Abstract
The gene for the mismatch-specific uracil DNA glycosylase (MUG) was identified in the Escherichia coli genome as a sequence homolog of the human thymine DNA glycosylase with activity against mismatched uracil base pairs. Examination of cell extracts led us to detect a previously unknown xanthine DNA glycosylase (XDG) activity in E. coli. DNA glycosylase assays with purified enzymes indicated the novel XDG activity is attributable to MUG. Here, we report a biochemical characterization of xanthine DNA glycosylase activity in MUG. The wild type MUG possesses more robust activity against xanthine than uracil and is active against all xanthine-containing DNA (C/X, T/X, G/X, A/X and single-stranded X). Analysis of potentials of mean force indicates that the double-stranded xanthine base pairs have a relatively narrow energetic difference in base flipping, whereas the tendency for uracil base flipping follows the order of C/U > G/U > T/U > A/U. Site-directed mutagenesis performed on conserved motifs revealed that Asn-140 and Ser-23 are important determinants for XDG activity in E. coli MUG. Molecular modeling and molecular dynamics simulations reveal distinct hydrogen-bonding patterns in the active site of E. coli MUG that account for the specificity differences between E. coli MUG and human thymine DNA glycosylase as well as that between the wild type MUG and the Asn-140 and Ser-23 mutants. This study underscores the role of the favorable binding interactions in modulating the specificity of DNA glycosylases.
Keywords: DNA Damage, DNA Repair, Enzyme Mutation, Mutagenesis Mechanisms, Site-directed Mutagenesis, DNA Glycosylase, Deamination, Uracil, Xanthine
Introduction
DNA is constantly assaulted by environmental and endogenous agents, causing various types of chemical damage. DNA bases are subject to deamination by hydrolytic or oxidative reactions due to the reactivity of the exocyclic amino groups (1–3). Uracil (U), xanthine (X), and oxanine (O), hypoxanthine (I), and thymine (T) are the corresponding deamination products derived from cytosine (C), guanine (G), adenine (A), and 5-methylcytosine, respectively. The amino-to-keto conversion of base deamination alters the hydrogen bond properties of the damaged bases from a hydrogen donor to a hydrogen bond acceptor, which may result in mutation during DNA replication.
Uracil DNA glycosylase (UDG),2 an enzyme present in organisms as simple as viruses or as complex as humans, initiates the repair of uracil in DNA. Five families, classified according to sequence and structural homologies, constitute a UDG superfamily (4, 5). Family 1 includes the extensively studied Escherichia coli, human, and herpes simplex virus 1 UNGs. Family 2 contains human thymine DNA glycosylase (hTDG) and E. coli mismatch-specific uracil DNA glycosylase (MUG). hTDG is unique in its ability to excise thymine from a G/T mismatch generated from 5-methylcytosine deamination (6). Family 3 is composed of SMUG1 (single-strand-selective monofunctional uracil DNA glycosylase) proteins found in vertebrates and some bacteria. Family 4 UDGs are iron-sulfur-containing enzymes found in prokaryotes. Family 5 are found in a limited number of species of prokaryotic organisms such as Archaea. It is not uncommon for an organism to possess more than one uracil DNA glycosylase. In addition to UDG, TDG, and SMUG1, humans also have another uracil DNA glycosylase called MBD4 that does not belong to the UDG superfamily (7).
Xanthine is now recognized as a stable lesion under physiological conditions (8, 9). As such, although repair of xanthine was noted in human lymphoblast cells in an earlier study (10), enzymes that may repair xanthine were not identified until recently. Both E. coli AlkA and its functional homolog human alkyladenine DNA glycosylase have xanthine DNA glycosylase activity (9, 11, 12). The SMUG1 enzymes from bacteria and humans are also active on xanthine-containing DNA (13). In addition, several homologs of bacterial endonuclease V exhibit deoxyxanthosine endonuclease activity (11, 14–16).
Identification of DNA repair activity in E. coli has led to the discovery of new repair enzymes or novel activities. DNA repair-deficient mutant strains have facilitated identification of functional homologs in eukaryotic systems (17–20). We are interested in achieving a comprehensive understanding of xanthine DNA repair in E. coli. Previous studies show that AlkA, endo VIII, and endo V in E. coli possess xanthine repair activities (11, 12, 16). Using an E. coli triple mutant strain (nfi nei alkA), we detected xanthine DNA glycosylase (XDG) activity in whole cell protein extracts. Further biochemical analysis led to the discovery of XDG activity in the mismatch-specific uracil DNA glycosylase MUG. Surprisingly, kinetic analysis revealed that the XDG activity from MUG was more robust than UDG activity. Rather than being active with only double-stranded mismatch uracil base pairs, MUG can excise xanthine from double-stranded base pairs as well as single-stranded DNA. Structural elements that are involved in determining the base recognition in MUG were probed by site-directed mutagenesis. Mutational effects on glycosylase activities of deaminated bases, X and U, were analyzed by activity assays and binding analyses. Thermodynamic properties associated with the flipping of deaminated base pairs were determined by calculating potentials of mean force. The base recognition specificity is discussed in light of molecular modeling and molecular dynamics simulations of MUG interactions with deaminated bases.
EXPERIMENTAL PROCEDURES
Preparation of E. coli Cell Extracts
Bacterial cells (BW1466 and BW1739) from 100-ml cultures grown to late exponential phase were harvested by centrifugation at 5000 rpm with GSA-10 rotor in RC5C Sorvall centrifuge (DuPont). The cell pellets were suspended in 5 ml of sonication buffer and sonicated 5 times with a burst duration of 1 min each. The lysates were centrifuged at 12,000 rpm at 4 °C for 20 min. The supernatants containing soluble proteins were transferred to fresh tubes, filtered with 0.45-μm syringe filters (Whatman, Clifton, New Jersey), and dialyzed at 4 °C overnight against a buffer containing 20 mm Tris-HCl (pH 8.0), 1 mm EDTA (pH 8.0), and 0.1 mm DTT. Protein concentrations were measured by the Bradford method using bovine serum albumin as a standard (21).
