Abstract
Wnt signaling pathways are involved in embryonic development and adult tissue maintenance and have been implicated in tumorigenesis. Dishevelled (Dvl/Dsh) protein is one of key components in Wnt signaling and plays essential roles in regulating these pathways through protein-protein interactions. Identifying and characterizing Dvl-binding proteins are key steps toward understanding biological functions. Given that the tripeptide VWV (Val-Trp-Val) binds to the PDZ domain of Dvl, we searched publically available databases to identify proteins containing the VWV motif at the C terminus that could be novel Dvl-binding partners. On the basis of the cellular localization and expression patterns of the candidates, we selected for further study the TMEM88 (target protein transmembrane 88), a two-transmembrane-type protein. The interaction between the PDZ domain of Dvl and the C-terminal tail of TMEM88 was confirmed by using NMR and fluorescence spectroscopy. Furthermore, in HEK293 cells, TMEM88 attenuated the Wnt/β-catenin signaling induced by Wnt-1 ligand in a dose-dependent manner, and TMEM88 knockdown by RNAi increased Wnt activity. In Xenopus, TMEM88 protein is sublocalized at the cell membrane and inhibits Wnt signaling induced by Xdsh but not β-catenin. In addition, TMEM88 protein inhibits the formation of a secondary axis normally induced by Xdsh. The findings suggest that TMEM88 plays a role in regulating Wnt signaling. Indeed, analysis of microarray data revealed that the expression of the Tmem88 gene was strongly correlated with that of Wnt signaling-related genes in embryonic mouse intestines. Together, we propose that TMEM88 associates with Dvl proteins and regulates Wnt signaling in a context-dependent manner.
Keywords: Microarray, NMR, Protein Structure, Protein-Protein Interactions, Signal Transduction
Introduction
Wnt signaling pathways play important roles in cell development, growth, and tumorigenesis (1–3). The cytoplasmic protein Dishevelled (Dvl/Dsh) (DVL-1, DVL-2, and DVL-3 in mammals) is a key component of Wnt-signaling pathways, which relays the signals from plasma membrane-bound Wnt receptors to downstream components (4–8). Dvl proteins consist of three highly conserved domains, the DIX (Dishevelled-axin), PDZ (post synaptic density-95, disc large and zonular occludens-1), and DEP (Dishevelled-EGL10-pleckstrin) domains (9–16). The PDZ domain modulates Wnt signaling by interacting with several proteins such as seven-transmembrane Wnt receptor Frizzled (Fz) proteins (12) and the single-transmembrane receptor Tyr kinase Ryk (3, 17–19), as well as Wnt signaling modulators such as the proteins Dapper (11) and Idax (20, 21). The Dvl PDZ domain is ∼100-amino acids-long and consists of six β-strands (βA–βF) and two α-helices (αA–αB) (22, 23). Although the binding mechanism of the Dvl PDZ domain and other proteins is not fully explored, we and others (11, 12, 21, 24) have reported that the Dvl PDZ domain recognizes the typical C-terminal PDZ-binding motifs or internal sequences of target proteins by using the same peptide-binding site on the surface of the Dvl PDZ domain, which is located between the αB-helix and the βB-strand structures. To identify additional peptide sequences that bind to the Dvl PDZ domain at the same peptide-binding site, we recently performed computational studies in combination with a NMR mapping assay (25). We showed that the VWV (Val-Trp-Val) motif is sufficient to recognize the Dvl PDZ domain. As the completion of genome sequencing projects in humans and other species have resulted in the discovery of many putative and uncharacterized proteins; we speculated that some of those proteins might have a Val-Trp-Val motif at their C termini and regulate the Wnt signaling pathways by binding Dvl proteins.
Here, we identified Dvl-binding molecules by combining bioinformatics, biophysics, and biochemistry approaches. In addition, microarray data deposited in a public database were analyzed to identify any correlation between the expression of the target TMEM88 gene and that of genes involved in Wnt signaling. As databases of gene expression profiles have recently increased dramatically in number and scope (26); we believe that a comprehensive analysis of microarray databases further facilitated the assessment of the function of a targeted gene.
Here, we report that the TMEM88 (transmembrane protein 88) interacts with the cytoplasmic Dvl protein through the PDZ domain and can attenuate Wnt/β-catenin signaling induced by the Wnt-1 ligand. The analysis of Gene Expression Omnibus data (26) from three different tissues revealed that the expression of the TMEM88 gene correlates with that of components of the Wnt pathways, suggesting that TMEM88 binds Dvl to modulate Wnt signaling.
EXPERIMENTAL PROCEDURES
Database Searches
Using the ScanProsite database, we searched for proteins containing a Val-Trp-Val motif at the C terminus in invertebrates and vertebrates (27) in December 2009. We searched for homologs by using Ensemble and HomoloGene from the National Center for Biotechnology Information (http://www.ncbi.nih.gov). The GEO database (http://www.ncbi.nlm.nih.gov/geo/) was analyzed to find any correlation between TMEM88 and Wnt signaling-related genes (26).
Expression and Purification of the 15N-labeled PDZ Domain of Mouse Dishevelled-1
The detailed method used to prepare the Dvl-1 PDZ domain has been described previously (12). In brief, to make a 15N-labeled protein, transformed cells were grown in MOPS-containing medium with 15NH4Cl (1 g/liter) as the source of 15N. Protein expression was induced by the addition of 1 mm isopropyl-1-thio-β-d-galactoside. When the A600 of the cells was ∼0.6, 16 h after induction at 16 °C, the cells were resuspended in lysis buffer (20 mm phosphate, 300 mm NaCl, pH 7.8) and sonicated. The lysate was centrifuged; the supernatant was then transferred to a column of nickel-nitrilotriacetic acid beads, and the column was washed with 50 mm imidazole. The protein was eluted by adding 200 mm imidazole in 20 mm phosphate buffer (pH 7.8) and 300 mm NaCl. The PDZ domain was further purified by chromatography on a Superdex® 75 preparative grade column (Amersham Biosciences Pharmacia) and was eluted by adding 100 mm phosphate buffer (pH 7.5) and 0.5 mm EDTA.
