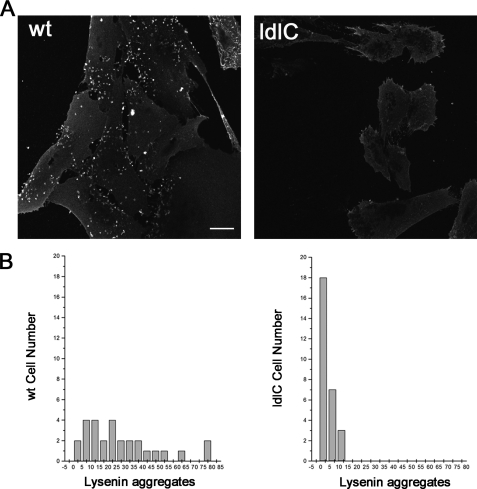
Lysenin is a pore-forming toxin from the earthworm Eisenia foetida that specifically binds SM clusters in membranes (30, 31). To see whether the decreased total SM content in lipid extracts of ldlC cells (Fig. 1) affects cell surface SM expression, the binding of lysenin was determined in fixed ldlC cells. By confocal microscopy, it was found that ldlC cell plasma membrane shows less lysenin aggregates than WT cells (Fig. 2A). Quantification of these aggregates indicates that although ∼70% of the WT cells show between 10 and 40 aggregates per cell, most ldlC cells show <10 aggregates (Fig. 2B), thus showing that the level of plasma membrane SM is also affected in ldlC cells.
FIGURE 2.
Lysenin binding reveals decreased levels of plasma membrane SM in ldlC cells. A, WT and ldlC CHO cells were fixed, treated with lysenin at room temperature for 1 h, and then treated with anti-lysenin antiserum followed by the fluorescent secondary antibody. Cells were observed in a confocal microscope. B, the number of lysenin aggregates per cell was determined by counting 30 cells in each condition using ImageJ software, as indicated under “Experimental Procedures.” Scale bar, 10 μm.
Sphingomyelin Synthase Activity Is Comparable in WT and ldlC Cells
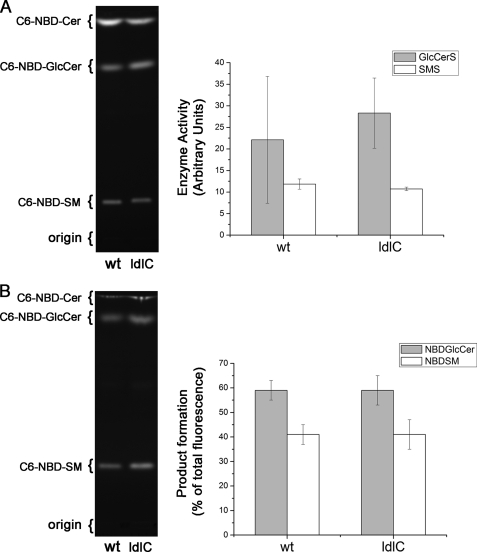
SM is synthesized in the Golgi complex by SMS1, a multipass membrane protein of the Golgi, and by SMS2, a plasma membrane protein. Both enzymes catalyze the transfer of phosphocholine moiety of PC to the acceptor ceramide to synthesize SM (24), but SMS1 acts as a major SM synthase activity (32). Recent immunoelectron microscopy data revealed that SMS1 was mostly present at the trans side of the Golgi of HeLa cells (33). We determined this activity in homogenates of WT and ldlC cells by measuring the conversion of the exogenous fluorescent ceramide analog C6-NBD-ceramide into fluorescent SM (Fig. 3A). Quantitative estimation of the formed product revealed that the SMS activity in ldlC cells was essentially as in WT cells (Fig. 3A). Formation of GlcCer, the product of glucosylceramide synthase activity also measured in the assay, was slightly increased in ldlC cells but the difference was not significant (Fig. 3A).
FIGURE 3.
In vitro SMS activity and in vivo C6-NBD-ceramide utilization. A, homogenates of WT and ldlC cells were assayed for the activities of SMS and glucosylceramide synthase in conditions of linearity with time and protein concentration. Lipids were extracted from incubates, separated by HPTLC, and formed fluorescent compounds visualized under UV light illumination. Quantification was carried out by densitometry of the fluorescent bands using ImageJ software. Enzyme activity values are given as mean arbitrary fluorescent units ± S.D. for three independent experiments. Other details were as described under “Experimental Procedures.” B, C6-NBD-ceramide (Cer) utilization in ldlC cells. WT and ldlC cells monolayers were cultured in the presence of 1 μg/ml C6-NBD-ceramide for 4 h. Cells were harvested, and lipids were extracted and separated by HPTLC. Formed fluorescent compounds were photographed under UV light illumination, and the percent contribution of the bands quantified by densitometry using ImageJ software. For A and B, the positions of lipid standards are indicated at the left of the chromatogram.