Oligodeoxynucleotide Substrates
The fluorescently labeled oligodeoxynucleotide substrates were prepared as described (22). The sequences of the oligonucleotides are shown in Fig. 1A. Oligodeoxyribonucleotides were ordered from IDT, purified by PAGE, and dissolved in Tris-EDTA buffer at a final concentration of 10 μm. The two complementary strands with the unlabeled strand in 1.2-fold molar excess were mixed, incubated at 85 °C for 3 min, and allowed to form duplex DNA substrates at room temperature for more than 30 min. The xanthine- and oxanine-containing oligonucleotide were constructed as previously described (15, 23).
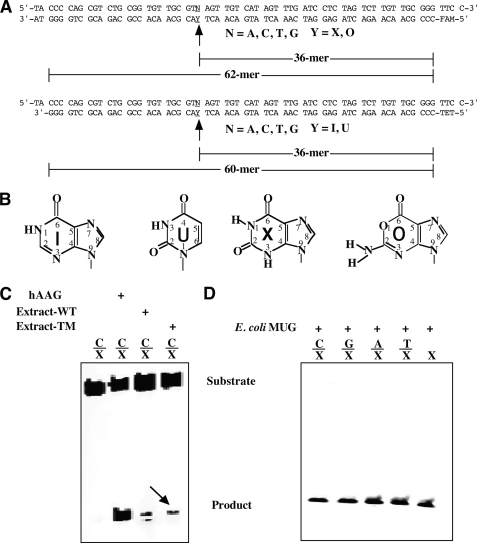
FIGURE 1.
Cleavage of deaminated base-containing DNA substrates by E. coli cell extracts and E. coli MUG from a commercial source. Cleavage reactions were performed as described under “Experimental Procedures” with 5 μl of cell extract or 1 unit of WT MUG protein (Trevigen) and 10 nm substrate. A, sequences of X- and O- and of I- and U-containing oligodeoxyribonucleotide substrates are shown. B, chemical structures of deaminated DNA bases are shown. C, DNA glycosylase activity on C/X in E. coli cell extracts is shown. D, DNA glycosylase activity of MUG on X-containing substrate is shown. WT, BW1466; TM, triple mutant, BW1739. Human alkyladenine DNA glycosylase (hAAG) was assayed as a control with 100 nm human alkyladenine DNA glycosylase protein and 10 nm substrate as described previously (23). Under the assay conditions the substrate was completely cleaved to product.
DNA Glycosylase Activity Assay
DNA glycosylase cleavage assays for E. coli MUG were performed at 37 °C for 60 min in a 10-μl reaction mixture containing 10 nm oligonucleotide substrate, an indicated amount of glycosylase protein, 20 mm Tris-HCl (pH 7.5), 100 mm KCl, 5 mm EDTA, and 2 mm 2-mercaptoethanol. The resulting abasic sites were cleaved by incubation at 95 °C for 5 min after adding 0.5 μl of 1 n NaOH. Reactions were quenched by the addition of an equal volume of GeneScan stop buffer. After incubation at 95 °C for 3 min, samples (3.5 μl) were loaded onto a 7 m urea, 10% denaturing polyacrylamide gel. Electrophoresis was conducted at 1500 V for 1.5 h using an ABI 377 sequencer (Applied Biosystems). Cleavage products and remaining substrates were quantified using GeneScan analysis software.
Gel Mobility Shift Assay
The binding reactions were performed on ice for 10 min in a 10-μl volume containing 50 nm DNA substrate, 20 mm Tris-HCl (pH 7.2), 50 mm NaCl, 5 mm EDTA, 1 mm DTT, 0.1 mg/ml bovine serum albumin, 10% glycerol, and the indicated amounts of E. coli MUG protein. Samples were supplemented with 2 μl of 100% glycerol and electrophoresed at 200 V on a 6% native polyacrylamide gel in 1× TB buffer (89 mm Tris base and 89 mm boric acid) supplemented with 5 mm EDTA. The bound and free DNA species were analyzed using a Typhoon 9400 Imager (GE Healthcare) with the following settings: photomultiplier tube at 600 V, excitation at 495 nm, and emission at 535 nm.
Molecular Modeling and Molecular Dynamics Simulations
Molecular models of the unbound and bound conformations of wild type (WT) E. coli MUG were used as initial structures for subsequent computational analyses. The crystal structure of E. coli MUG (pdb accession code 1mug) was used as a model for the unbound MUG enzyme. The molecular model of the WT E. coli MUG complexed with a DNA decamer sequence containing uracil was constructed based on the crystal structure of UDG bound to a DNA decamer (pdb accession code 1emh).
Molecular dynamics simulations were performed on the bound MUG structures using the CHARMM 32b1 molecular mechanics software package (24) and the CHARMM 27 force field (25, 26). Interaction energies consisting of Coulomb and van der Waals potential energies were calculated over the molecular dynamics trajectory between the active site residues and the substrates using the “coor inter” module in CHARMM.
Potentials of mean force (PMF) describe free energy changes along a predefined reaction coordinate while averaging over the remaining degrees of freedom. Here, potentials of mean force are used to describe the free energy changes associated with rotating a nucleotide from the interior of the DNA double helix into the aqueous solvent. A detailed description of the computational methods is provided in the supplemental data.
