Abstract
Agonist-induced internalization of G protein-coupled receptors plays an important role in signal regulation. The underlying mechanisms of the internalization of the human neuropeptide Y2 receptor (hY2R), as well as its desensitization, endocytosis, and resensitization are mainly unknown. In the present study we have investigated the role of carboxyl-terminal (C-terminal) Ser/Thr residues and acidic amino acids in regulating receptor internalization, arrestin interaction, and recycling by fluorescence microscopy, cell surface enzyme-linked immunosorbent assay, and bioluminescence resonance energy transfer in several cell lines. Strikingly, C-terminal truncation mutants revealed two different internalization motifs. Whereas a distal motif 373DSXTEXT379 was found to be the primary regulatory internalization sequence acting in concert with arrestin-3, the proximal motif 347DXXXSEXSXT356 promoted ligand-induced internalization in an arrestin-3-independent manner. Moreover, we identified a regulatory sequence located between these internalization motifs (357FKAKKNLEVRKN368), which serves as an inhibitory element. We found that hY2R recycling is also governed by structural determinants within the proximal internalization motif. In conclusion, these results indicate that the hY2R C terminus is involved in multiple molecular events that regulate internalization, interaction with arrestin-3, and receptor resensitization. Our findings provide novel insights into complex mechanisms of controlled internalization of hY2R, which is likely applicable to other GPCRs.
Keywords: G Protein-coupled Receptors (GPCR), Hormone Receptors, Neuropeptide, Peptide Hormones, Receptor Endocytosis
Introduction
G protein-coupled receptors (GPCRs)2 constitute the largest family of cell-surface receptors with ∼800 known human subtypes. Members of this family share a common architecture of seven membrane-spanning α-helices connected by extra- and intracellular loops. GPCRs can be activated by a wide variety of extracellular stimuli and regulate diverse physiological processes (1). Because they are responsible for a multitude of cellular responses and their dysfunction can result in many diseases (2–4), this receptor family represents highly important pharmaceutical targets. More than one-third of the currently available therapeutics act on GPCRs (1). Whereas antagonists mainly prevent receptor interactions with endogenous ligands, agonist drugs might lead to distinct effects in addition to receptor activation, e.g. functional antagonism by receptor removal from the surface (5), direct intracellular cross-signaling (6), and may behave as biased ligands (7). Current information about the complex intracellular network of GPCRs regulation is rather limited. Therefore, it is of great interest to unravel the mechanisms and regulation modalities of receptor internalization and subsequent resensitization processes.
The Y2R is one of four human neuropeptide Y (NPY) receptor subtypes (hY1R, hY2R, hY4R, and hY5R) and belongs to the rhodopsin-like superfamily (class A) of GPCRs. YRs together with their three native ligands, NPY, pancreatic polypeptide (PP), and peptide YY (PYY), form a multiligand/multireceptor system (8), which is involved in many important physiological processes, such as regulation of food intake (9), control of blood pressure, and regulation of pancreatic and gastric secretion (10). The Y2R is predominantly expressed as a 381-amino acid protein in the brain cortex (11, 12), hippocampus, intestine, and certain blood vessels (8, 13, 14) and is involved in the inhibition of neurotransmitter release (15), regulation of memory retention (16), circadian rhythm, and angiogenesis (13), which makes it an attractive target for drug development. Potent Y2R agonists were suggested as therapy for epilepsy, because Y2R selective ligands have been shown to reduce epileptic seizures (17). Y2R selective peptides have also been shown to reduce food intake and lead to a significant loss in body weight (18). Because it has been reported that not only Y1Rs but also Y2Rs are expressed in distinct tumors, e.g. renal cell carcinomas, ovarian cancers, adrenal gland, and related tumors, the Y2R is a promising target for tumor diagnostics and therapy by using selectively labeled agonistic peptides (19, 20). Previous findings showed that, like Y1R and Y4R, Y2R undergoes rapid agonist-induced internalization (21).
In the current study we generated a series of C-terminal truncated hY2R mutants to investigate the impact of C-terminal sequences on receptor internalization properties. We identified novel regulatory motifs within the hY2R C-terminal domain, which contribute to receptor internalization and arrestin-3 (arr-3) association. Interestingly, our findings revealed arr-3-dependent and -independent hY2R internalization, and also led to the identification of a sequence that modulates receptor recycling.
EXPERIMENTAL PROCEDURES
Materials
Porcine NPY (pNPY) was produced by automated solid phase peptide synthesis using the Fmoc/tBu (9-fluorenylmethoxycarbonyl-tert-butyl) strategy (22). Propionylated [3H]pNPY ([3H]pNPY) was purchased from GE Healthcare. Materials for tissue culture were supplied by PAA Laboratories GmbH (Pasching, Austria). 8-well μ-slides were obtained from ibidi GmbH (Martinsried, Germany), 48-well plates were from Greiner Bio-One GmbH (Frickenhausen, Germany), and white/black opaque 96-well plates were from Nunc (Rochester, NY). Coelenterazine-h was obtained from DiscoveRx (Fremont, CA). LipofectamineTM 2000 transfection reagent and Opti®-MEM I reduced serum media were purchased from Invitrogen. MetafecteneTM transfection reagent was obtained from Biontex Laboratories GmbH (Martinsried/Planegg, Germany) and poly-d-lysine from Sigma. Anti-HA-peroxidase was purchased from Roche Diagnostics GmbH and TMB solution was from Calbiochem (Darmstadt, Germany).
Generation of YR Mutants and Arrestin-3 Variants
For fluorescence detection the cDNA of the hY2R was C-terminal fused to the cDNA of the enhanced yellow fluorescent protein (EYFP), as previously described (23, 24). Mutations, deletions, and truncations were introduced by using QuikChangeTM site-directed mutagenesis (Stratagene, La Jolla, CA). To perform enzyme-linked immunosorbent assays (ELISA) for quantification of receptor internalization rates, an N-terminal HA tag (25) was inserted downstream of the start codon of the hY2R. To visualize EYFP-tagged receptors in parallel with arr-3 in a fluorescence-based arrestin recruitment assay, bovine arr-3 was C-terminal fused to the red fluorescent protein Cherry (pCherry-NE/S vector was a kind gift of M. Hatzfeld; Martin-Luther University, Halle-Wittenberg). Pre-activated, phosphorylation-independent arr-3 constructs were obtained by distinct amino acid substitutions, as described elsewhere (26). A dominant positive variant of arr-3, bearing the triple alanine substitution I386A,V387A,F388A (arr-3(3A)), was generated by using arr-3-Cherry in a QuikChange site-directed mutagenesis.