Incubation of ldlC Cells with C6-NBD-ceramide Results in Normal Formation of C6-NBD-sphingomyelin
The fact that in SM-defective ldlC cells the in vitro activity of SMS1 is essentially normal (Fig. 3A) suggests that in vivo, some of the two participating substrates of the SM synthesis reaction, PC, and/or ceramide, may be not available to SMS1. To test this possibility and considering that the PC level was normal in ldlC cells (Fig. 1B), they were tested for their capacity to incorporate exogenous C6-NBD-ceramide into SM when cultured in its presence. This fluorescent ceramide analog permeates the cellular membranes and accumulates in intracellular structures, concentrating particularly in the Golgi complex (34); it has been suggested that this is due to its trapping in the lumen as a consequence of the addition of the phosphorylcholine head group in the conversion into SM by Golgi SMS activity (32). Fig. 3B shows the HPTLC analysis of the lower phase of a lipid extract from these cells. The percentage of fluorescent SM relative to the total fluorescent products formed was essentially the same in WT and ldlC cells. GlcCer formation from C6-NBD-ceramide was also similar in both cells. This result indicates that SMS is catalytically active also in vivo provided the substrate ceramide is given exogenously, suggesting that other factors may be affecting the normal supply of endogenous ceramide to SMS1.
ldlC Cells Have Increased Ceramide Content
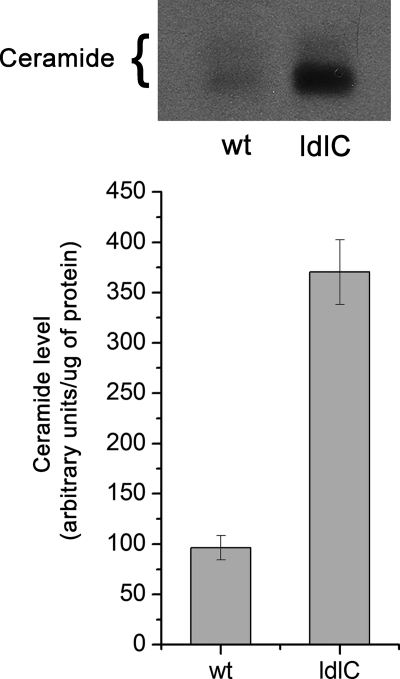
The results of Fig. 3B suggest that reduced SM synthesis in ldlC cells may be the consequence of reduced ceramide levels in these cells. To investigate this possibility, WT and ldlC cells were cultured overnight in the presence of [3H]palmitic acid, and the level of endogenous ceramide was determined after HPTLC separation of the alkali-resistant fraction of the lower phase lipids. Contrary to what was expected, the ceramide level in ldlC cells was found increased ∼3-fold with respect to WT cells (Fig. 4). Direct mea- surement of ceramide mass in lipid extracts from both cells types gave essentially the same value of increase in favor of ldlC cells (supplemental Fig. 2). The simplest interpretation of this experiment is that reduced ceramide content is not the cause for the reduced level of SM in ldlC cells. Rather, the increase suggests a reduced consumption of ceramide due to the block in SM synthesis.
FIGURE 4.
ldlC cells show increased ceramide levels. Lipids from WT and ldlC cells cultured overnight in the presence of [9,10-3H]palmitic acid were extracted with chloroform:methanol (2:1, by volume) and Folch-partitioned into a lower and an upper phase. Lower phase lipids were subjected to mild alkaline hydrolysis and partitioned, and the chloroform phase was separated by TLC using chloroform:acetic acid (9:1, by volume). Other details were as described under “Experimental Procedures.”