Peptide Synthesis and Purification
The peptide GSRSVPTPGKVWV (residues 147–159), which is referred to as TMEM88C, was derived from the C-terminal tail of human TMEM88 and chemically synthesized by staff in the Hartwell Center for Bioinformatics & Biotechnology at St. Jude Children's Research Hospital. TMEM88C was then purified by reverse-phase HPLC. Analytical HPLC was performed using a Waters 2695 Separations Module equipped with a Waters 2487 UV detector. Peptide TMEM88C was dissolved in acetonitrile: water (50:50) at a concentration of 2 mg/ml and injected onto a VYDAC® C18 5 μm column. A gradient profile was used to run the analysis starting with 100% buffer A (0.1% trifluoroacetic acid in water) to 100% buffer B (80% acetonitrile, 20% water, 0.1% trifluoroacetic acid) in 45 min. Analysis was performed at 220 nm. The peptide was 84% pure and was used without any further purification.
NMR Spectroscopy
All NMR experiments were performed at 25 °C using a Varian INOVA 600-MHz spectrometer equipped with an actively shielded xyz-axis gradient triple-resonance probe head. Samples consisted of the 15N-PDZ domain (0.2–3 mm) in 100 mm phosphate (pH 7.5) and 0.5 mm EDTA. The 15N heteronuclear single quantum coherence (HQSC)2 experiments were performed to detect chemical shift perturbations in the spectra of uniformly 15N-labeled PDZ domains of Dvl-1 with the addition of TMEM88C peptide. NMR spectra were processed using NMRpipe software (28) and analyzed by Sparky (version 3.1; T. D. Goddard and D.G. Kneller, University of California, San Francisco). Chemical shift perturbations (Δδbinding) of 1H and 15N resonances were obtained and weighted according to the following equation: Δδbinding = ((Δδ1H)2 + (Δδ15N × 0.2)2)1/2.
Fluorescence Spectroscopy
A Fluorolog-3 spectrofluorometer (Jobin-Yvon, Inc.) was used for the binding assay. A 10 × 4 mm quartz cell (Hellma®) with a minimagnetic stirring bar was used for equilibration during the titration experiments with continuous stirring to equilibrate the temperature of the sample. To record the Trp fluorescent polarization of the Val-Trp-Val tripeptide and TMEM88C, we used an excitation wavelength of 280 nm and an emission wavelength of 360 nm. The 1.4-ml samples contained ∼0.5 μm peptides in 0.1 m potassium phosphate buffer (pH 7.5). The Trp fluorescence polarization data were measured during the titration of the Dvl-1 PDZ domain and analyzed by Prism software (GraphPad, Inc.).
Cell Culture, Transfection, and Luciferase Assay
Most experimental methods used have been described previously (13). In brief, the full cDNA clone of human TMEM88 (catalog no. IOH29129) was purchased from Invitrogen. The expression of Wnt-1 and TMEM88 was driven by a cytomegalovirus promoter. HEK293 cells were maintained in 24-well plates. Cells in each well were co-transfected with 0.01 μg Lef-1 expression plasmid, 0.075 μg Lef-1 luciferase reporter plasmid, 0.05 μg GFP expression plasmid, 0.003 μg Wnt-1 expression plasmid, and various amounts of TMEM88. Cells were transfected without Wnt-1 expression plasmid as basal control. Additional lacZ plasmid was added to to equilibrate the total amount of DNA to 0.25 μg per transfection. After 1 day of growth, the cells were lysed, and GFP levels was measured by using a Wallac multicounter, and luciferase activity levels were measured using a Luciferase Reporter Gene Assay kit (high sensitivity, Roche Applied Science) according to the manufacturer's instructions; luciferase activity was normalized against the levels of GFP expression. Each experiment was carried out in duplicate, and the S.D. was calculated.
TMEM88 Knockdown Experiment
HEK293T cells were seeded in 24-well plate. 24 h later, cells in each well were transfected with 0.19 μg TOPflash, 0.01 μg GFP, and 0.14 μg TMEM88 RNAi (catalog no. TMEM88-HSS190049 from Invitrogen) by using Lipofectamine 2000 according to the manufacturer's instructions. For the control, cells in each well were transfected with 0.19 μg TOPflash (T-cell factor reporter plasmid), 0.01 μg GFP, and 0.14 μg lacZ. 66 h after transfection, cell culture medium was replaced by DMEM. After 6 h cultured in DMEM, cells were lysed and subjected to GFP and luciferase measurement according to a method described previously (29).
Xenopus Embryo Studies
Xenopus eggs were obtained from female frogs that had been injected with 500 international units of human chorionic gonadotropin (Sigma-Aldrich) and fertilized artificially. Synthesis and microinjection of mRNAs were carried out as described previously (30, 31).
To investigate the subcellular distribution of TMEM88, we generated a fusion of the Myc epitope to the N-terminal region of TMEM88, Myc-tagged TMEM88, and injected the Myc-tagged mRNA into the animal pole region of Xenopus embryos at the four-cell stage. Animal cap explants were dissected at the late blastula stage and fixed. Immunofluorescent staining was performed using an anti-Myc antibody and an FITC-conjugated secondary antibody. In addition, the effect of TMEM88 on the subcellular distribution of the GFP-tagged Xdsh (Dsh-GFP) in Xenopus was examined. Dsh-GFP mRNA was injected alone or co-injected with mRNA of human TMEM88 or TMEM88-ΔC that lacks the PDZ-binding motif at the last C-terminal tail into animal blastomerase at the four to eight cell stage. GFP fluorescence was assessed in animal explants. Localization of myc-TMEM88 or Dsh-GFP protein in Xenopus was examined by fluorescence confocal microscopy.