Plasmid Construction for Bioluminescence Resonance Energy Transfer (BRET)
For BRET assays bovine arr-3 was N-terminal tagged with Venus (27) (Venus plasmid was a generous gift from Dr. J. A. Javitch, Columbia University). The sequence of the wild type and mutant HA-tagged hY2Rs were C-terminal fused with Renilla luciferase variant 8 (Rluc8) (28) (a plasmid harboring Rluc8 was a generous gift of Dr. Nevin A. Lambert, Medical College of Georgia). For the overexpression of human G protein-coupled receptor kinase 2 (GRK2) in COS-7 cells, the GRK2 sequence was included in hY2R-Rluc8 (GRK2 sequence was cloned from a vector kindly provided by Dr. Antonio De Blasi, University of Rome “Sapienza,” Rome, Italy) (for details, see supplemental data). All constructs were verified by DNA sequencing.
Cell Culture, Transfection, and Functional Studies
HEK293 cells (human embryonic kidney) and COS-7 cells (African green monkey) were cultured, as previously described (25). SMS-KAN and MHH-NB-11 cells (both human neuroblastoma cells that express the hY2R endogenously) were grown, as described (29). For live cell imaging HEK293 and SMS-KAN cells were plated into sterile 8-well μ-slides and MHH-NB-11 cells were plated in collagen-coated 8-well μ-slides. For cell surface ELISA studies HEK293 cells were seeded onto 48-well plates pre-treated with 0.01% (w/v) poly-d-lysine in phosphate-buffered saline (PBS). Cells were grown to 70% confluence and transfected with 1 μg of plasmid DNA encoding hY2R mutants C-terminal fused to EYFP or co-transfected with either 0.1 μg of plasmid DNA coding for bovine arr-3-Cherry or 0.2 μg of phosphorylation-independent bovine arr-3(3A)-Cherry using LipofectamineTM 2000 transfection reagent. For signal transduction assays (inositol phosphate accumulation in response to agonist stimulation) COS-7 cells were seeded into 24-well plates and transiently co-transfected with 0.32 μg of plasmid DNA encoding for the hY2R mutants C-terminal fused to EYFP and 0.08 μg of plasmid DNA coding for the chimeric G-protein Gαqi4 (kindly provided by E. Kostenis) using Metafectene transfection reagent. Signal transduction assays were performed as described previously (25, 30). Radioligand binding studies were performed using COS-7 cells, seeded into 25-cm2 cell culture flasks and transfected with 4 μg of DNA encoding for the hY2R wild type or mutants C-terminal fused to EYFP using Metafectene transfection reagent. Displacement radioligand binding assays were carried out with [3H]pNPY (radioactive concentration: 7.4 MBq/ml; specific activity: 3.55 TBq/mmol) and increasing concentrations of pNPY as described elsewhere (31).
BRET Assays
BRET assays (32–35) were used to characterize the binding of Venus-arr-3 to wild type hY2R-RLuc8 or the luciferase-fused Δ357, Δ369, Δ342–356, ST1* (S351A,S354A,T356A), and ST2* (S374A,T376A,T379A) mutants. COS-7 cells were transfected with increasing amounts of Venus-arr-3 (0–12 μg) with 35–200 ng of plasmids encoding wild type and mutant hY2R using Lipofectamine 2000, according to the manufacturer's protocol. Twenty-four hours after transfection, cells were re-seeded into white (luminescence measurements) or black (fluorescence determination) opaque 96-well microplates. Forty-eight hours post-transfection, the medium was replaced with HEPES-buffered PBS. Eight min after agonist stimulation (10−6 m pNPY), 5 μm coelenterazine-h was added and luminescence was measured 15 min after stimulation using a POLARstar Optima dual channel luminometer and fluorimeter microplate reader (BMG Labtech, Cary, NC). Luminescence was measured through 465–485 and 522.5–547.5 nm bandpass filters. The net BRET ratio was calculated as the long wavelength emission divided by the short wavelength emission and expressed as the relative change compared with unstimulated cells. The expression of Venus-arr-3 was evaluated using fluorescence at 535 nm upon excitation by 485 nm. The Venus-arrestin fluorescence was normalized by the basal luminescence from the corresponding wild type or mutant hY2R-RLuc8 to account for variations in cell numbers and expression levels. The resulting curves were fit by non-linear regression to a one-site hyperbola model using Prism 5.04 (GraphPad Software, San Diego, CA). To assess the difference between the respective wild type and mutant arrestin curves, a global fit algorithm based on the extra sum-of-squares F test was used.
Quantification of Receptor Cell Surface Density and Internalization by Cell Surface ELISA
To quantify plasma-membrane receptors prior and after stimulation, a cell surface ELISA with N-terminal HA-tagged wild type or mutant receptors (36) was carried out as described previously (37), with slight modifications. To investigate the influence of arr-3 overexpression on hY2R internalization, cells were co-transfected with arr-3-Cherry. Briefly, 24 h post-transfection cells were starved followed by ligand stimulation (10−6 m pNPY) for different time periods (to determine cell surface density, cells were not stimulated). To stop the internalization process cells were fixed, blocked overnight at 4 °C in Opti-MEM, and then probed with anti-HA-peroxidase conjugate (1:1000). Quantification of the bound peroxidase was performed as described (37).
Live Cell Imaging with Fluorescence Microscopy
Fluorescence microscopy studies were performed using a Zeiss Axio Observer microscope with an ApoTome Imaging System equipped with a Heating Insert P Lab-Tek S1 unit. Fluorescence images from living cells were taken as described previously (21) at 37 °C on a heated microscope stage using the AxioVision Rel. 4.6 software. To characterize the internalization properties of hY2R constructs as well as to follow arr-3 translocation in living cells, cells were starved in Opti-MEM for 30 min and treated with 10−6 m pNPY at 37 °C for the indicated time periods. To study receptor recycling in living cells the ligand was removed, cells were washed several times with PBS, and receptors were allowed to recover for 30–120 min.