ldlC Cells Show Altered Subcellular Localization of SMS
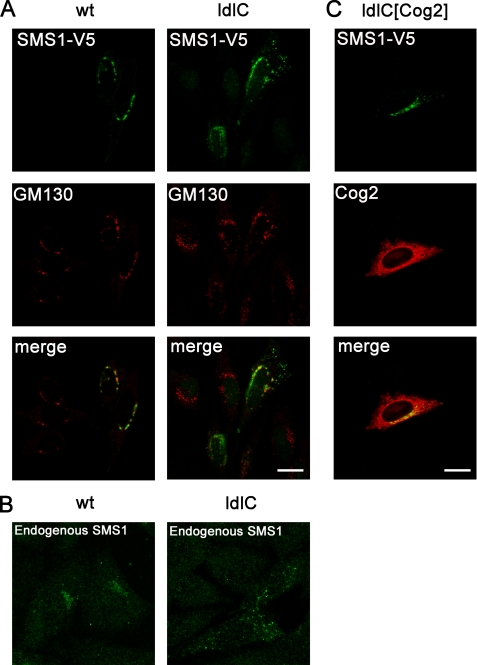
Considering the possibility that the structural misorganization of the Golgi complex of ldlC cells (35) may affect the coupling of biochemical reactions leading to SM formation, the localization of SMS1 was examined in these cells. WT and ldlC cells were transfected with V5-tagged SMS1, and the subcellular localization was compared with that of the endogenous Golgi marker GM130, a Cog-insensitive protein (16). Whereas in 90 ± 2% of WT cells, SMS1-V5 and the Golgi marker GM130 localized in typical perinuclear, Golgi-like structures (Fig. 5A), only 14 ± 4% of ldlC cells showed SMS1 with the typical Golgi perinuclear localization. Instead, in most ldlC cells, a considerable fraction of SMS1 localizes in vesicular structures scattered throughout the cytoplasm. The difference in localization is most impressive when the endogenous SMS1 was immunostained with a specific antibody (Fig. 5B). Co-transfection of ldlC cells with Cog2 cDNA, re-established the localization of SMS1-V5 to a pattern characteristic of the Golgi complex in 88 ± 4% of the co-transfected cells (Fig. 5C); Cog2-HA itself was just slightly concentrated in the Golgi region, at variance with the typical Golgi localization of endogenous COG complex subunits. It was already reported for overexpressed Cog1-HA that it also localizes to the ER without impairment of its function (19). These results point to SMS1 mislocalization due to Cog2 absence as a possible cause of the observed defective SM synthesis.
FIGURE 5.
Cog2 transfection restores Golgi localization of SMS1 in ldlC cells. A, WT and ldlC cells transiently expressing SMS1-V5 were immunostained with appropriated antibodies for V5 tag (green) and the endogenous cis Golgi marker GM130 (red). Data were obtained from three independent experiments, each with n = 50 cells. B, endogenous SMS1 was immunostained with its specific antibody, and controls without primary antibody for WT and ldlC cells were made. Phenotype observed in three independent experiments, each with n = 100 cells. C, ldlC cells transiently co-expressing SMS1-V5 (green) and Cog2-HA (red) were immunostained with the appropriated antibodies. Data were obtained from three independent experiments, each with n = 30 cells. Scale bar, 10 μm.
Cog2 Transfection Restores SM Levels of ldlC Cells to Near Normal Values
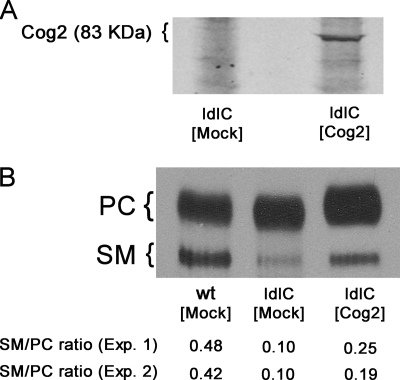
If the reduced SM content of ldlC cells actually results from Cog2 deficiency, transfecting Cog2 cDNA to ldlC cells should revert the SM-deficient phenotype. Cells were transfected with Cog2-HA, and 24 h after transfection, they were pulsed overnight with [3H]palmitate. Transfection was verified by Western blot, which showed Cog2-HA protein present in the transfected cell population (Fig. 6A). Lipids were extracted from WT, mock transfected, and transfected ldlC cells, separated as in Fig. 1B, and the PC to SM ratio was determined by densitometric scanning of the x-ray autoradiograms. In two independent experiments, nontransfected ldlC cells show a SM/PC labeling ratio of ∼0.1, which was reverted to ∼0.20 upon transfection with Cog2 (Fig. 6B). Determination of the SM mass in the alkali-resistant lower phase lipids of these cells gave similar values of increase (∼1.8 fold) in the SM content in Cog2-HA-transfected ldlC cells with respect to nontransfected ldlC cells (supplemental Fig. 3). These values were ∼50% of the corresponding value in WT cells, but because in our hands, the efficiency of transfection is ∼40%, the values indicate that SM synthesis was practically restored in Cog2-transfected ldlC cells. Taken together, the results of Figs. 5 and 6 and supplemental Fig. 3 indicate a close causal relationship between Cog2 deficiency, SMS1 mislocalization, and SM synthesis deficiency.