RESULTS
Detection of XDG Activity in E. coli Cell Extracts
Studies of uracil repair in E. coli has led to the discovery of a uracil DNA glycosylase and a MUG. Previous investigations using purified enzymes revealed that two glycosylases (AlkA and endo VIII) and endo V in E. coli possess xanthine DNA glycosylase activity or deoxyxanthosine endonuclease activity (11, 12, 16). However, XDG activities from the two glycosylases, particularly the latter, are quite low. To survey whether E. coli contains additional XDG enzymes, we examined the cleavage activity in cell extracts of a triple mutant strain that had alkA, nei (endo VIII), and nfi (endo V) deleted. The assays were performed using fluorescently labeled xanthine-containing deoxyoligonucleotide substrates (Fig. 1, A and B). As expected, we detected cleavage of C/X in the wild type cell extract (Fig. 1C). Surprisingly, we also detected xanthine DNA glycosylase activity in the triple mutant strain that eliminated the activity of all previously known XDG enzymes (Fig. 1C). This result indicated that the E. coli genome contained an additional XDG enzyme. Given that a previous study had investigated XDG activity in purified E. coli AlkA, endo III, endo V, endo VIII, Fpg/MutM, MutY, and UDG (11), we surmised that MUG, which was not included in the previous study, may be accountable for the observed XDG activity in the triple mutant cell extract. A quick test was performed using purified MUG from a commercial source (Trevigen). As shown in Fig. 1D, MUG was found to be active on all five xanthine-containing substrates, including the single-stranded substrate. We also tested XDG activity using a MUG single deletion strain and a quadruple deletion strain. XDG activity was not detected in either case, indicating that MUG is the predominant activity in cell extracts.
XDG Activity in Purified E. coli MUG
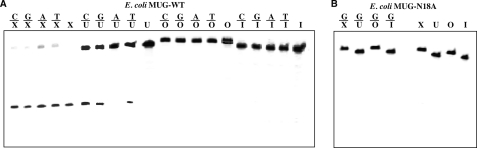
To further confirm the novel XDG activity, we set out to clone, express, and purify the WT MUG and an active site mutant MUG-N18A. MUG was expressed in the ung mug knock-out strain BH214 to avoid contaminating activity from the host. We tested deaminated base repair activity on X-, U-, O-, and I-containing substrates (Fig. 2A). The purified WT MUG again showed cleavage of all five X-containing substrates (Fig. 2A). The uracil DNA glycosylase activity observed in the WT MUG was essentially identical to that reported in previous studies, i.e. active on C/U, G/U, and T/U mismatched uracil base pairs (27, 28). The XDG activity was noticeably more robust than the UDG activity as indicated by close-to-complete cleavage of the xanthine substrates (Fig. 2A). No oxanine or hypoxanthine DNA glycosylase activities were detected from the WT enzyme under our assay conditions (Fig. 2A). To further verify that the xanthine DNA glycosylase activity was authentic to MUG, we performed the same assay using an active site mutant MUG-N18A, which abolished the ability of the enzyme to activate a water molecule to attack the N-glycosidic bond (29, 30). No glycosylase activity on deaminated bases was detected, confirming that the XDG activity was native to the MUG protein (Fig. 2B).
FIGURE 2.
Cleavage of X-, U-, O-, and I- containing DNA substrates by E. coli MUG. Cleavage reactions were performed as described under “Experimental Procedures” with 100 nm WT E. coli MUG protein and 10 nm substrate. A, cleavage by WT E. coli MUG is shown. B, cleavage by N18A mutant of E. coli MUG is shown.
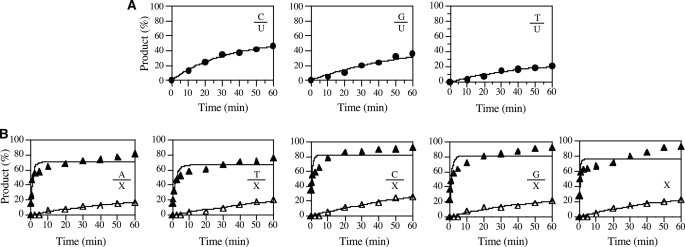
The robust XDG activity in MUG prompted us to quantitatively determine the deaminated base repair activity in MUG. Under the condition that the enzyme was in excess (E:S ratio = 10:1), removal of uracil in the three mismatched base pairs was less than 50% (Fig. 3A). Alternatively, excision of xanthine is significantly more efficient, resulting in a close-to-complete cleavage (Fig. 3B). The apparent rate constants were in general comparable among the double-stranded xanthine substrates but appeared to be slightly more active on C/X and G/X substrates (Table 1). However, MUG exhibited a much more robust DNA glycosylase activity on xanthine-containing DNA than uracil-containing DNA. For example, the cleavage efficiency of C/X was about 13-fold higher than that of C/U (Table 1). Interestingly, the XDG activity on C/X base pair, the likely biological substrate resulting from the direct deamination of a guanine base, is slightly stronger than other base pairs. Within the uracil-containing substrates, MUG is most active on C/U followed by G/U and then by T/U (Table 1). No activity was detected on A/U or single-stranded U (Fig. 2 and Table 1). Under the condition that the substrate was in excess (E:S ratio = 1:10), cleavage of xanthine-containing substrates reached a level of ∼20% (Fig. 3B). However, no cleavage of uracil-containing substrates was detected (data not shown). Although MUG was discovered as a uracil DNA glycosylase, these results suggested that MUG was more efficient as an XDG than as a UDG.
FIGURE 3.
Kinetic analysis of glycosylase activity of WT E. coli MUG on X- and U-containing substrates. A, shown is a time course analysis of cleavage activity on U-containing substrates. Cleavage reactions were performed as described under “Experimental Procedures” with 100 nm WT E. coli MUG protein and 10 nm substrate. E:S = 10:1. B, shown is a time course analysis of cleavage activity on X-containing substrates. S = 10 nm. ▲, E:S = 10:1; △, 1:10.