RESULTS
Internalization of hY2R in Living Cells
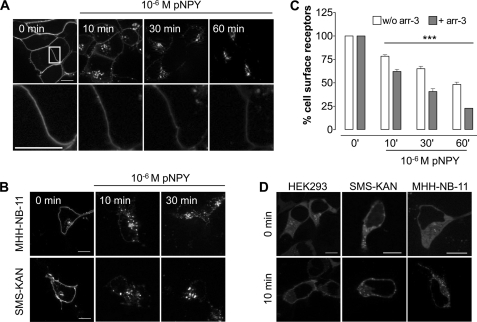
To investigate agonist-dependent hY2R internalization in living cells, HEK293 cells that transiently express hY2R-EYFP were incubated with 10−6 m pNPY for 10, 30, and 60 min. The internalization was analyzed by fluorescence microscopy. Prior to agonist stimulation hY2Rs are localized entirely in the cell membrane. Upon agonist stimulation the receptors are rapidly internalized and visible in vesicular compartments (Fig. 1A, upper panel). To visualize the remaining receptors at the cell surface, pictures were taken by focusing exclusively on the cell membrane with a constant exposure time (Fig. 1A, lower panel).
FIGURE 1.
Time-dependent arrestin3-promoted hY2R internalization upon agonist stimulation in different cell lines. Visualization (A) and quantification (C) of the time course for hY2R internalization in HEK293 cells transiently expressing hY2R-EYFP in response to stimulation with 10−6 m pNPY. A, representative images showing hY2R-EYFP fluorescence before (0 min) and after stimulation (10, 30, and 60 min). Lower panels show the magnified area indicated by the white box on the image 0 min. The decrease in remaining cell surface hY2R-EYFP fluorescence intensity in an individual cell was observed by fluorescence microscopy over the indicated time in response to agonist stimulation by using a constant exposure time of 3500 ms. B, time course for hY2R internalization in SMS-KAN and MHH-NB-11 cells transiently transfected with hY2R-EYFP. Representative images show hY2R-EYFP fluorescence before (0 min) and after (10 min and 30 min) 10−6 m pNPY treatment. C, quantification of cell surface receptors by ELISA with or without (w/o) arr-3-Cherry overexpression. The amount of cell surface receptors was measured as described under “Experimental Procedures.” Mean ± S.E. of at least three independent experiments, each performed in triplicate, are shown. Differences were significant (***, p < 0.001 by one-way analysis of variance) between nonstimulated and stimulated hY2R expressing HEK293 cells. D, distribution pattern of arr-3-Cherry fluorescence in HEK293, SMS-KAN, and MHH-NB-11 cells. Cells were co-transfected with hY2R-EYFP and arr-3-Cherry. Pictures were taken before (0 min) and after 10 min 10−6 m pNPY stimulation. Representative images show the changes in subcellular arr-3-Cherry fluorescence. All microscopic images were taken from living cells and are representative of at least three independent experiments. Scale bar, 10 μm.
To exclude the possibility that the internalization properties of the hY2R subtype are due to the exogenous expression in the HEK293 cell system, we also investigated the internalization in two human cell lines known to express the receptor endogenously, SMS-KAN (14) and MHH-NB-11. As no suitable specific Y2R antibodies are available, we transiently transfected hY2R-EYFP for visualization. Upon agonist stimulation the receptor is internalized in a comparable mode in all tested cell lines (Fig. 1B). Thus, hY2R internalization is not cell type dependent and occurs in cells that endogenously express the receptor.
To address the impact of arrestins on the internalization processes co-transfection of arr-2-Cherry or arr-3-Cherry with hY2R-EYFP was performed in all three cell lines and analyzed by fluorescence microscopy. Whereas arr-3-Cherry showed a clear redistribution to endocytic vesicles after stimulation with pNPY (Fig. 1D), we did not detect a significant recruitment of arr-2-Cherry (data not shown). Thus, hY2R internalization is preferentially mediated by arr-3 in all three cell lines.
To quantify the observed internalization process in HEK293 cells, we performed a modified cell surface ELISA. After stimulation of receptor internalization by exposure to 10−6 m agonist for 10, 30, and 60 min, respectively, the percentage of the remaining cell surface receptors in cells transfected with hY2R-EYFP decreased with time (Fig. 1C, white bars). Additional overexpression of arr-3 enhanced the internalization, as indicated by the reduction of remaining cell surface receptors from 78 ± 2 to 62 ± 2% after 10 min, from 65 ± 3 to 41 ± 3% after 30 min, and from 48 ± 2 to 23 ± 0.1% after 60 min (Fig. 1C, gray bars).