For the luciferase assay, the Siamois promoter-driven reporter DNA construct (Sialuc, 400 pg) (12, 31, 32) was injected alone or coinjected with mRNAs of Xdsh, β-catenin, and Myc-tagged TMEM88 (100 pg each) into the animal pole region of Xenopus embryos at the two-cell stage. Injected embryos were cultured, and animal cap explants were dissected at the late blastula stage, and luciferase assays (Promega) were performed by preparing cell lysates from 10 explants and measuring luciferase activity in a Lumat LB 9507 luminometer. For the secondary axis assay, Xdsh mRNA (500 pg) or Xdsh mRNA (500 pg) together with Myc-tagged TMEM88 (500 pg) were injected into the ventrovegetal region of embryos at the four-cell stage, and injected embryos were then cultured until they reached the larval stage.
Analysis of GEO Profiling
Three microarray data sets were selected from GEO that had high TMEM88 expression (replicates) and were performed on current Affymetrix GeneChip mouse genome 430 2.0 arrays (26). To stabilize variance, the MAS 5.0 signal was natural log-start transformed by the following formula: ln(MAS 5.0 signal + 20). The transformed expression of each probe set on the array was correlated linearly with the transformed signal of TMEM88 by using Pearson's coefficient (ρ) using a script written for STATA software (version 10.1, College Station, TX) for each of the experiments. In brief, a correlation sums the gene expression differences between any two genes across multiple arrays and then divides that sum by the total number of array experiments examined to give the Pearson's ρ. ρ values, which measure the degree of association between variables, range from −1 to 1.
RESULTS
Identification of Proteins That Have a Val-Trp-Val Motif at the C-terminal Tail
The Dvl PDZ domain recognizes the Val-Trp-Val tripeptide (25); thus, we hypothesized that by interacting with the Dvl PDZ domain, some of the proteins containing the motif at the C terminus would be novel Wnt-signaling modulators. We, therefore, searched the ScanProsite database for such proteins and identified 294 proteins from invertebrates and vertebrates (protein fragments were excluded, data not shown). Among these proteins, six are human proteins (Table 1). To ascertain possible binding partners, we searched for data about these proteins on the protein-protein interaction databases (33–35), but the results were negative. We then used the on-line database Ensemble to examine the expression patterns and subcellular localizations of the five proteins (Table 1). Because the human DVL proteins are located in the cytoplasm, ubiquitously expressed in fetal and adult tissues and not secreted (10, 36), we focused on TMEM88, a transmembrane protein, as the most likely binding partner of the Dvl PDZ domain. TMEM88 is a two-transmembrane-type protein; residues 44–66 comprise the first transmembrane domain, and residues 86–108 comprise the second (Fig. 1A). Additional database searches revealed that, in humans, TMEM88 (Q6PEY1) is expressed in placenta (28.8%), embryonic tissue (10.02%), spleen (9.66%), muscle (8.48%), heart (6.64%), bone (6.50%), eye (5.65%), kidney (4.75%), and other tissues (4.67%). (NCBI Gene ID 92162). TMEM88 is also well conserved in mammals (Fig. 1, B and C). Furthermore, ensemble database mining showed that TMEM88 (Q9D0N8) is expressed throughout the mouse embryo (data not shown).
TABLE 1.
Human proteins that contain the VWV motif at the C terminus
Accession numbers are from the ScanProsite database. aa, amino acid; N/A, not available.
| Accession no. | Description | Tissue | Subcellular location | Length (aa) | C-terminal sequence |
|---|---|---|---|---|---|
| P02743 | Serum amyloid P-component precursor (9.5 S α-1 glycoprotein) | Serum and urine | Extracellular | 223 | VIIKPLVWV |
| Q5H8X8 | Urotensin 2 (hormone activity) | N/A | Extracellular | 139 | IFKNYVWV |
| Q6PEY1 | Hypothetical protein TMEM88 | Placenta | Membrane | 159 | PTSGKVWV |
| Q6ZNZ5 | Hypothetical protein FLJ26833 | N/A | N/A | 121 | CVMVIVWV |
| Q9NTI7 | Hypothetical protein C1or f183 | Brain | N/A | 297 | DINTAVWV |
FIGURE 1.
Identification and characterization of the TMEM88 protein. A, schematic of the TMEM88 protein. The two transmembrane domains, TM1 and TM2, are indicated in gray, and the PDZ-binding domain is indicated in black. B, amino acid sequence alignments of hTMEM88, mTMEM88, rTMEM88, cTMEM88, and bTMEM88 by using CLUSTW. h, m, r, c, and b represent human, mouse, rat, Canis familiaris (dog), and Bos taurus (cow), respectively. The asterisks indicate identical residues in alignment; the colons indicate that conserved substitutions have been observed; and periods indicate semiconserved substitutions have been observed. C, percent identity of TMEM88 protein among the species. TM, transmembrane; C, C terminus; N, N terminus.