TABLE 1.
Apparent rate constants for cleavage of X and U substrates by E. coli MUG and mutants (min−1)
The reactions were performed as described under “Experimental Procedures” with 100 nm MUG and 10 nm substrates. Data are an average of two independent experiments. NA, no activity was detected under assay conditions. ss, single-stranded.
| MUG | Bottom strand | Top strand |
||||
|---|---|---|---|---|---|---|
| C | G | A | T | ss | ||
| WT | X | 0.24 | 0.23 | 0.21 | 0.21 | 0.36 |
| I17L | X | 0.024 | 0.016 | 0.013 | 0.015 | 0.022 |
| S23A | X | 0.071 | 0.046 | 0.024 | 0.027 | 0.059 |
| L144S | X | 0.012 | 0.010 | 0.011 | 0.011 | 0.012 |
| WT | U | 0.018 | 0.014 | NA | 0.0094 | NA |
| S23A | U | 0.032 | 0.031 | NA | 0.024 | NA |
| N140H | U | 0.0029 | 0.0021 | NA | 0.00034 | NA |
Identification of XDG Activity Determinants
To identify amino acid residues that may play a role in recognition of deaminated bases, we selected eight positions in motifs I and II that define the base recognition pocket for a site-directed mutagenesis study (supplemental Fig. S1). Six positions are located in motif 1, and two positions are located in motif 2. Ser-23 and Asn-140 were identified as major determinants of the XDG activities.
Alanine substitution at the S163 position in Schizosaccharomyces pombe TDG results in the complete loss of XDG activity (31). The same substitution in the equivalent Ser-23 position in E. coli MUG exhibited an interesting effect. The UDG activity on C/U, G/U, and T/U substrates was enhanced (supplemental Fig. S2B and Table 1). On the other hand, the XDG activity was reduced, ranging from 3-fold for C/X to 5-fold for G/X to 9-fold for A/X to 8-fold for T/X, and to 6-fold for single-stranded X-containing DNA (supplemental Fig. S2B and Table 1). The role that Ser-23 may play in xanthine recognition is discussed later in light of modeled MUG-X complex structure.
Two mutations were constructed at the Asn-140 position of motif 2. N140M, which mimicked human TDG, resulted in a complete loss of XDG and UDG activity (data not shown). N140H, which mimicked family 1 UDGs, only maintained a reduced UDG activity toward C/U and G/U and a substantially reduced activity toward T/U (supplemental Fig. S2C and Table 1). Strikingly, no xanthine DNA glycosylase activity was detected in N140H under the assay conditions (supplemental Fig. S2C). The Asn-140 position in motif 2 is a His residue in all UDG superfamily proteins except for the MUG/TDG family (supplemental Fig. S1). This His residue forms a hydrogen bond with the C2-carbonyl oxygen of uracil in UNG and SMUG1 (32–34). However, in the MUG structure, Asn-140 is too far (>4 Å) to make a direct contact with the C2-carbonyl oxygen (30). The effect of substitutions at the Asn-140 position was further studied by molecular modeling as described later.
In addition, I17L and L144S had a similar effect, i.e. a significant reduction in XDG activity and a loss of UDG activity (supplemental Fig. S2 and Table 1). A detailed description of these two and other mutants is provided in the supplemental data.
Binding Analysis
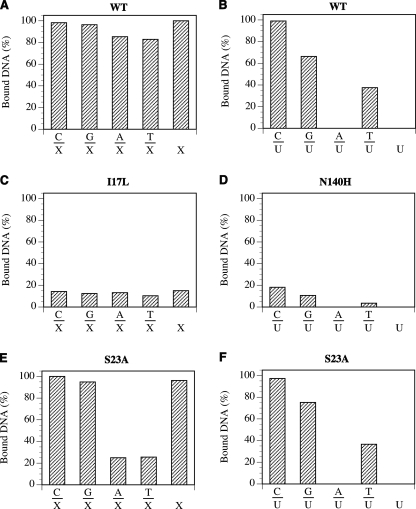
To better understand the mutational effects on the binding affinities to deaminated base-containing substrates, we performed gel mobility shift analyses. Consistent with the results obtained through the activity assays, the WT MUG interacted with all xanthine-containing substrates and C/U, G/U, and T/U to form a stable complex (Fig. 4, A and B). In keeping with its glycosylase activity, I17L was able to bind to all xanthine-containing substrates but not any uracil-containing substrates (Fig. 4C and data not shown). For S23A, the binding to uracil-containing substrates remained similar to the WT enzyme; however, binding to some xanthine-containing substrates such as A/X and T/X was reduced by ∼4-fold (Fig. 4, E and F). Consistent with the loss of XDG activity and the reduction of UDG activity, N140H showed no detectable binding affinity toward all xanthine-containing substrates and much reduced affinity toward substrates containing C/U, G/U, and T/U base pairs (Fig. 4D and data not shown). N140M showed no binding to either X- or U-containing substrates (data not shown).
FIGURE 4.
Binding of X- and U-containing DNA substrates by E. coli MUG. Gel mobility shift analysis was performed as described under “Experimental Procedures” with 500 nm protein and 50 nm substrate. Data are an average of two independent experiments. A, shown is WT MUG with X-containing DNA. B, shown is WT MUG with U-containing DNA. C, shown is I17L with X-containing DNA. D, shown is N140H with U-containing DNA. E, shown is S23A with X-containing DNA. F, shown is S23A with U-containing DNA.