C-terminal Truncations Reveal Different Internalization Phenotypes
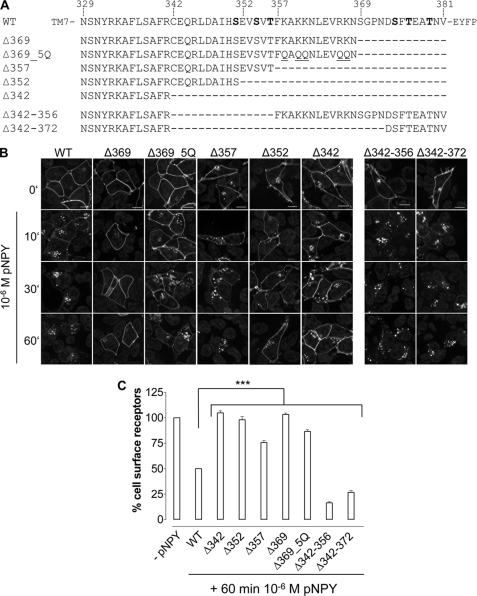
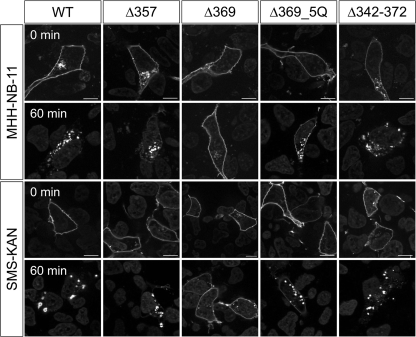
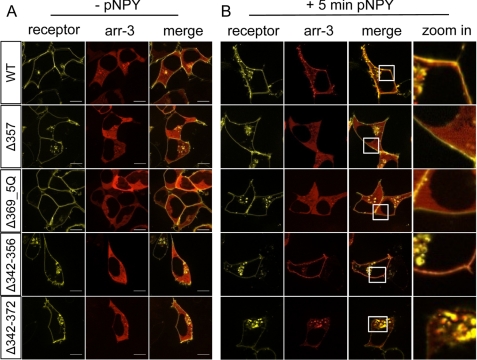
To clarify the role of the C terminus for hY2R internalization, a series of truncation mutants were generated, which were C-terminal shortened by 13 to 40 of the total 53 residues (Fig. 2A, for lower magnification images, see supplemental Fig. S1). All truncated variants are normally transported to the plasma membrane and are not significantly different when compared with wild type as indicated by fluorescence microscopy (Fig. 2B, 0′) and ELISA (Table 1). Moreover, they bind pNPY normally (wild type: IC50 = 1.2 ± 0.01 nm; Δ342: IC50 = 1.8 ± 1.1 nm) and inositol phosphate accumulation assays revealed EC50 values and efficacies (Emax) in the same range as wild type hY2R (Table 1). Internalization of these mutants was studied by fluorescence microscopy (Fig. 2B). Surprisingly, in contrast to all other deletion variants (Δ342, Δ352, and Δ369), only the mutant Δ357, lacking 25 C-terminal residues, internalized only slightly slower than wild type receptor. Taking into account that Δ369 did not internalize, whereas Δ357 did, we generated the mutant Δ369_5Q with five Lys and Arg residues (K358Q,K360Q,K361Q,R366Q,K367Q) simultaneously substituted to Gln to neutralize the positive charge of the 358–369 sequence. Interestingly, in contrast to Δ369, internalization of Δ369_5Q was observed, albeit with a much slower rate than wild type. These data show that the distal C terminus is important for internalization, whereas the positively charged segment, 357FKAKKNLEVRKN368, is an inhibitory element. Next, two mutants were generated that lack the central part of the C terminus, Δ342–356 and Δ342–372. Interestingly, both mutants internalized rapidly, similar to wild type. Mutant Δ357 and Δ342–372 have an identical sequence up to residue 341, but carry different C-terminal extensions. As both variants internalize after agonist stimulation, in contrast to Δ342, these results suggest that the C terminus of hY2R contains two distinct internalization motifs, located within residues 343–356 and 373–381. In addition to microscopy studies, internalization kinetics by means of the remaining cell surface receptors of hY2R mutants were examined by cell surface ELISA (Fig. 2C). Absence of internalization after 60 min stimulation was confirmed for mutants Δ342, Δ352, and Δ369 (Δ342, 105 ± 2%; Δ352, 98 ± 3%; Δ369, 103 ± 1). Δ357 and Δ369_5Q internalized, although not as rapidly as wild type (Δ357, 76 ± 2%; Δ369_5Q, 87 ± 2%; wild type, 48 ± 2), whereas Δ342–356 and Δ342–372 displayed considerably faster internalization properties compared with wild type (Δ342–356, 17 ± 1%; Δ342–372, 27 ± 2%). These data are consistent with the internalization properties obtained by fluorescence microscopy (Fig. 2B). To confirm that mutant hY2R internalization properties are not limited to HEK293 cells, the internalization of selected mutants (Δ357, Δ369, Δ369_5Q, and Δ342–372) were studied in MHH-NB-11 and SMS-KAN cells (Fig. 3). Internalization properties of all studied mutants are comparable with HEK293 cells, demonstrating that internalization of hY2R variants is not cell type dependent. Next, we tested whether both motifs can recruit arr-3 in HEK293 cells. To this end, all internalizing mutants, namely Δ357, Δ369_5Q, Δ342–356, and Δ342–372, were co-transfected with arr-3-Cherry. Prior to agonist treatment receptor-EYFP fluorescence was mainly localized in the plasma membrane and arr-3-Cherry fluorescence was homogenously distributed in the cytoplasm. Agonist stimulation led to a rapid redistribution of arr-3-Cherry from a diffuse cytoplasmic to a membrane-associated vesicular pattern only for wild type, Δ342–356, and Δ342–372, indicated by the orange hot spots in the membrane area (Fig. 4, right panel). For mutants Δ357 and Δ369_5Q no arr-3-Cherry translocation was detected, as indicated by the sustained diffuse distribution of red fluorescence throughout the cytoplasm after ligand exposure and receptor internalization. Thus, the distinct internalization motifs behave differently with respect to arr-3 recruitment.
FIGURE 2.
Sequence alignment and internalization properties of the C-terminal truncated hY2R mutants in HEK293 cells. A, sequence comparison of the COOH domains of the wild type and the generated hY2R constructs. The deleted amino acids are indicated with a hyphen (−). The Ser/Thr residues shown in bold are part of the internalization clusters ST1 and ST2. Underlined Gln residues indicate simultaneously introduced point mutations. B, time-dependent agonist-induced receptor internalization as revealed by fluorescence microscopy. HEK293 cells were transiently transfected with 1 μg of receptor DNA of the hY2R wild type and the following hY2R mutants C terminally fused to EYFP: Δ342, Δ352, Δ357, Δ369, Δ369_5Q, Δ342–356, or Δ342–372, respectively. Cells were incubated with 10−6 m pNPY for 10 to 60 min. Fluorescence images were taken from living cells and are representative of at least three independent experiments (scale bar, 10 μm). C, quantification of receptor internalization after 60 min 10−6 m pNPY stimulation as determined by cell surface ELISA. The amount of cell surface receptors was measured as described under “Experimental Procedures.” Mean ± S.E. of two independent experiments, each performed in triplicate, are shown. Differences in response to 60 min agonist stimulation were significant between wild type and hY2R mutant expressing HEK293 cells (***, p < 0.001 by one-way analysis of variance).
TABLE 1.