Direct Interaction between C Terminus of TMEM88 and Dvl PDZ Domain
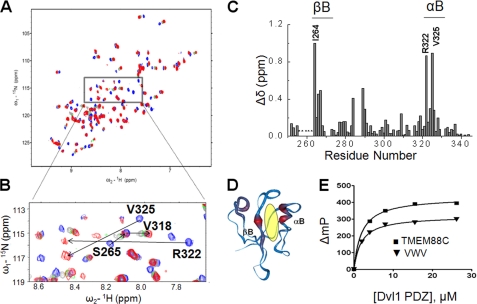
Although the Val-Trp-Val tripeptide is bound to the Dvl PDZ domain (25), interactions with other upstream amino acid residues at the C terminus of TMEM88 may be important to regulate the interaction of the Dvl PDZ domain. Therefore, we characterized the interaction between the Dvl PDZ domain and a 13-mer peptide from the C terminus of human TMEM88 (TMEM88C) by performing NMR titration experiments in which the 1H,15N-HSQC spectra of the uniformly 15N-labeled PDZ domain of Dvl were recorded, whereas unlabeled TMEM88C was added gradually (Fig. 2A) (37). The expanded region of the 15N-HSQC in Fig. 2B represents the “fingerprint” region corresponding to the important binding-site residues in the Dvl-1 PDZ domain. During titration, some peaks disappeared and reappeared, indicating that the binding was in the intermediate state on the NMR time scale (37). Based on the peak assignments of the Val-Trp-Val-bound state of Dvl-1 PDZ (25), we were able to determine the resonance signals of the 15N Dvl-1 PDZ domain bound to the TMEM88C peptide. The largest perturbation of signals was found in residue Ile-264 (Δδ binding = 0.591 ppm) on the βB-strand and in residues Arg-322 (Δδ binding = 0.513 ppm) and Val-325 (Δδ binding = 0.527 ppm) on the αB-helix of the Dvl-1 PDZ domain at the saturated concentration (Fig. 2B). The normalized chemical shift changes (Δδbinding) for each backbone amide are shown in Fig. 2C. In addition, the backbone worm structure of the Dvl-1 PDZ domain was generated to show the effect of binding the TMEM88C peptide (Fig. 2D) and indicated that TMEM88C binds to the conventional peptide-binding groove between the αB-helix and βB-sheet structures in the PDZ domain of Dvl-1 as expected. We observed little difference in the chemical shift perturbations of the Val-Trp-Val tripeptide and TMEM88C caused by binding to the Dvl PDZ domain (25). The difference of Δδbinding (ΔΔδbinding) of the Dvl-1 PDZ domain for the two bound peptides was <0.03, indicating that the last three residues at the extreme C terminus of TMEM88 would be crucial to determine the binding.
FIGURE 2.
Direct binding of the TMEM88C peptide to the PDZ domain of Dvl. A, 1H,15N HSQC-spectra of the Dvl-1 PDZ domain and with varying concentrations of the 13-mer TMEM88C peptide. B, the extended 1H,15N HSQC spectra of the Dvl-1 PDZ domain as a function of the concentration of the 13-mer TMEM88C peptide (blue, 1:0; gray, 1:1; green, 1:3; purple, 1:5; and red, 1:8). C, normalization of the chemical shift perturbation by Ile-264 (Δδ = 0.591 ppm). D, the worm representation of the backbone structure of the Dvl-1 PDZ domain (Protein Data Bank code 1MC7). The thickness of the worm structure was normalized to the chemical shift perturbation datum of Ile-264 (red, high; blue, low). Insight II software was used to prepare this model. E, the binding affinity (KD) of the Val-Trp-Val tripeptide (triangles) and that of the TMEM88C peptide (squares) to the Dvl-1 PDZ domain. The Trp fluorescence polarization of both peptides was measured during the titration of the Dvl-1 PDZ domain. mP, polarization × 1000.
To further examine the binding between TMEM88C and the Dvl-1 PDZ domain, we measured Trp fluorescence polarization of the peptide during its titration with Dvl-1 PDZ domain, which does not contain a Trp residue. Changes in Trp fluorescence polarization were used to estimate the binding affinity (KD) of the tripeptide and TMEM88C for the Dvl-1 PDZ domain (Fig. 2E). As predicted by results from the NMR experiments, the binding affinity of TMEM88C (KD = 2.0 ± 0.2 μm) and that of the tripeptide Val-Trp-Val (KD = 1.9 ± 0.3 μm) to the Dvl-1 PDZ domain were comparable.
TMEM88 Attenuates Wnt1 Ligand-induced Signaling
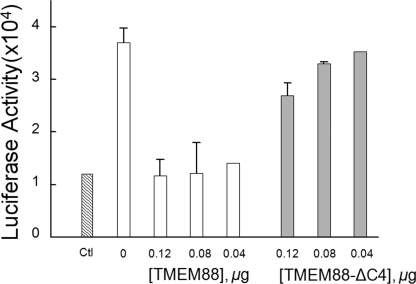
Because TMEM88 binds to the Dvl PDZ domain in vitro, we examined its effect on Wnt/β-catenin signaling. Because a direct interaction of the Dvl PDZ domain with the internal sequence of Fz protein is essential to activate Wnt signaling (12, 16, 38), we reasoned that if TMEM88 binds to the Dvl PDZ domain in cells, then overexpressing TMEM88 might disrupt the Dvl-Fz interaction at the plasma membrane region, which, in turn, would suppress Wnt/β-catenin signaling. To test this hypothesis, we employed a luciferase-based cell assay (29). In HEK293 cells transfected with the Wnt-1 plasmid, canonical Wnt signaling was activated, indicated by increased luciferase activity (Fig. 3) (13). When TMEM88 was co-expressed with Wnt-1 in the cells, Wnt-1-induced activation of luciferase was attenuated (Fig. 3). The increased amount of TMEM88 expression plasmid resulted in the substantial attenuation of Wnt-1-induced luciferase activity, indicating that the TMEM88 protein blocks Wnt signaling, presumably by disrupting the Dvl-Fz interaction in cells. To confirm this result, we performed the same experiment with TMEM88ΔC4, which lacks four amino acid residues at the extreme C terminus (Fig. 3). The loss of the PDZ-binding motif of TMEM88 causes little inhibitory effect on Wnt-1-induced signaling in a dose-dependent manner, indicating that the binding of TMEM88 to the Dvl-1 PDZ domain is essential to block Wnt signaling.