DISCUSSION
MUG as a Xanthine DNA Glycosylase
E. coli MUG was initially discovered as a mismatch-specific double-stranded uracil DNA glycosylase that shared significant sequence homology with the glycosylase domain of human TDG (35). Its substrate specificity has been broadened to include 3,N4-ethenocytosine and 8-(hydroxymethyl)-3,N4-ethenocytosine (28, 36), thymine (28, 29), 5-hydroxymethyluracil (37), and more interestingly, the guanine derivative 1,N2-ethenoguanine (38). Xanthine, as a deaminated product of guanine, can be viewed structurally as a fusion of a uracil ring with an imidazole ring (Fig. 1B). Our biochemical analysis using E. coli cell extracts and purified proteins provides evidence that MUG can also act as a xanthine DNA glycosylase. Notably different from other glycosylase activities, MUG excises xanthine from both single-stranded and double-stranded DNA. It was reported that MUG was more effective in removing 3,N4-ethenocytosine than uracil (28). However, MUG did not seem to be active on single-stranded 3,N4-ethenocytosine-containing DNA (28). Another surprising finding is that the efficiency of removing xanthine is at least 10-fold greater than that of removing uracil (Table 1), suggesting that MUG is more robust as an XDG than a UDG. These results reinforce the question of whether the biological role MUG plays in a cell is more than just a uracil repair enzyme; rather, it may help repair xanthine or other damaged bases as well (5, 27, 28, 39). Although human TDG does not have XDG activity, in contrast to E. coli MUG, mammalian systems may utilize alkyladenine DNA glycosylase, SMUG1, or other DNA glycosylases to excise xanthine (9, 13).
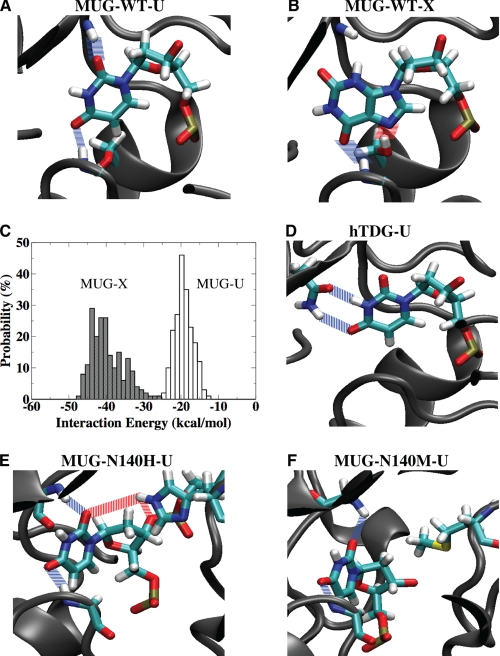
Given the highly robust xanthine DNA glycosylase activity, we set out to understand how the active site of E. coli MUG accommodates a xanthine base. Molecular models were constructed to characterize how a xanthine base may fit into the active site of E. coli MUG. Both uracil and xanthine are accommodated in the active site without significant distortion of the enzyme structure (Fig. 5, A and B). Uracil forms two hydrogen bonds with the main chains of Asn-18 and Phe-30 (Fig. 5A). On the other hand, xanthine is stabilized by a main-chain interaction with Phe-30 and a side-chain interaction with Ser-23 (Fig. 5B). The side-chain interaction of Ser-23 with N7 of xanthine apparently plays a role in xanthine recognition. Alanine substitution at the Ser-23 position, which eliminates the side-chain interaction, resulted in reduced binding affinity to A/X and T/X substrates (Fig. 4E). More profoundly, the XDG activity was also reduced, in particular with A/X and T/X substrates (supplemental Fig. S2B and Table 1). Although substitutions at the Ser residue in MUG does not cause a complete loss of XDG activity as seen in S. pombe TDG (31), the side chain of Ser-23 in MUG appears to provide a favorable interaction that facilitates the recognition of xanthine base.
FIGURE 5.
Molecular modeling of base recognition by E. coli MUG. A, interactions between WT E. coli MUG and uracil are shown. Main-chain hydrogen bonding between Asn-18, Phe-30, and uracil are shown in blue. B, interactions between WT E. coli MUG and xanthine are shown. Main-chain hydrogen bonding between Phe-30 and uracil is shown in blue. Side-chain hydrogen bonding between Ser-23 and N7 of xanthine is shown in red. C, energetics of WT E. coli MUG interactions with G/X (solid bars) and G/U base pairs (blank bars) are shown. D, interactions between human TDG and uracil are shown. Side-chain hydrogen bonding between Asn-191 and uracil are shown in blue. E, interactions between E. coli MUG-N140H and uracil are shown. Hydrogen bonding between the side chain of N140H and the uracil and that of the 3′-phosphate are shown in red. Main-chain hydrogen bonding between Asn-18, Phe-30, and uracil is shown in blue. F, interactions between E. coli MUG-N140M and uracil are shown. Main-chain hydrogen bonding between Asn-18, Phe-30, and uracil are shown in blue.
To further understand the interactions of MUG in the bound state, structural ensembles were constructed for MUG bound to uracil and xanthine through molecular dynamics simulations. Electrostatic and van der Waals interaction energies between the deaminated base (xanthine or uracil) and active site residues (within ∼8 Å of substrate) were calculated over the 400 ps of production trajectory. Based on these calculations, MUG is capable of interacting favorably in the bound state with the xanthine substrate (Fig. 5C). The average interaction energies calculated between MUG and xanthine are stronger than those between MUG and uracil (Fig. 5C). The ability to accommodate xanthine is consistent with previous studies, which show that ethenocytosine derivatives can comfortably fit into the binding pocket (29, 36). However, the activity on deaminated purine bases seems to be limited to the xanthine base. The favorable interactions with xanthine as shown in Fig. 5C may determine its specificity as a xanthine DNA glycosylase.