Functional characterization of wild type and C-terminal truncated hY2R variants
COS-7 cells (signal transduction assays) or HEK293 cells (ELISA) were transfected with wild type or mutant hY2Rs. EC50 values were obtained from concentration-response curves using GraphPad Prism 3.02. Emax values are presented as -fold over basal IP levels of wild type receptor (1306 ± 113 dpm/well) at 10−6 m pNPY. Data are given as mean ± S.E. of at least two independent experiments each performed in duplicate (EC50) or triplicate (Emax). Cell surface expression levels were measured by cell surface ELISA. Data are given as percentage of wild type HA-tagged hY2R. The corresponding receptors without HA tagged were used as controls and subtracted to account for unspecific antibody binding. ELISA data are given as mean ± S.E. of at least two to five independent experiments each performed in quadruplicate.
| Receptor variant | Signal transduction assay |
ELISA, cell surface expression | ||
|---|---|---|---|---|
| EC50 | EC50 | Emax | ||
| nm | -fold over WT | -fold over basal | % WT | |
| WT | 6.7 ± 2.8 | 1.0 | 14.1 ± 0.5 | 100 |
| Δ342 | 5.6 ± 3.9 | 0.3 | 17.2 ± 3.4 | 50 ± 3 |
| Δ352 | 8.4 ± 0.6 | 1.3 | 16.3 ± 2.1 | 82 ± 3 |
| Δ357 | 19.8 ± 0.1 | 3.0 | 14.2 ± 0.8 | 100 ± 4 |
| Δ369 | 1.9 ± 0.6 | 0.3 | 15.3 ± 1.7 | 93 ± 4 |
| Δ369_5Q | 25.2 ± 7.4 | 3.8 | 16.0 ± 0.1 | 99 ± 2 |
| Δ342–356 | 2.9 ± 0.2 | 0.4 | 13.8 ± 3.0 | 58 ± 4 |
| Δ342–372 | 3.4 ± 1.0 | 0.5 | 12.8 ± 0.6 | 58 ± 1 |
FIGURE 3.
Internalization properties of the C-terminal truncated hY2R mutants in MHH-NB-11 and SMS-KAN cells. Time-dependent agonist-induced receptor internalization as revealed by fluorescence microscopy. MHH-NB-11 and SMS-KAN cells were transiently transfected with 1 μg of receptor DNA of hY2R wild type and the following hY2R mutants C-terminal fused to EYFP: Δ357, Δ369, Δ369_5Q, and Δ342–372, respectively. Cells were incubated with 10−6 m pNPY for 60 min. Representative fluorescence images were taken from living cells from at least two independent experiments. Scale bar, 10 μm.
FIGURE 4.
Agonist stimulation of truncated hY2R mutants leads to different subcellular distribution of arr-3-Cherry. Visualization of arr-3-Cherry redistribution in response to receptor activation with 10−6 m pNPY. HEK293 cells were co-transfected with hY2R mutants C-terminal fused to EYFP (WT, Δ357, Δ369_5Q, Δ342–356, and Δ342–372) and arr-3-Cherry. A, prior to agonist stimulation receptor-EYFP fluorescence (yellow) is at the membrane, whereas arr-3-Cherry (red) is in the cytoplasm. B, subcellular distribution of the receptor mutants (yellow), arr-3-Cherry (red), and their co-localization (orange) following 5 min exposure to 10−6 m pNPY. The zoomed in section represents the indicated white boxes with a higher magnification. Representative images of living cells from at least three independent experiments are shown. Scale bar, 10 μm.
Residues in the Distal C-terminal Tail Promote Arrestin-3-mediated Internalization
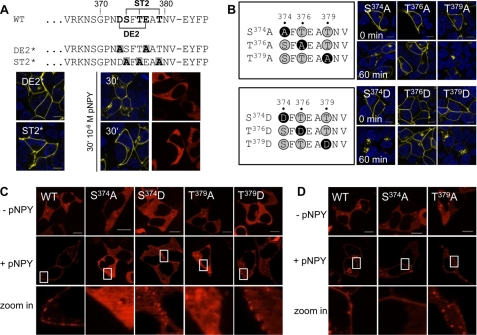
In contrast to the central Δ342–372 deletion, deletion of the distal C-terminal 13 residues in Δ369 completely abolished internalization. Therefore we set out to characterize the distal C-terminal sequence 373DSFTEATNV381 in more detail, as this sequence appears to be a typical internalization motif containing numerous Ser/Thr and Asp/Glu residues. Consequently, all Ser/Thr or Asp/Glu residues were mutated simultaneously to Ala (ST2* and D373A,E377A, referred to as DE2*). All generated hY2R variants within this section as well as all variants used at later stages in this study displayed EC50 values comparable with wild type as revealed by inositol phosphate formation in signal transduction assays (Table 2). After co-transfection of the EYFP-fused ST2* or DE2* with arr-3-Cherry in HEK293 cells, neither internalization nor arr-3-Cherry translocation was observed (Fig. 5A). Thus, both mutations prevented arr-3 recruitment. Next, we characterized the Ser/Thr cluster ST2 by generating a series of individual point mutants. First, combinations of double Ala mutations were generated: S374A,T376A; S374A,T379A, and T376A,T379A, all of which completely prevented agonist-induced internalization (data not shown). Next, we introduced single point mutations: S374A, T376A, and T379A. Interestingly, agonist treatment of S374A and T379A led to a fast internalization, whereas T376A did not internalize at all (Fig. 5B, upper panel). However, no arr-3-Cherry recruitment could be observed for S374A and T379A (Fig. 5C). By mutating each Ser or Thr residue in ST2 individually to Asp (referred to as S374D, T376D, and T379D), we observed fast internalization for mutants S374D and T379D, whereas T376D did not internalize (Fig. 5B, lower panel). In contrast to S374A and T379A, a weak clustering of arr-3-Cherry at the cell membrane was observed for S374D and T379D (indicated by the white boxes in Fig. 5C; see supplemental Fig. S3) after ligand stimulation. When phosphorylation-independent arr-3(3A)-Cherry and S374A or T379A were co-transfected in HEK293 cells, agonist treatment led to a rapid redistribution of arr-3(3A)-Cherry to the plasma membrane, as indicated by the red punctual fluorescence pattern, similar to that obtained with wild type receptor (Fig. 5D; see supplemental Fig. S3). Thus, the distal sequence motif 373DSXTEXT379 triggers arr-3-mediated internalization.
TABLE 2.