FIGURE 3.
TMEM88 attenuates Wnt-1-induced β-catenin signaling. HEK293 cells were seeded in 24-well plates 24 h before transfection, and the cells in each well were co-transfected with 0.01 μg Lef-1 expression plasmid, 0.075 μg Lef-1 luciferase reporter plasmid, 0.05 μg GFP expression plasmid, 0.003 μg Wnt-1 expression plasmid, and various amounts of either TMEM88 (blank boxes) or TMEM88ΔC4 (light gray boxes). As controls (Ctl), the cells were transfected with the same preparation in the absence of the Wnt-1 expression plasmid. The lacZ plasmid was added to equilibrate the total amount of 0.25 μg DNA per transfection. After 1 day of growth, the cells were lysed, and GFP levels and luciferase activities were determined. Each experiment was carried out in duplicate, and error bars represent the S.D.
Knockdown of TMEM88 Increases Wnt Activity
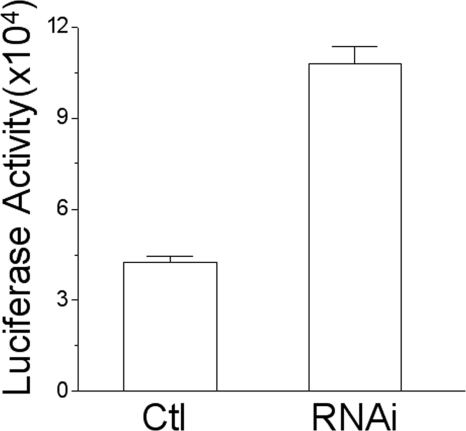
Because TMEM88 is expressed in kidney and can inhibit the Wnt-1-induced signaling in HEK293 cells, we used RNAi to suppress the endogenous expression of TMEM88 in HEK293 cells to investigate the effect of TMEM88 knockdown on Wnt activity. By using the luciferase report assay (29), we found that, comparing to the control, the Wnt activity was increased >2-fold in the TMEM88-suppressed cells (Fig. 4), indicating that endogenous TMEM88 inhibits canonical Wnt signaling in HEK293 cells.
FIGURE 4.
TMEM88 knockdown increases Wnt activity. HEK293T cells were seeded in a 24-well plate. 24 h later, cells in each well were transfected with 0.19 μg TOPFlash, 0.01 μg GFP, and 0.14 μg TMEM88 RNAi by using Lipofectamine 2000. For the control (Ctl), cells in each well were transfected with 0.19 μg TOPFlash, 0.01 μg GFP, and 0.14 μg lacZ. 66 h after transfection, cell culture medium were replaced by DMEM. After 6 h of culture in DMEM, cells were lysed and subjected to GFP and luciferase measurement. The luciferase activity was increased about two times by knockdown of TMEM88 by RNAi.
Subcellular Localization of TMEM88 Protein and Its Effect on Wnt Signaling in Xenopus
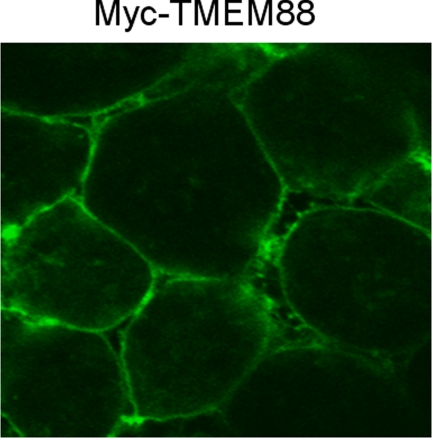
Because TMEM88 was predicted to be a membrane protein, we decided to investigate the subcellular distribution of TMEM88 in a Xenopus embryo system. We first generated a fusion of the Myc epitope to the N-terminal region of TMEM88, Myc-tagged TMEM88, and injected the mRNA of Myc-tagged into the animal pole region of Xenopus embryos at the four-cell stage. Animal cap explants were dissected at the late blastula stage and fixed. Then, immunofluorescent stainings were performed and localization of Myc-TMEM88 protein in Xenopus was examined by fluorescence confocal microscopy. As showed in Fig. 5, Myc-TMEM88 proteins were essentially localized to the cell membrane; indeed, TMEM88 is a membrane protein.
FIGURE 5.
Subcellular localization of TMEM88 in Xenopus. Representative fluorescence confocal microscopy image of Myc-TMEM88 in the animal cap explants after they were dissected at the late blastula stage and fixed; Myc-TMEM88 is localized in the cell membrane.
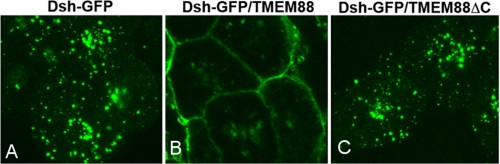
We then examined the effect of TMEM88 on the subcellular distribution of Xdsh-GFP in Xenopus. Xdsh-GFP mRNA was injected alone or co-injected with TMEM88 mRNA into animal blastomerase at the four to eight cell stages. Xdsh-GFP was found in a punctuate cytoplasm distribution in animal cap cells (Fig. 6A) (39). Co-injection of TMEM88 led to the recruitment of Xdsh-GFP to the cell membrane (Fig. 6B). Remarkably, TMEM88ΔC that lacks the PDZ binding motif at the C terminus failed to recruit Xdsh-GFP to the cell membrane (Fig. 6C). Together, the data suggest that TMEM88 binds to Dvl protein through its last C terminus residues and the binding recruits Dvl to the membrane.
FIGURE 6.