Comparison of E. coli MUG and Human TDG
To understand the structural differences that may underlie the functional distinction, we created bound models of hTDG to compare the differences between how hTDG interacts with uracil and xanthine. Although uracil was stabilized by side-chain interactions provided by Asn-191 (Fig. 5D), xanthine appeared to have fewer favorable interactions in the active site (data not shown). The favorable side-chain hydrogen bonding between Ser-23 of MUG and xanthine is not available because the position is occupied by an Ala residue (Ala-145) in hTDG (supplemental Fig. S1). The reduction of XDG activity observed in MUG-S23A mutant illustrates the role of this interaction in xanthine recognition.
To further investigate the different activities in the WT MUG and S23A mutant, differences in protein-DNA interaction energies were examined within canonical ensembles generated using molecular dynamics. Unsurprisingly, the results indicate that the WT MUG has stronger electrostatic and van der Waals interactions with xanthine than the S23A mutant, as a result of the loss of a hydrogen bond between the side-chain hydroxyl of Ser-23 and N7 in xanthine (supplemental Fig. S3A). Interestingly, the S23A mutant has a higher level of catalytic activity against C/U, G/U, and T/U base pairs than the WT enzyme (Table 1). This is also consistent with the outcome of MD simulation, indicating a stronger interaction between S23A and uracil (supplemental Fig. S3B). To better understand the origin of the enhancement, we performed per residue decomposition of the interaction energies. It appears that the main chains of Phe-30 and Asn-18 form stronger hydrogen bonds with uracil in the S23A mutant (supplemental Fig. S3C). To examine the impact of the Ala substitution at the Ser-23 position on the dynamic motion of the protein, we calculated the mean square fluctuation differences (ΔMSF) between the WT MUG and the S23A mutant. One change observed during the analysis was that motif 2 became more rigid in the S23A mutant (supplemental Fig. S3D). Given that motif 2 provides a wedge to occupy the space vacated by the flipped base, it is possible that a more rigid wedge can be more effective in keeping the damaged base in a flipped out conformation. This could be the result of a reduction in the entropic penalty, resulting from the wedge interaction with the DNA. These analyses are consistent with the experiments showing that the S23A mutant has a higher level of catalytic activity against C/U, G/U, and T/U base pairs than the WT enzyme (Table 1).
Asn-140 and Xanthine DNA Glycosylase Activity
The role of individual amino acids in base recognition was probed by site-directed mutagenesis. Most of the mutants still maintain activity on xanthine-containing DNA. In stark contrast, two of the Asn-140 mutants we constructed (N140M and N140H) showed no detectable XDG activity (supplemental Fig. S2C and data not shown). Substitution with Met also results in the loss of UDG activity, whereas substitution with His reduces the UDG activity. Given that the WT MUG is much more robust on xanthine than uracil, the complete loss of XDG activity while still maintaining some UDG activity is dramatic (Fig. 2 and supplemental Fig. S2C). These data underscore the role that Asn-140 may play in modulating XDG activity. Molecular models of the Asn-140 mutants bound to uracil and xanthine were constructed to investigate the interactions at position 140. In the modeled MUG-uracil complex structure, Asn-140 in MUG interacts with the phosphate backbone through hydrogen bonding (supplemental Fig. S4A), which may enhance the DNA binding. Although Asn-140 in MUG is sequentially aligned with M269 in hTDG, the structural alignment of these enzymes, performed with SPDBV (40), superimposes Asn-140 of MUG with Ser-271 of hTDG. Likewise, Ser-271 of hTDG could form equivalent hydrogen bonds with the phosphate backbone (supplemental Fig. S4B). In the modeled N140H-uracil structure, N140H appears capable of forming a hydrogen bond with the C2-keto of uracil and a weak hydrogen bond with the 3′-phosphate (Fig. 5E). The presence of these favorable interactions may underscore the weak but observable UDG activity of the N140H mutant (supplemental Fig. S2C and Fig. 4D). However, these potential interactions are lost when the uracil is substituted by xanthine (supplemental Fig. S4C), which may explain the loss of XDG activity. The loss of both XDG and UDG activity in N140M can be viewed as due to the loss of DNA backbone interactions as seen in Asn-140 of MUG and Ser-271 of hTDG or loss of direct hydrogen bonding to uracil as seen in N140H. The lack of favorable interactions with the backbone or the base may lead to the complete loss of both XDG and UDG activity (Fig. 5F).
Base Pair Stability and DNA Repair Activity
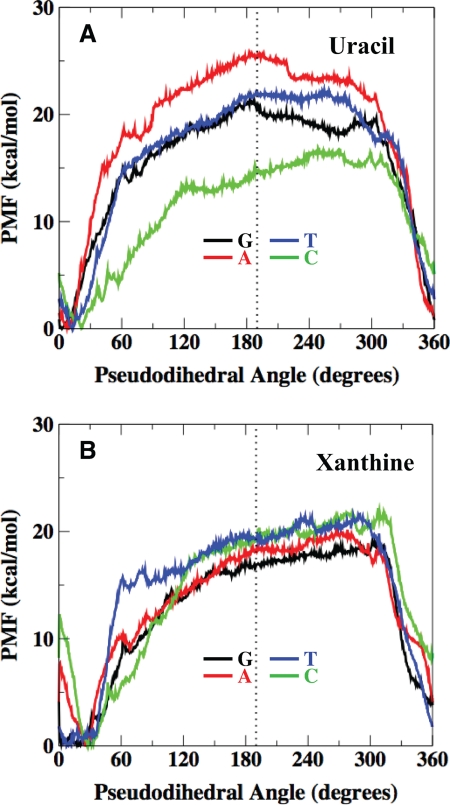
An obvious difference between the XDG and UDG activity on the double-stranded DNA is that E. coli MUG is active on all xanthine-containing DNA but only active on C/U, G/U, and T/U substrates (Figs. 2–3). It is known that mismatched uracil-containing base pairs such as G/U are thermodynamically less stable than the A/U base pair (41). However, data on the stability of xanthine-containing base pairs are limited (42). To understand whether the difference in activity is related to conformational stability of the DNA, the thermodynamic stabilities of xanthine- and uracil-containing base pairs were determined by constructing the corresponding PMF profiles of the base flipping mechanism. These PMFs provide a clear description of the thermodynamic tendencies associated with DNA base flipping.