Functional characterization of hY2R point mutants by signal transduction assays
COS-7 cells were transfected with wild type or mutant hY2Rs. EC50 values were obtained from concentration-response curves using GraphPad Prism 3.02. Data are given as mean ± S.E. of at least two independent experiments each performed in duplicate.
| Receptor variant | EC50 |
|
|---|---|---|
| nm | -Fold over WT | |
| WT | 6.7 ± 2.8 | 1.0 |
| ST1* | 1.8 ± 0.6 | 0.3 |
| ST2* | 6.4 ± 0.4 | 1.0 |
| DE1* | 2.7 ± 1.5 | 0.4 |
| DE2* | 11.4 ± 1.5 | 1.7 |
| Δ357_DE1* | 8.7 ± 1.1 | 1.3 |
| Δ357_ST1* | 3.2 ± 1.3 | 0.5 |
| S374A/T376A | 8.6 ± 4.0 | 1.3 |
| S374A/T379A | 10.8 ± 6.8 | 1.6 |
| T376A/T379A | 12.6 ± 1.2 | 1.9 |
| S374A | 11.7 ± 3.7 | 1.7 |
| S374D | 9.5 ± 0.8 | 1.4 |
| T376A | 8.2 ± 2.0 | 1.2 |
| T376D | 8.5 ± 1.4 | 1.3 |
| T379A | 12.7 ± 5.3 | 1.9 |
| T379D | 7.7 ± 3.5 | 1.2 |
FIGURE 5.
Characterization of the distal C-terminal hY2R internalization motif. A, sequence alignment of DE2* and ST2*. Important Ser/Thr/Asp/Glu residues are shown in bold. Introduced Ala substitutions are shown in bold and highlighted by a gray box. HEK293 cells were transiently co-transfected with hY2R mutants, C-terminal tagged with EYFP, and arr-3-Cherry. Internalization was documented prior to (left panel) and in response to 30 min agonist exposure (middle row). Arr-3-Cherry distribution after 30 min agonist stimulation is shown in the right panel. B, schematic of introduced point mutations. White letters on black indicate the introduced mutations to Ala or Asp. These hY2R mutants, C-terminal fused to EYFP, were transfected in HEK293 cells and internalization was studied by fluorescence microscopy. Fluorescence distribution was visualized prior to (upper row, 0 min) and after 60 min stimulation with 10−6 m pNPY (lower row, 60 min). C, HEK293 cells were co-transfected with hY2R-EYFP (WT), hY2R-EYFP mutants (S374A, S374D, T379A, and T379D) and arr-3-Cherry (D) or hY2R-EYFP (WT), hY2R-EYFP mutants (S374A and T379A) and phosphorylation-independent arr-3(3A)-Cherry, respectively. Subcellular distribution of arr-3-Cherry (C) and arr-3(3A)-Cherry (D) was studied prior to (upper row, −pNPY) and upon receptor stimulation ((C) 5 min, (D) 15 min) with 10−6 m pNPY (middle row, +pNPY). Lower row (zoomed in section) represents the indicated white boxes from the middle row at higher magnification. Representative images of living cells from at least three independent experiments are shown. Scale bar, 10 μm.
Residues in the Proximal C-terminal Tail Promote Arrestin-3-independent Internalization
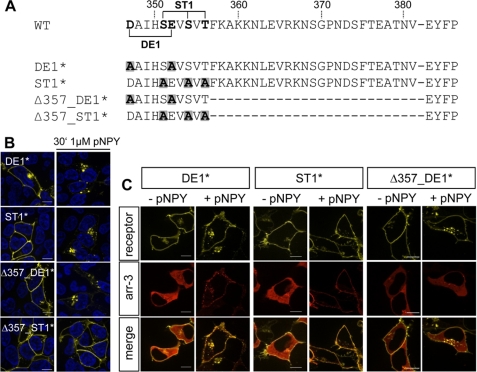
As Δ357 internalizes (in contrast to Δ342), we studied the 343–356 segment in more detail. 347DAIHSEVSVT356 contains many features of typical internalization motifs, such as numerous Ser/Thr and Asp/Glu residues. First, either Ser/Thr or Asp/Glu residues within this sequence were simultaneously mutated in the wild type receptor to Ala (ST1* and D347A,E352A, referred to as DE1*, Fig. 6A). ST1* or DE1* C-terminal fused to EYFP were co-transfected with arr-3-Cherry in HEK293 cells. Internalization and arr-3-Cherry translocation, visualized by fluorescence microscopy, revealed behavior indistinguishable from wild type (Fig. 6, B and C, upper and middle row). Next, the same mutations were introduced into Δ357 (referred to as Δ357_DE1* and Δ357_ST1*, Fig. 6A). Mutations D347A,E352A did not affect internalization as Δ357_DE1* was internalized in response to agonist stimulation, with a rate comparable with Δ357. Like Δ357, Δ357_DE1* was internalized in an arr-3-independent fashion (Fig. 6C, lower row). Interestingly, for Δ357_ST1* no agonist-induced internalization could be observed, suggesting Ser/Thr residues play an important role (Fig. 6B). These data indicate that the Ser/Thr residues, 351SXXSXT356, in the proximal C-terminal motif are important for internalization, which is arr-3-independent.
FIGURE 6.
Characterization of the proximal C-terminal internalization motif. A, sequence alignment of the generated point and deletion mutants DE1*, ST1*, Δ357_DE1*, and Δ357_ST1*. Mutated Ser/Thr/Asp/Glu residues are shown in bold. Introduced Ala substitutions are shown in bold and highlighted by a gray box. B, HEK293 cells were transiently transfected with hY2R mutants C-terminal tagged with EYFP. Receptor distribution is shown before (left panel) and after 30 min agonist exposure (right panel). Shown are representative images of at least three independent experiments. C, subcellular distribution of DE1*, ST1*, Δ357_DE1* (yellow), and arr-3-Cherry (red) in the absence or presence of agonist. HEK293 cells were co-transfected with the hY2R mutant C-terminal fused to EYFP and arr-3-Cherry. Fluorescence images were taken prior to (−pNPY) and following (+pNPY) 5 min agonist stimulation. Co-localization is indicated by orange membrane areas (merge, +pNPY for DE1* and ST1*). Representative images of living cells from at least three independent experiments are shown. Scale bar, 10 μm.