Direct interaction of Xdsh-GFP and TMEM88 in Xenopus. Dsh-GFP RNA (0.5 ng) was injected alone or with TMEM88 RNA (1 ng) or TMEM88ΔC RNA(1 ng) into animal blastomerase at the four to eight cell stage. GFP fluorescence was assessed in animal cap explants. A, Xdsh-GFP was found in a punctuate cytoplasmic distribution. B, co-injection of TMEM88 leads to the recruitment of Dsh-GFP to the cell membrane. C, co-injection of TMEM88ΔC did not change the subcellular distribution of Dsh-GFP, showing that the last C-terminal residue of TMEM88 is important to the subcellular distribution of Dsh-GFP.
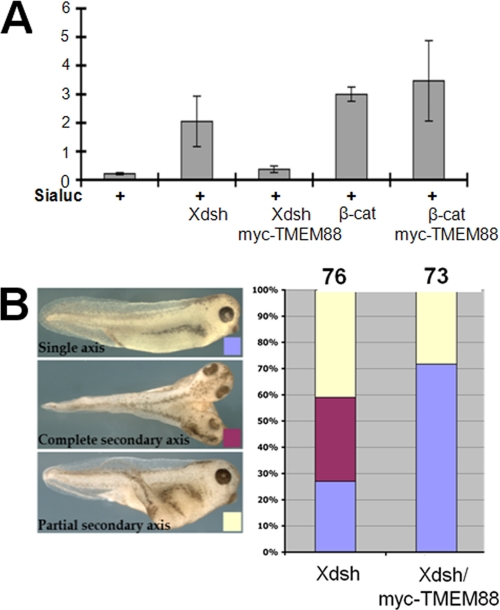
To assess the role of TMEM88 in the Wnt signaling in vivo, we used a previously established luciferase assay (12, 31, 32). Xenopus Wnt target gene Siamois promoter-driven luciferase reporter construct was injected alone or co-injected with synthetic Xdsh mRNA, Xdsh mRNA together with Myc-tagged TMEM88 mRNA, β-catenin mRNA, and β-catenin mRNA plus Myc-tagged TMEM88 mRNA, respectively, in the animal pole region at the two-cell stage (31). Injected embryos were cultured and animal cap explants were dissected at the late blastula stage, and luciferase assays (Promega) were performed by preparing cell lysates from 10 explants and measuring luciferase activity in a Lumat LB 9507 luminometer. Consistent with the results in the cell assay, TMEM88 inhibited Siamois promoter-driven luciferase activity induced by Xdsh but not by β-catenin (Fig. 7A). This result indicates that TMEM88 inhibits canonical Wnt signaling at the level of Xdsh.
FIGURE 7.
The effect of TMEM88 on canonical Wnt signaling in Xenopus. A, Siamois reporter construct and synthetic mRNAs corresponding to Xdsh (1pg) and β-catenin (β-cat; 100 pg) were injected into the animal pole region of four-cell stage Xenopus embryos, and ectodermal explants were dissected at the late blastula stage for a luciferase activity assay. Luciferase activity induced by Xdsh was examined. TMEM88 inhibits canonical Wnt signaling induced by Xdsh but not by β-catenin, indicating that TMEM88 blocks the signaling at the level of Xdsh. B, summary of the effect of TMEM88 on the formation of the secondary axis induced by Xdsh. TMEM88 efficiently blocks the formation of the secondary axis induced by Xdsh in Xenopus embryos. Sialuc, plasmid encoding luciferase driven by the Siamois promoter.
To further examine the role of TMEM88, we also employed the Xenopus secondary axis assay (12). As showed in Fig. 7B, Xdsh mRNA injected into the ventro-vegetal region of a Xenopus ectodermal explant at the four-cell stage induced a complete secondary axis in ∼30% of Xenopus embryos and a partial secondary axis in ∼40% of Xenopus embryos. Co-injection of embryos with equal amounts of mRNAs of Xdsh and Myc-tagged TMEM88 abolished the formation of a complete secondary axis, and only ∼30% of embryos formed a partial secondary axis. The data also suggest that TMEM88 can regulate Wnt signaling pathway.
TMEM88 Gene Expression Profile
Given that TMEM88 suppresses Wnt-1-induced β-catenin signaling in cells and in Xenopus embryos, we wondered whether the expression of the TMEM88 gene is correlated with that of other genes involved in Wnt signaling. To determine this, we reanalyzed the TMEM88 gene expression profile on the GEO database (26). We downloaded raw data from three embryonic mouse tissues: cardiac (GEO accession no. GSE1479), colon (GEO accession no. GSE5261), and small intestines (GEO accession no. GSE6383) and examined the Pearson correlations of TMEM88 with the upstream components of Wnt signaling.
We detected strong Pearson correlations of TMEM88 expression with that of several Wnt signaling-related genes in the embryonic small intestines: Fzd1 (ρ = 1.00), Fzd2 (ρ = 1.00), Fzd7 (ρ = 0.86), Fzd9 (ρ = 0.90), Frzb (ρ = 0.89), Dvl-1 (ρ = 0.82), Dvl-2 (ρ = 0.89), and Dvl-3 (ρ = 0.92) (supplemental Table S1). We also observed modest Pearson correlations between the expression of TMEM88 and that of the following Wnt signaling-related genes in other tissues: embryonic mouse cardiac tissue, ROR1 (ρ = 0.61); and the colon, Fzd1 (ρ = −0.67), Fzd2 (ρ = −0.49), and Frzb (ρ = −0.63) (supplemental Table S1). These results suggest that TMEM88 conditionally modulates Wnt signaling-related gene expression.