FIGURE 6.
Cog2 cDNA transfection increases SM level of ldlC cells. A, ldlC cells were transfected with Cog2 cDNA, and the expression of Cog2 (83 KDa) was visualized by Western blot using anti-HA monoclonal antibody in both mock-transfected and Cog2-transfected cells, as indicated. B, transfected ldlC cells were cultured overnight in the presence of [3H]palmitate. Radioactive lipids from mock-transfected WT and ldlC cells and from Cog2-transfected ldlC cells (ldlC [Cog2]) were extracted with chloroform:methanol (2:1, by volume), the extract was partitioned, and the lower phase was analyzed by HPTLC as described under “Experimental Procedures.” Chromatograms were exposed to radiographic films, and the percent contribution of the bands was quantified by densitometry using ImageJ software. The SM/PC ratio determined for each cell type in two independent experiments (Exp. 1 and Exp. 2) is indicated at the bottom of the autoradiogram.
CERT and SMS1 Localize to Different Membranes in ldlC Cells
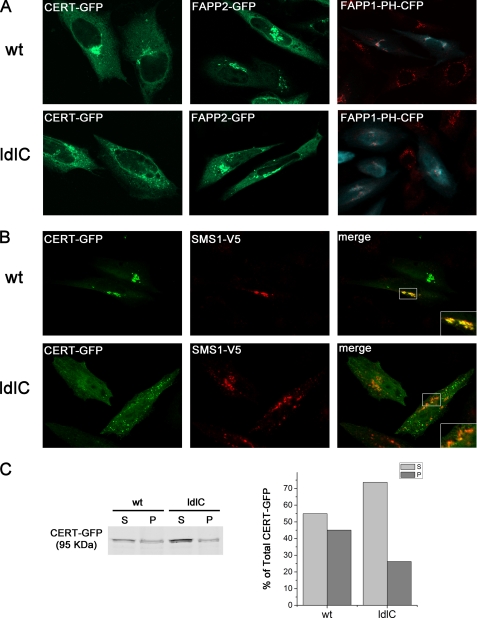
Ceramide for SM synthesis is synthesized in the ER and transported from the ER to the trans Golgi, where it is converted to SM. The cytosolic protein CERT mediates the ER-to-Golgi trafficking of ceramide. It has been suggested that CERT extracts ceramide from the ER and transfers it to the Golgi apparatus in a nonvesicular manner at close apposition sites between the ER and the Golgi apparatus membranes (36). It is possible that in ldlC cells with abnormal Golgi organization, CERT binding and ceramide delivery to SMS1-containing compartments do not operate properly. To address this point, WT and ldlC cells were transfected with a GFP-tagged version of CERT, and 18 h later, cells were fixed and observed under a confocal microscope. As described previously (37), CERT was found concentrated in the perinuclear region of ∼75% of WT cells (Fig. 7A). Conversely, most ldlC cells show that CERT is distributed throughout the cytoplasm. Only 23% of ldlC cells show CERT concentrated in vesicular structures dispersed in the cytoplasm, reminiscent of Golgi fragments (Fig. 7A). Determination of the amount of CERT-GFP in the supernatant and membrane-bound fractions of WT and ldlC cells by Western blot, evidenced that the membrane bound fraction of CERT in ldlC cells relative to the unbound fraction, was lower than in WT cells (0.33 versus 0.82, respectively) (Fig. 7C). As shown in Fig. 5, A and B, most SMS1 also localize to vesicular structures scattered throughout the cytoplasm in ldlc cells (Fig. 7B). To investigate whether SMS1 and CERT co-localize in the vesicular structures, WT and ldlC cells were co-transfected with CERT-GFP and SMS1-V5, fixed with paraformaldehyde, permeabilized with Triton X-100, and observed under the microscope. This procedure may allow the release of a fraction of the cytoplasmic CERT-GFP (see Fig. 7C) and hence explain the apparent lower fluorescence in cells of Fig. 7B with respect to Fig. 7A. Results indicate that most of the membranes containing SMS1 in ldlC cells do not bind CERT. Only 17.8% of co-transfected ldlC cells show CERT and SMS1 partially co-localizing in vesicular structures (Pearson's coefficient = 0.58 ± 0.04). A deficit of PI(4)P in membranes containing SMS1 in ldlC cells is probably the cause of reduced CERT binding, but if so, the deficit is selective for SMS1-containing structures because the binding of a GFP-tagged version of FAPP2 (four-phosphate adaptor protein 2), that is also mediated by PI(4)P was essentially as in WT cells, with the fluorescence concentrated in perinuclear Golgi-like structures in both cells (Fig. 7A) (see “Discussion”). In addition, examining the localization of PI(4)P by visualizing the localization of transfected FAPP1-PH-CFP (Fig. 7A, cyan), shows that in WT cells, the binding occurs mainly in the Golgi region marked by GM130 (red). On the other hand, in ldlC cells, a wide subcellular distribution of PI(4)P is observed. Even so, a faint specific binding of FAPP1-PH-CFP (cyan) in the Golgi region marked by GM130 (red) is still observed.