The Distal C-terminal Tail Promotes GRK2-dependent hY2R/Arrestin-3 Interaction
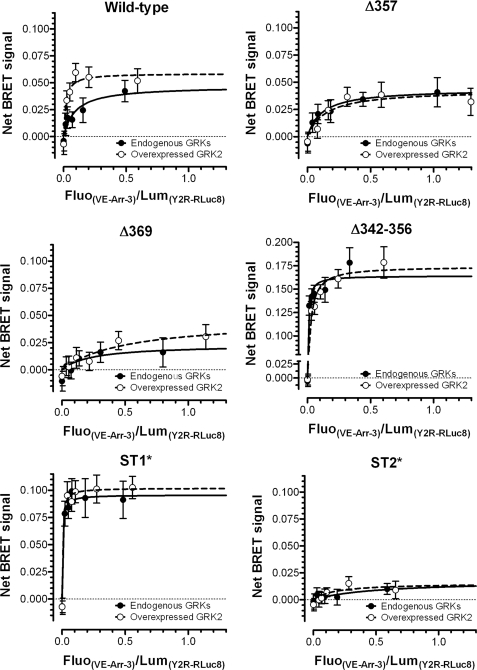
To further clarify if arr-3 recruitment and subsequent hY2R internalization depends on GRK2 activity, BRET assays with wild type and several mutant hY2R constructs in the absence or presence of overexpressed GRK2 were performed in COS-7 cells (Fig. 7). BRETmax values were not affected by cellular GRK2 levels for all receptor variants (see supplemental Table S1). However, GRK2 overexpression increased the apparent affinity for Venus-arr-3 for wild type hY2R ∼10-fold (EC50 = 0.1195 ± 0.0487 (−GRK2); 0.0231 ± 0.0087 (+GRK2)). Next we compared the internalization patterns of different hY2R mutants with their ability to recruit arr-3 in a GRK2-dependent manner. For Δ369 and ST2*, receptor variants that do not internalize, no receptor/arr-3 interaction was observed as evidenced by the very low BRETmax values. For receptor variants that internalize and recruit arr-3 rapidly, Δ342–356 and ST1*, an enhanced interaction was detected along with considerably increased BRETmax values compared with wild type ((+)GRK2: Δ342–356, BRETmax = 0.1854 ± 0.0111; ST1*, BRETmax = 0.1019 ± 0.0020; wild type, BRETmax = 0.0589 ± 0.0075). A high apparent affinity for Δ342–356 in concert with a high BRETmax is in agreement with the substantially faster internalization kinetics compared with wild type (Fig. 2C). These data are consistent with internalization and arr-3 recruitment properties as determined by fluorescence microscopy (Fig. 2 and 4–6). Δ357, which internalizes independently of arr-3, showed greatly reduced apparent affinity for arr-3 ((−GRK2) EC50 = 0.0944 ± 0.0187; (+GRK2) EC50 = 0.2002 ± 0.0573) as compared with wild type. The BRETmax values (0.0588 ± 0.016 (+GRK2)) similar to those observed for wild type, in contrast to undetectable arr-3-mCherry recruitment, are most likely due to much higher cellular concentrations of arr-3 in BRET experiments. Overall, BRET data clearly show that the wild type hY2R/arr-3 interaction is facilitated by GRK2. Most likely Ser and Thr residues upstream of residue 369 within the distal C-terminal tail are phosphorylated by GRK2 and participate in arr-3 recruitment.
FIGURE 7.
GRK2-dependent recruitment of arrestin-3 by wild type and mutant hY2Rs in COS-7 cells as measured by BRET. BRET signal as a function of Venus-arr-3 in the absence (filled circles, solid line) or presence of human GRK2 (open circles, dashed line) co-expression with the indicated wild type or mutant hY2R-RLuc8 in COS-7 cells. Shown is the difference between BRET signals in the presence and absence of 10−6 m pNPY, which reflects the agonist-induced increase in Venus-arr-3 interaction with fixed amounts of each of the hY2R-RLuc8 tested. Mean ± S.E. of a representative experiment performed in sextuplicate from 2–10 independent experiments.
hY2R Recycling Is Regulated by Its Proximal C-terminal Domain
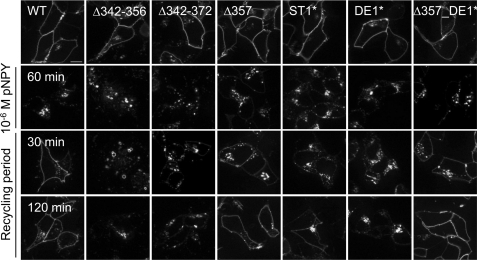
All hY2R variants that have been shown to internalize were next studied with respect to their recycling properties in HEK293 cells. After a 60-min ligand stimulation, the respective receptor-EYFP fusion proteins were observed exclusively in endocytic vesicles (Fig. 8, 10−6 m pNPY). Upon removal of the agonist, internalized wild type receptors recycled back to the plasma membrane after 30 min (Fig. 8, left panel). Recycling was not detected for variants Δ342–356 and Δ342–372. As a matter of interest, Δ357 recycled comparably to wild type, suggesting that the sequence 343EQRLDAIHSEVSVT356 acts as a structural determinant for rapid recycling. Therefore ST1* and DE1* were investigated and revealed a moderate (ST1*) or no (DE1*) recycling for up to 120 min. However, Δ357_DE1* displayed recycling properties comparable with wild type and Δ357 rather than to DE1*. These results strongly indicate that the proximal C-terminal motif 343EQRLDAIHSEVSVT356 is not only involved in regulation of internalization, but also dictates whether receptors internalized in an arr-3-dependent manner recycle back to the plasma membrane or are degraded. Unexpectedly, we found that all hY2R mutants that internalize independently of arr-3 rapidly recycle. This result suggests that arrestin dissociation from hY2R is the rate-limiting step in the recycling of receptors internalized via a arrestin-dependent mechanism, as was previously proposed for the β2-adrenergic receptor (38). Additionally recycling of wild type, Δ357, and Δ342–372 was investigated in SMS-KAN and MHH-NB-11 cells to assure that recycling properties are not cell type dependent. Importantly, recycling properties of all studied receptor mutants in these cells were comparable with HEK293 cells (supplemental Fig. S2), demonstrating that changes in the receptor C terminus, rather than cell type, define the recycling pattern.
FIGURE 8.
Recycling properties of the hY2R variants. HEK293 cells were transiently transfected with hY2R variants C-terminal tagged with EYFP. Receptor localization before (upper row) and after a 60-min agonist exposure (10−6 m pNPY panel) is shown. Recycling was documented after 30 and 120 min (recycling period panels). Shown are representative images of at least two independent experiments. Scale bar, 10 μm.