DISCUSSION
During the course of this work, Stiffler et al. (40) suggested that TMEM88 protein might be a Dvl-3 PDZ-binding candidate. In this report, we demonstrated that TMEM88 is a novel Dvl-1-binding protein that modulates Wnt signaling. The chemical shift perturbations of the 15N-labeled PDZ domain of Dvl-1 show that the C terminus of TMEM88 occupies the binding groove of the Dvl-1 PDZ domain, as do the known Dvl PDZ-binding partners Fz and Dapper (12, 25, 41). The binding affinity of TMEM88C to the Dvl-1 PDZ domain obtained by analyzing fluorescence data is similar to that of known Dvl PDZ-binding partners (12, 25, 41), suggesting that TMEM88 may compete with known Dvl-binding partners.
Sequence analysis suggested that TNEM88 has two transmembrane domains, and our studies in Consistent In Xenopus confirmed that TMEM88 is a membrane protein. We showed that the interaction between TMEM88 and Dvl protein could recruit Dvl to the membrane.
Both the cell-based and Xenopus assays showed that TMEM88 disrupts canonical Wnt signaling. During canonical Wnt/β-catenin signaling, the Wnt ligand binds to its co-receptors, Fz and LRP5/6 (a single-span transmembrane LDL receptor-related protein 5/6), resulting in the activation of cytoplasmic Dvl proteins (7). The activated Dvl proteins then relay the Wnt signals to the downstream components (5). Moreover, we reported previously that the protein-protein interaction between the membrane-bound Fz receptor and the PDZ domain of Dvl plays a role in conveying Wnt signals downstream (12, 22). Therefore, the attenuation of Wnt-1-induced signaling by TMEM88 implies that the C-terminal region of TMEM88 locates in the cytoplasmic region and binds to the Dvl PDZ domain, which leads to the disruption of the Fz-LRP5/6-Dvl complex, thus affecting canonical Wnt signaling. Consistent with this hypothesis, TMEM88 knockdown in HEK293 cells increased Wnt activity. Additionally, the reanalysis of microarray data provided evidence that the expression of TMEM88 was closely associated with the genes involved in Wnt signaling in embryonic mouse intestines.
The Dvl proteins are key players in Wnt signaling pathways, and >20 binding partners of these proteins have been reported (4, 5). For example, Dapper, Idax, and Naked cuticles inhibit canonical Wnt signaling by binding to Dvl proteins (4, 5, 11, 20); however, Ryk and Diversin function in the noncanonical Wnt-signaling pathway by interacting with Dvl (18, 42). Because Dvl protein-protein interactions regulate a variety of biological processes (5, 43), identifying novel Dvl-binding partners is of great importance. This study is a useful example of how a combinatorial approach can be broadly applied to reduce the number of candidate proteins when searching for novel Dvl-binding partners.
Finally, TMEM88 may play an important role in diseases, including cancers. Human TMEM88 gene is mapped to chromosome 17p13.1, from position 7,699,108 to 7,700,142 (Ensemble database) (supplemental Fig. S1). Several studies have shown that the loss of heterozygosity at 17p13.1 appears to be associated with an adverse disease course (44–48). Although most of these analyses addressed the role of TP53, further study of TMEM88 may be interesting because the two genes between TP53 and TMEM88 are separated by only ∼200 kb.
Supplementary Material
Acknowledgments
We thank Drs. Weixing Zhang and the late Charles Ross for support with NMR instruments and computer resources; Dr. Patrick Rodrigues and Robert Cassell for peptide synthesis; and Drs. Cherise Guess and Angela J. McArthur for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM081492 and by National Institutes of Health, NCI Cancer Center Support Grant CA21765. This work was also supported by the American Lebanese Syrian Associated Charities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- HQSC
- heteronuclear single quantum coherence
- GEO
- Gene Expression Omnibus.
REFERENCES
- 1.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Gen. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 2.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 3.Angers S., Moon R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 4.Wharton K. A., Jr. (2003) Dev. Biol. 253, 1–17 [DOI] [PubMed] [Google Scholar]
- 5.Wallingford J. B., Habas R. (2005) Development 132, 4421–4436 [DOI] [PubMed] [Google Scholar]
- 6.Malbon C. C., Wang H. Y. (2006) Curr. Top. Dev. Biol. 72, 153–166 [DOI] [PubMed] [Google Scholar]
- 7.Zeng X., Huang H., Tamai K., Zhang X., Harada Y., Yokota C., Almeida K., Wang J., Doble B., Woodgett J., Wynshaw-Boris A., Hsieh J. C., He X. (2008) Development 135, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan W., Choi S. C., Wang H., Qin Y., Volpicelli-Daley L., Swan L., Lucast L., Khoo C., Zhang X., Li L., Abrams C. S., Sokol S. Y., Wu D. (2008) Science 321, 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz-Romond T., Fiedler M., Shibata N., Butler P. J., Kikuchi A., Higuchi Y., Bienz M. (2007) Nat. Struct. Mol. Biol. 14, 484–492 [DOI] [PubMed] [Google Scholar]
- 10.Semënov M. V., Snyder M. (1997) Genomics 42, 302–310 [DOI] [PubMed] [Google Scholar]
- 11.Cheyette B. N., Waxman J. S., Miller J. R., Takemaru K., Sheldahl L. C., Khlebtsova N., Fox E. P., Earnest T., Moon R. T. (2002) Dev. Cell 2, 449–461 [DOI] [PubMed] [Google Scholar]