FIGURE 7.
CERT, SMS1, FAPP2, and FAPP1-PH domain localization in WT and ldlC cells. A, CERT-GFP and FAPP1-PH-CFP but not FAPP2-GFP localization are altered in ldlC cells. WT and ldlC cells were transfected with CERT-GFP (green), FAPP2-GFP (green) or transfected wit FAPP1-PH-CFP (cyan) and stained for the endogenous Golgi market GM130 (red). Cells were fixed and observed under a confocal microscope. B, CERT and SMS1 localized to different membranes in ldlC cells. WT and ldlC cells were co-transfected with CERT-GFP and SMS1-V5. Cells were fixed with 3% paraformaldehyde in PBS for 20 min and permeabilized with Triton X-100 0.1% glycine (200 mm) for 10 min at room temperature and immunostained with monoclonal mouse anti-V5 and the appropriate secondary antibody conjugated with Alexa Fluor 546. WT cells show significant co-localization (Pearson's coefficient = 0.86 ± 0.05), whereas ldlC cells only show partial co-localization (Pearson's coefficient = 0.58 ± 0.04) between CERT and SMS1 in three independent experiments, with each analyzing 30 cells. Insets are a higher magnification of the boxed areas in the cells. C, WT and ldlC CERT-GFP-transfected cells were homogenized, and supernatant (S) and pellet (P) fractions were obtained by ultracentrifugation at 400,000 × g as described under “Experimental Procedures.”
DISCUSSION
Cog2 null mutant ldlC cells are CHO cells characterized by having deficient LDL receptor activity due to its decreased stability associated to the abnormal processing of its oligosaccharides in the late Golgi (11). This phenotype was corrected by a cDNA coding for an 83-kDa Golgi-associated, brefeldin A-sensitive peripheral protein, referred to as Cog2 (12). Subsequent work, led to the identification of mutations in different proteins of the COG complex in patients with congenital disorders of glysolation type II (13).
We have examined the lipid composition of ldlC cells, with particular attention to members of the phospholipid family and have found that a lipid compound identified as SM was decreased to ∼25% of WT cells (Fig. 1B) and that such a reduction is also detectable at the plasma membrane level by measuring the binding of lysenin (Fig. 2). Noticeably, the enzyme activity of SMS in cell homogenates was similar in WT and ldlC cells (Fig. 3A) and also was similar in the metabolic conversion of exogenous C6-NBD-ceramide into C6-NBD-SM (Fig. 3B). Lack of endogenous ceramide was discarded as the main cause for the defective synthesis of SM because ceramide was found increased 3-fold in ldlC cells (Fig. 4).
SM synthesis is mainly catalyzed by SMS1, although a plasma membrane-localized SMS2 also contributes to total cellular SM synthesis (24, 32). Fine ultrastructural localization studies in HeLa cells mapped the residence of SMS1 to the trans cisternae of the Golgi (33). Microscopic examination of transfected SMS1-V5 to WT CHO cells showed this enzyme in a compact perinuclear distribution comparable with that of the Golgi marker GM130. However, in ldlC cells, SMS1-V5 was found in vesicular structures scattered throughout the cytoplasm (Figs. 5A and 7B). Cog2 transfection was effective in restoring SMS1 localization to the Golgi (Fig. 5C) and also in restoring the SM synthesis level to nearly normal levels in ldlC cells metabolically labeled with [3H]palmitate (Fig. 6).
Altogether, these results strongly suggest that structural alterations of the Golgi complex associated to the Cog2 deficiency in ldlC cells restrict proper coupling between SMS1 and its substrates. Because ldlC cells show an increased level of ceramide (Fig. 4), no difference in PC content with respect to WT cells, and metabolic conversion of exogenous C6-NBD-ceramide into SM was essentially as in WT cells, results point to a failure in the usage of endogenous ceramide by SMS1.