The potentials of mean force generated through umbrella sampling indicate a greater thermodynamic tendency for deaminated bases to flip out of an isolated B-form DNA double helix relative to undamaged bases. Considering uracil flipping first, the calculated potentials of mean force demonstrate that the thermodynamic tendency for uracil to flip is significantly reduced when paired with adenine (Fig. 6A). This has been noted previously and is expected given that uracil forms a stable Watson-Crick base pair with adenine through two hydrogen bonds (43). Results further indicate that uracil has the greatest tendency to flip when paired with cytosine followed by G/U and T/U base pairs (Fig. 6A). Interestingly, the PMF data are quite consistent with the UDG activity profile reported here (Fig. 3A). These results indicate that the tendency of the mismatched uracil-containing base pairs to flip out of the helix greatly facilitates their recognition by E. coli MUG. In contrast, the umbrella sampling results indicate that the paired base has relatively little influence on the thermodynamic tendency of xanthine to flip out of the DNA. As a consequence, the four xanthine-containing base pairs show a relatively narrow difference in the free energy of flipping (Fig. 6B). The XDG activity profile is, in general, consistent with similar flipping tendencies of the xanthine-containing base pairs (Fig. 3B). The role that poor base stacking and base flipping may play in DNA lesion recognition has been discussed recently (44–46). The data presented here on the WT E. coli MUG are in general in accord with the hypothesis that spontaneous base flipping plays a role in determining the catalytic efficiency. However, one should keep in mind that how a glycosylase interacts with the damaged base in the base recognition pocket or the wedge region will also influence the catalytic efficiency of the base removal.
FIGURE 6.
PMF of uracil- and xanthine-containing base pairs along the pseudodihedral angle coordinate. Watson-Crick base pairing is ∼10°-30° pseudodihedral angle, and the flipped out state is ∼190°. A, uracil-containing base pairs are shown. B, xanthine-containing base pairs are shown.
In summary, this work reports that MUG is a robust xanthine DNA glycosylase despite the fact that it is generally considered as a uracil DNA glycosylase in the extensively studied organism E. coli. The correlation of the activity profiles with base flipping energetics underlies the role of spontaneous base flipping in initial damaged base recognition and subsequent catalysis. The ability to recognize both a deaminated pyrimidine base and a purine base underscores the plasticity of the active site, a feature that distinguishes E. coli MUG from human TDG in the same family and UNGs in family 1 of the UDG superfamily. The ability to favorably interact with a DNA base lesion provides a means to determine the specificity of DNA glycosylases.
Supplementary Material
Acknowledgments
We thank Dr. Bernard Weiss at Emory University, Dr. Ashok Bhagwat at Wayne State University, and Dr. Richard Cunningham at State University of New York for providing E. coli strains.
This work was supported, in whole or in part, by National Institutes of Health Grant GM090141 (to W. C.). This project was also supported in part by the Cooperative State Research, Education, and Extension Service/United States Department of Agriculture (SC-1700274, technical contribution no. 5805), Department of Defense Grants W911NF-05-1-0335, W911NF-07-1-0141, and W81XWH-10-1-0385 (to W. C.), National Science Foundation Grant MCB-0953783 (to B. N. D.), and SC-Life.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. 1–4.
- UDG
- uracil DNA glycosylase
- MUG
- mismatch-specific uracil DNA glycosylase
- TDG
- thymine DNA glycosylase
- hTDG
- human TDG
- XDG
- xanthine DNA glycosylase
- U
- uracil
- X
- xanthine
- O
- oxanine
- I
- hypoxanthine
- T
- thymine
- C
- cytosine
- G
- guanine
- A
- adenine
- SMUG1
- single-strand-selective monofunctional uracil DNA glycosylase
- PMF
- potentials of mean force
- UNG
- uracil N-glycosylase.