- 12.Wong H. C., Bourdelas A., Krauss A., Lee H. J., Shao Y., Wu D., Mlodzik M., Shi D. L., Zheng J. (2003) Mol. Cell 12, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong H. C., Mao J., Nguyen J. T., Srinivas S., Zhang W., Liu B., Li L., Wu D., Zheng J. (2000) Nat. Struct. Biol. 7, 1178–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan W. J., Pang S. Z., Huang T., Guo H. Y., Wu D., Li L. (2004) Cell Research 14, 324–330 [DOI] [PubMed] [Google Scholar]
- 15.Park T. J., Gray R. S., Sato A., Habas R., Wallingford J. B. (2005) Curr. Biol. 15, 1039–1044 [DOI] [PubMed] [Google Scholar]
- 16.Simons M., Gault W. J., Gotthardt D., Rohatgi R., Klein T. J., Shao Y., Lee H. J., Wu A. L., Fang Y., Satlin L. M., Dow J. T., Chen J., Zheng J., Boutros M., Mlodzik M. (2009) Nat. Cell Biol. 11, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshikawa S., McKinnon R. D., Kokel M., Thomas J. B. (2003) Nature 422, 583–588 [DOI] [PubMed] [Google Scholar]
- 18.Lu W., Yamamoto V., Ortega B., Baltimore D. (2004) Cell 119, 97–108 [DOI] [PubMed] [Google Scholar]
- 19.Kim G. H., Her J. H., Han J. K. (2008) J. Cell Biol. 182, 1073–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hino S., Kishida S., Michiue T., Fukui A., Sakamoto I., Takada S., Asashima M., Kikuchi A. (2001) Mol. Cell Biol. 21, 330–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.London T. B., Lee H. J., Shao Y., Zheng J. (2004) Biochem. Biophys. Res. Commun. 322, 326–332 [DOI] [PubMed] [Google Scholar]
- 22.Wang N. X., Lee H. J., Zheng J. J. (2008) Drug News Perspect. 21, 137–141 [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H. J., Zheng J. J. (2010) Cell Commun. Signal. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Appleton B. A., Wiesmann C., Lau T., Costa M., Hannoush R. N., Sidhu S. S. (2009) Nat. Chem. Biol. 5, 217–219 [DOI] [PubMed] [Google Scholar]
- 25.Lee H. J., Wang N. X., Shao Y., Zheng J. J. (2009) Bioorg. Med. Chem. 17, 1701–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Muertter R. N., Edgar R. (2009) Nucleic Acids Res. 37, D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hulo N., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P. S., Pagni M., Sigrist C. J. (2006) Nucleic Acids Res. 34, D227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29.Li L., Yuan H., Xie W., Mao J., Caruso A. M., McMahon A., Sussman D. J., Wu D. (1999) J. Biol. Chem. 274, 129–134 [DOI] [PubMed] [Google Scholar]
- 30.Shan J., Shi D. L., Wang J., Zheng J. (2005) Biochemistry 44, 15495–15503 [DOI] [PubMed] [Google Scholar]
- 31.Grandy D., Shan J., Zhang X., Rao S., Akunuru S., Li H., Zhang Y., Alpatov I., Zhang X. A., Lang R. A., Shi D. L., Zheng J. J. (2009) J. Biol. Chem. 284, 16256–16263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umbhauer M., Djiane A., Goisset C., Penzo-Méndez A., Riou J. F., Boucaut J. C., Shi D. L. (2000) EMBO J. 19, 4944–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader G. D., Betel D., Hogue C. W. (2003) Nucleic Acids Res. 31, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han K., Park B., Kim H., Hong J., Park J. (2004) Bioinformatics. 20, 2466–2470 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki H., Saito R., Kanamori M., Kai C., Schönbach C., Nagashima T., Hosaka J., Hayashizaki Y. (2003) Genome Res. 13, 1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray R. S., Bayly R. D., Green S. A., Agarwala S., Lowe C. J., Wallingford J. B. (2009) Dev. Dyn. 238, 2044–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellecchia M., Sem D. S., Wüthrich K. (2002) Nature Reviews Drug Discovery 1, 211–219 [DOI] [PubMed] [Google Scholar]
- 38.Schwarz-Romond T., Merrifield C., Nichols B. J., Bienz M. (2005) J. Cell Sci. 118, 5269–5277 [DOI] [PubMed] [Google Scholar]
- 39.Rothbächer U., Laurent M. N., Deardorff M. A., Klein P. S., Cho K. W., Fraser S. E. (2000) EMBO J. 19, 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H. J., Wang N. X., Shi D. L., Zheng J. J. (2009) Angew. Chem. Int. Ed Engl. 48, 6448–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenny A., Reynolds-Kenneally J., Das G., Burnett M., Mlodzik M. (2005) Nat. Cell Biol. 7, 691–697 [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Hamblet N. S., Mark S., Dickinson M. E., Brinkman B. C., Segil N., Fraser S. E., Chen P., Wallingford J. B., Wynshaw-Boris A. (2006) Development 133, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang H., Sloan S., Li D., Keith Stewart A. (2004) Br. J. Haematol. 127, 280–284 [DOI] [PubMed] [Google Scholar]
- 45.Jung H. L., Wang K. C., Kim S. K., Sung K. W., Koo H. H., Shin H. Y., Ahn H. S., Shin H. J., Cho B. K. (2004) J. Neurooncol. 67, 41–46 [DOI] [PubMed] [Google Scholar]
- 46.Flavin R. J., Smyth P. C., Laios A., O'Toole S. A., Barrett C., Finn S. P., Russell S., Ring M., Denning K. M., Li J., Aherne S. T., Sammarae D. A., Aziz N. A., Alhadi A., Sheppard B. L., Lao K., Sheils O. M., O'Leary J. J. (2009) Mod. Pathol. 22, 197–205 [DOI] [PubMed] [Google Scholar]
- 47.Dolan K., Garde J., Walker S. J., Sutton R., Gosney J., Field J. K. (1999) Hum. Pathol. 30, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 48.Cousin P., Billotte J., Chaubert P., Shaw P. (2000) Genomics 63, 60–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.