Ceramide for SM synthesis is transported from the ER to the Golgi by a nonvesicular mechanism involving the cytoplasmic ceramide transfer protein CERT (37). Disruption of Golgi morphology of HeLa cells by the microtubule-destabilizing agent nocodazole reduced SM synthesis by ∼50% (38). This was attributed to disruption of zones of close proximity between membranes of trans ER trans Golgi used by CERT for delivery of ceramide for synthesis of SM. However, dispersion of the Golgi seems not to be an obvious cause of defective SM synthesis, because ilimaquinone, which also promotes Golgi disorganization, does not affect SM synthesis (38).
The morphological alterations of the trans Golgi region in Cog2-deficient ldlC cells, where SMS1 resides (33), are reminiscent of the effects of the pharmacological agents mentioned above. We found CERT concentrated in the Golgi region of the majority of WT cells, but only in 23% of ldlC cells (Fig. 7A), the majority of which present CERT mainly spread throughout the cytoplasm. When CERT was co-expressed with SMS1 in ldlC cells, both proteins were found localized to vesicular structures in the cytoplasm; however, they scarcely co-localize in these structures (Fig. 7B). CERT is targeted to the Golgi by the binding to PI(4)P, present in Golgi membranes, through its PH domain. Among several possibilities for the lack of co-localization, one is that SMS1-containing structures are topologically segregated from those bearing CERT-binding capacity; another possibility is that SMS1-containing membranes have particularly reduced PI(4)P content. PI(4)P synthesis depends on PI4K IIIβ activation by protein kinase D (39), which is in turn activated by diacylglycerol. As diacylglycerol is a by-product of SMS1 activity, diacylglycerol and consequently PI(4)P may be reduced in these membranes, affecting binding of CERT, but this deserves further investigation.
ldlC cells show a slight accumulation of GlcCer, a marked accumulation of LacCer and a reduced content of GM3 (Fig. 1A) (11). These results indicate that the supply of ceramide for GlcCer synthesis and the coupling of GlcCer for LacCer synthesis are not heavily affected in ldlC cells, if at all affected. FAPP2 mediates the delivery of GlcCer from proximal to distal Golgi where LacCer synthesis occurs (40), or to the ER where it enters the secretory pathway and then reaches the Golgi lumen for further elongation to LacCer (33). Transfected FAPP2 show a perinuclear staining in a majority of WT and ldlC cells (Fig. 7A), indicating that localization of FAPP2 is not heavily affected. FAPP2 binds GlcCer and PI(4)P at the Golgi complex by its glycolipid transfer protein and PH domains, respectively (40). Therefore, we cannot distinguish whether FAPP2 was localized to the Golgi via binding to PI(4)P or via binding to GlcCer. In addition, because the existence of PI4K IIIβ and the binding of the PH domain of FAPP1 in proximal Golgi membranes was reported (41; for a review, see Ref. 42), the possibility also exists that the binding of FAPP2 that we observed occurred on proximal Golgi membranes. In fact, determination of PI(4)P levels by examining the localization of FAPP1-PH-CFP evidenced a major fraction of the probe spread over the cytoplasm, and also a detectable fraction localizing to the Golgi region of ldlC cells. Thus, at difference with CERT that is mainly spread in the cytoplasm (Fig. 7A), both FAPP2 and FAPP1-PH-CFP localization assays suggest the existence of an available PI(4)P pool, presumably in proximal Golgi membranes, which may allow the extraction of GlcCer by FAPP2 and therefore the provision of GlcCer for LacCer synthesis (33, 40). The marked increase of LacCer and reduction of GM3 and the essentially normal activity of sialyltransferase 1, which converts LacCer into GM3 in ldlC cells (43), suggests that the compartment containing sialyltransferase 1 in ldlC cells is either unable to acquire LacCer for further sialylation to GM3 or does not provide the optimal milieu for the reaction to occur.
Although it is well established that defects in glycan terminal glycosylation observed in congenital disorders of glycosylation type II disorders can be ascribed to late Golgi dysfunction, it is less clear whether the clinical phenotype of the patients results only from the abnormal glycosylation pattern of membrane-bound or secreted glycoconjugates or whether other products requiring a functional late Golgi for their synthesis may add to this phenotype. SM has important cell biological implications as a structural component of plasma membranes and plasma membrane microdomains (44) and as co-regulator of cholesterol homeostasis (45). In addition, its synthesis by SMS1 modulates the cellular levels of diacylglycerol and ceramide, which are lipid messengers involved in the control of cell proliferation (32, 46). Therefore, defective levels of SM and imbalances of diacylglycerol to ceramide ratios may be considered as potential contributors to the clinical phenotype of patients with congenital disorders of glycosylation type II. Future investigation will be needed to understand how ldlC cells cope with the affected SM synthesis.