REFERENCES
- 1.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2.Shapiro R. (1981) Damage to DNA caused by hydrolysis. in Chromosome Damage and Repair (Seeberg E., Kleppe K. eds) pp. 3–18, Plenum Press, New York [Google Scholar]
- 3.Suzuki T., Yamaoka R., Nishi M., Ide H., Makino K. (1996) J. Am. Chem. Soc. 118, 2515–2516 [Google Scholar]
- 4.Huffman J. L., Sundheim O., Tainer J. A. (2005) Mutat. Res. 577, 55–76 [DOI] [PubMed] [Google Scholar]
- 5.Pearl L. H. (2000) Mutat. Res. 460, 165–181 [DOI] [PubMed] [Google Scholar]
- 6.Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. (2007) DNA Repair 6, 489–504 [DOI] [PubMed] [Google Scholar]
- 7.Hendrich B., Hardeland U., Ng H. H., Jiricny J., Bird A. (1999) Nature 401, 301–304 [DOI] [PubMed] [Google Scholar]
- 8.Vongchampa V., Dong M., Gingipalli L., Dedon P. (2003) Nucleic Acids Res. 31, 1045–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wuenschell G. E., O'Connor T. R., Termini J. (2003) Biochemistry 42, 3608–3616 [DOI] [PubMed] [Google Scholar]
- 10.Jaruga P., Dizdaroglu M. (1996) Nucleic Acids Res. 24, 1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong M., Vongchampa V., Gingipalli L., Cloutier J. F., Kow Y. W., O'Connor T., Dedon P. C. (2006) Mutat. Res. 594, 120–134 [DOI] [PubMed] [Google Scholar]
- 12.Terato H., Masaoka A., Asagoshi K., Honsho A., Ohyama Y., Suzuki T., Yamada M., Makino K., Yamamoto K., Ide H. (2002) Nucleic Acids Res. 30, 4975–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi R., Dong L., Kaulgud T., Hackett K. W., Dominy B. N., Cao W. (2009) J. Mol. Biol. 385, 761–778 [DOI] [PubMed] [Google Scholar]
- 14.Feng H., Dong L., Klutz A. M., Aghaebrahim N., Cao W. (2005) Biochemistry 44, 11486–11495 [DOI] [PubMed] [Google Scholar]
- 15.Feng H., Klutz A. M., Cao W. (2005) Biochemistry 44, 675–683 [DOI] [PubMed] [Google Scholar]
- 16.He B., Qing H., Kow Y. W. (2000) Mutat. Res. 459, 109–114 [DOI] [PubMed] [Google Scholar]
- 17.Chakravarti D., Ibeanu G. C., Tano K., Mitra S. (1991) J. Biol. Chem. 266, 15710–15715 [PubMed] [Google Scholar]
- 18.Chen J., Derfler B., Maskati A., Samson L. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7961–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samson L., Derfler B., Boosalis M., Call K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9127–9131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Kemp P. A., Thomas D., Barbey R., de Oliveira R., Boiteux S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5197–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. (1998) Current Protocols in Protein Science, John Wiley & Sons, Inc., New York [Google Scholar]
- 22.Huang J., Lu J., Barany F., Cao W. (2001) Biochemistry 40, 8738–8748 [DOI] [PubMed] [Google Scholar]
- 23.Hitchcock T. M., Dong L., Connor E. E., Meira L. B., Samson L. D., Wyatt M. D., Cao W. (2004) J. Biol. Chem. 279, 38177–38183 [DOI] [PubMed] [Google Scholar]
- 24.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comput. Chem. 4, 187–217 [Google Scholar]
- 25.MacKerell A. D., Banavali N. K. (2000) J. Comput. Chem. 21, 105–120 [Google Scholar]
- 26.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 27.O'Neill R. J., Vorob'eva O. V., Shahbakhti H., Zmuda E., Bhagwat A. S., Baldwin G. S. (2003) J. Biol. Chem. 278, 20526–20532 [DOI] [PubMed] [Google Scholar]
- 28.Saparbaev M., Laval J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T. E., Savva R., Panayotou G., Barlow T., Brown T., Jiricny J., Pearl L. H. (1998) Cell 92, 117–129 [DOI] [PubMed] [Google Scholar]
- 30.Barrett T. E., Schärer O. D., Savva R., Brown T., Jiricny J., Verdine G. L., Pearl L. H. (1999) EMBO J. 18, 6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong L., Mi R., Glass R. A., Barry J. N., Cao W. (2008) DNA Repair 7, 1962–1972 [DOI] [PubMed] [Google Scholar]
- 32.Savva R., McAuley-Hecht K., Brown T., Pearl L. (1995) Nature 373, 487–493 [DOI] [PubMed] [Google Scholar]
- 33.Wibley J. E., Waters T. R., Haushalter K., Verdine G. L., Pearl L. H. (2003) Mol. Cell 11, 1647–1659 [DOI] [PubMed] [Google Scholar]
- 34.Drohat A. C., Stivers J. T. (2000) Biochemistry 39, 11865–11875 [DOI] [PubMed] [Google Scholar]
- 35.Gallinari P., Jiricny J. (1996) Nature 383, 735–738 [DOI] [PubMed] [Google Scholar]
- 36.Hang B., Downing G., Guliaev A. B., Singer B. (2002) Biochemistry 41, 2158–2165 [DOI] [PubMed] [Google Scholar]
- 37.Baker D., Liu P., Burdzy A., Sowers L. C. (2002) Chem. Res. Toxicol. 15, 33–39 [DOI] [PubMed] [Google Scholar]
- 38.Saparbaev M., Langouët S., Privezentzev C. V., Guengerich F. P., Cai H., Elder R. H., Laval J. (2002) J. Biol. Chem. 277, 26987–26993 [DOI] [PubMed] [Google Scholar]
- 39.Lutsenko E., Bhagwat A. S. (1999) J. Biol. Chem. 274, 31034–31038 [DOI] [PubMed] [Google Scholar]
- 40.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 41.Liu P., Theruvathu J. A., Darwanto A., Lao V. V., Pascal T., Goddard W., 3rd, Sowers L. C. (2008) J. Biol. Chem. 283, 8829–8836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T., Matsumura Y., Ide H., Kanaori K., Tajima K., Makino K. (1997) Biochemistry 36, 8013–8019 [DOI] [PubMed] [Google Scholar]
- 43.Kawase Y., Iwai S., Inoue H., Miura K., Ohtsuka E. (1986) Nucleic Acids Res. 14, 7727–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker J. B., Bianchet M. A., Krosky D. J., Friedman J. I., Amzel L. M., Stivers J. T. (2007) Nature 449, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W. (2006) DNA Repair 5, 654–666 [DOI] [PubMed] [Google Scholar]
- 46.Yang W. (2008) Cell Res. 18, 184–197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.