Supplementary Material
Acknowledgments
We thank Javier Valdez-Taubas and Mariana Ferrari for critical reading of the manuscript. We also thank Gabriela Schachner and Susana Deza for technical assistance with cell cultures and Carlos Mas and María Cecilia Sampedro for assistance with confocal microscopy.
This work was supported in part by Grant PICT 2006-01239 from Agencia Nacional de Promocion Cientifica y Tecnologica, Universidad Nacional de Córdoba, y MinCyT Córdoba.
- SM
- sphingomyelin
- SMS
- sphingomyelin synthase
- HPTLC
- high performance thin layer chromatography
- CERT
- ceramide transfer protein
- PI4K IIIβ
- phosphatidylinositol 4-kinase IIIβ
- PH
- pleckstrin homology domain
- PC
- phosphatidylcholine
- COPI
- coat protein complex I
- GM3
- NeuAcα2,3Galβ1,4Glc-ceramide
- CFP
- cyan fluorescent protein
- GFP
- green fluorescent protein
- FAPP1
- four-phosphate adaptor protein 1
- FAPP2
- four-phosphate adaptor protein 2
- LacCer
- lactosylceramide
- GlcCer
- glucosylceramide
- PI(4)P
- phosphatidylinositol 4′-monophosphate.
REFERENCES
- 1.Bonifacino J. S., Glick B. S. (2004) Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2.Whyte J. R., Munro S. (2002) J. Cell Sci. 115, 2627–2637 [DOI] [PubMed] [Google Scholar]
- 3.Ungar D., Oka T., Krieger M., Hughson F. M. (2006) Trends Cell Biol. 16, 113–120 [DOI] [PubMed] [Google Scholar]
- 4.Ungar D., Oka T., Brittle E. E., Vasile E., Lupashin V. V., Chatterton J. E., Heuser J. E., Krieger M., Waters M. G. (2002) J. Cell Biol. 157, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oka T., Vasile E., Penman M., Novina C. D., Dykxhoorn D. M., Ungar D., Hughson F. M., Krieger M. (2005) J. Biol. Chem. 280, 32736–32745 [DOI] [PubMed] [Google Scholar]
- 6.Vasile E., Oka T., Ericsson M., Nakamura N., Krieger M. (2006) Exp. Cell Res. 312, 3132–3141 [DOI] [PubMed] [Google Scholar]
- 7.Richardson B. C., Smith R. D., Ungar D., Nakamura A., Jeffrey P. D., Lupashin V. V., Hughson F. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruinsma P., Spelbrink R. G., Nothwehr S. F. (2004) J. Biol. Chem. 279, 39814–39823 [DOI] [PubMed] [Google Scholar]
- 9.Krieger M., Brown M. S., Goldstein J. L. (1981) J. Mol. Biol. 150, 167–184 [DOI] [PubMed] [Google Scholar]
- 10.Krieger M., Reddy P., Kozarsky K., Kingsley D., Hobbie L., Penman M. (1989) Methods Cell Biol. 32, 57–84 [DOI] [PubMed] [Google Scholar]
- 11.Kingsley D. M., Kozarsky K. F., Segal M., Krieger M. (1986) J. Cell Biol. 102, 1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podos S. D., Reddy P., Ashkenas J., Krieger M. (1994) J. Cell Biol. 127, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterton J. E., Hirsch D., Schwartz J. J., Bickel P. E., Rosenberg R. D., Lodish H. F., Krieger M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sztul E., Lupashin V. (2009) FEBS Lett. 583, 3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shestakova A., Zolov S., Lupashin V. (2006) Traffic 7, 191–204 [DOI] [PubMed] [Google Scholar]
- 16.Oka T., Ungar D., Hughson F. M., Krieger M. (2004) Mol. Biol. Cell 15, 2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steet R., Kornfeld S. (2006) Mol. Biol. Cell 17, 2312–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X., Steet R. A., Bohorov O., Bakker J., Newell J., Krieger M., Spaapen L., Kornfeld S., Freeze H. H. (2004) Nat. Med. 10, 518–523 [DOI] [PubMed] [Google Scholar]
- 19.Foulquier F., Vasile E., Schollen E., Callewaert N., Raemaekers T., Quelhas D., Jaeken J., Mills P., Winchester B., Krieger M., Annaert W., Matthijs G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynders E., Foulquier F., Leão Teles E., Quelhas D., Morelle W., Rabouille C., Annaert W., Matthijs G. (2009) Hum. Mol. Genet. 18, 3244–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kranz C., Ng B. G., Sun L., Sharma V., Eklund E. A., Miura Y., Ungar D., Lupashin V., Winkel R. D., Cipollo J. F., Costello C. E., Loh E., Hong W., Freeze H. H. (2007) Hum. Mol. Genet. 16, 731–741 [DOI] [PubMed] [Google Scholar]
- 22.Paesold-Burda P., Maag C., Troxler H., Foulquier F., Kleinert P., Schnabel S., Baumgartner M., Hennet T. (2009) Hum. Mol. Genet. 18, 4350–4356 [DOI] [PubMed] [Google Scholar]
- 23.Foulquier F. (2009) Biochim. Biophys. Acta 1792, 896–902 [DOI] [PubMed] [Google Scholar]
- 24.Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. (2004) EMBO J. 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsley D. M., Krieger M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 5454–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolte S., Cordelieres F. P. (2006) J. Microscopy 244, 213–232 [DOI] [PubMed] [Google Scholar]
- 27.Daniotti J. L., Landa C. A., Maccioni H. J. (1994) J. Neurochem. 62, 1131–1136 [DOI] [PubMed] [Google Scholar]
- 28.Veldman R. J., Mita A., Cuvillier O., Garcia V., Klappe K., Medin J. A., Campbell J. D., Carpentier S., Kok J. W., Levade T. (2003) FASEB J. 17, 1144–1146 [DOI] [PubMed] [Google Scholar]
- 29.Daniotti J. L., Landa C. A., Rösner H., Maccioni H. J. (1991) J. Neurochem. 57, 2054–2058 [DOI] [PubMed] [Google Scholar]
- 30.Yamaji A., Sekizawa Y., Emoto K., Sakuraba H., Inoue K., Kobayashi H., Umeda M. (1998) J. Biol. Chem. 273, 5300–5306 [DOI] [PubMed] [Google Scholar]
- 31.Yamaji-Hasegawa A., Makino A., Baba T., Senoh Y., Kimura-Suda H., Sato S. B., Terada N., Ohno S., Kiyokawa E., Umeda M., Kobayashi T. (2003) J. Biol. Chem. 278, 22762–22770 [DOI] [PubMed] [Google Scholar]
- 32.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. (2007) J. Biol. Chem. 282, 17537–17547 [DOI] [PubMed] [Google Scholar]
- 33.Halter D., Neumann S., van Dijk S. M., Wolthoorn J., de Mazière A. M., Vieira O. V., Mattjus P., Klumperman J., van Meer G., Sprong H. (2007) J. Cell Biol. 179, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipsky N. G., Pagano R. E. (1985) J. Cell Biol. 100, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolov S. N., Lupashin V. V. (2005) J. Cell Biol. 168, 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanada K., Kumagai K., Tomishige N., Yamaji T. (2009) Biochim. Biophys. Acta 1791, 684–691 [DOI] [PubMed] [Google Scholar]
- 37.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 38.Chandran S., Machamer C. E. (2008) Traffic 9, 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fugmann T., Hausser A., Schöffler P., Schmid S., Pfizenmaier K., Olayioye M. A. (2007) J. Cell Biol. 178, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., Platt F. M., Hannun Y. A., Polishchuk R., Mattjus P., De Matteis M. A. (2007) Nature 449, 62–67 [DOI] [PubMed] [Google Scholar]
- 41.Weixel K. M., Blumental-Perry A., Watkins S. C., Aridor M., Weisz O. A. (2005) J. Biol. Chem. 280, 10501–10508 [DOI] [PubMed] [Google Scholar]
- 42.D'Angelo G., Vicinanza M., Di Campli A., De Matteis M. A. (2008) J Cell Sci. 121, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 43.Spessott W., Uliana A., Maccioni H. J. (2010) Neurochem. Res., in press [DOI] [PubMed] [Google Scholar]
- 44.Van der Luit A. H., Budde M., Zerp S., Caan W., Klarenbeek J. B., Verheij M., Van Blitterswijk W. J. (2007) Biochem. J. 401, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridgway N. D. (2000) Biochim. Biophys. Acta 1484, 129–141 [DOI] [PubMed] [Google Scholar]
- 46.Hannun Y. A., Obeid L. M. (2002) J. Biol. Chem. 277, 25847–25850 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.