DISCUSSION
Here we analyzed the role of the hY2R C terminus in receptor internalization and recycling. We identified key regulatory motifs responsible for internalization, arr-3 recruitment, and receptor recycling. The issue of Y2R internalization is controversial. It has been reported that this YR subtype hardly internalizes (39–42). However, in previous studies we and others showed that the hY2R rapidly internalizes after agonist treatment in HEK293 cells, with a rate comparable with hY1R and hY4R (21, 43). Furthermore, here we confirmed by fluorescence microscopy that hY2R internalization is not limited to HEK293 cells, but also occurs comparably in SMS-KAN and MHH-NB-11 cells expressing this receptor endogenously (Fig. 1, A and B). Therefore, we explored the mechanisms of regulation of hY2R internalization and subsequent recycling.
A regulatory mechanism shared by many GPCRs involves receptor phosphorylation by GRKs, followed by arrestin binding and arrestin-mediated internalization via coated pits (44). Indeed, we observed rapid arr-3 recruitment to the plasma membrane upon hY2R stimulation in all cell lines tested (Fig. 1D). Consistent with the data obtained by fluorescence microscopy, quantification of receptor internalization in the presence or absence of arr-3 overexpression by cell surface ELISA, as well as analysis of hY2R/arr-3 interaction by BRET strongly suggested an arr-3-dependent internalization process. These data represent the first direct demonstration of hY2R interaction with arr-3 and are in good agreement with BRET2 studies showing slow association rates of the rhesus Y2R with arr-3 in HEK293 cells (39).
Extensive effort has been invested into the identification of specific receptor motifs participating in GPCR endocytosis. The contribution of intracellular elements to internalization is well described for several GPCRs. In addition to the third intracellular loop these studies revealed a major role of the C terminus in internalization, particularly for the human neuropeptide Y1 receptor (45), angiotensin II type one receptor (46), human P2Y1 receptor (47), A2B adenosine receptor (48), TRHR (49), and the N-formyl peptide receptor (50). However, so far nothing is known about receptor domains involved in hY2R internalization. Our previous studies with hY5/hY2 chimeric receptors showed that the hY2R C terminus plays a key role in the internalization of these receptors, because its exchange between hY2R and hY5R led to a rapid internalization of the usually non-internalizing hY5R (21). In all cellular systems used we identified two internalization motifs: a distal DSXTEXT (373DSFTEAT379) and proximal DXXXSEXSXT (347DAIHSEVSVT356).
Interestingly, these two motifs mediate distinct internalization pathways. In contrast to wild type receptor where both motifs are intact, Δ357, with only the proximal motif, internalizes slower and is notably defective in agonist-induced arr-3 binding. This is reminiscent of the A2B adenosine receptor, where deletion of seven C-terminal amino acids eliminating several Ser/Thr residues prevented arrestin recruitment (48). The loss of the distal C-terminal motif STXSXS of the receptor, which is homologous to the distal hY2R motif SXTXXT, switched the internalization from an arrestin-dependent to an -independent mechanism (48). In general, motifs mediating arrestin-dependent internalization include clusters of Ser/Thr residues usually targeted by GRKs (51). The direct connection of phosphorylation with internalization has been documented for angiotensin type II AT1A (52), β2-adrenergic receptor (53, 54), follitropin receptor (55), somatostatin sst2A (56), and CRTH2 receptor (57). Usually GPCR phosphorylation by GRKs facilitates arrestin recruitment, which connects the receptor to the endocytic machinery (51). The presence of acidic amino acids in close proximity to these clusters can guide GRKs to the phosphorylation sites (57–59). The distal C-terminal internalization motif 373DSFTEAT379 in the hY2R is a typical example, containing multiple Ser/Thr flanked by Asp and Glu residues. The substitution of either all Ser/Thr or Asp/Glu residues by Ala (ST2*, DE2*) in the distal hY2R internalization motif blocks its function, completely preventing internalization (Fig. 5A). Here Thr376 appears to be particularly important, because no substitution was tolerated. Notably, only a weak arr-3 clustering was observed for S374D and T379D (Fig. 5C). Apparently, Asp acts as partial phosphomimetic (61), mimicking phosphorylated Ser/Thr residues involved in binding of the arr-3 recognition domain that has multiple positive charges (44). The recruitment of only phosphorylation independent arr-3 obtained for S374A and T379A strongly indicates that Ser374 and Thr379 are potential phosphorylation sites. Thus, phosphorylation of the distal C-terminal residues Ser374 and Thr379 is likely required for a high affinity interaction with wild type arr-3.
We also identified a second internalization motif. This proximal sequence 347DAIHSEVSVT356 was found to be unique for Y1R and Y2R subtypes within the YR family. This conserved C-terminal motif (φ-H-(S/T)-(E/D)-V-(S/T)-X-T) was implicated in Y1R internalization (41, 45). Our data provide the first evidence that this motif participates in internalization of the hY2R as well. The most striking finding is the mutant Δ357 internalized, in contrast to wild type, in an arr-3-independent manner. To further characterize Δ357 and the proximal motif associated with its internalization, we analyzed Δ357_DE1* and Δ357_ST1* mutants and obtained different internalization phenotypes. Δ357_DE1* retained internalization capability, whereas Δ357_ST1* lost the ability to internalize. Thus, in addition to Ser/Thr residues in the distal C-terminal motif, Ser/Thr residues within 347DAIHSEVSVT356 seem to be crucial for internalization. The distal arr-3-sensitive motif is likely the major phosphorylation site. In contrast, the proximal internalization motif appears to be phosphorylation-independent, as it does not confer arrestin sensitivity and does not bind phosphorylation-independent arrestins. BRET assays monitoring receptor/arr-3 interaction in a GRK2-dependent manner support hY2R phosphorylation in the distal internalization motif. Whereas all mutants lacking the proximal tail display a high apparent affinity for receptor/arr-3 interaction (ST1* and Δ342–356), mutants without the distal motif (ST2* and Δ369) show little to no arrestin-3 recruitment. Thus, the distal motif upstream of amino acid 369 is the key GRK phosphorylation site. In contrast to microscopy-based arr-3 recruitment, BRET-based assays are performed with severalfold excess of arrestin over receptor, which likely accounts for differences in arr-3 recruitment as observed for Δ357. Nonetheless, the apparent affinity of Δ357 for arr-3 was extremely low, as compared with wild type, which is consistent with arr-3-independent internalization. Apart from the hY2R several other activated receptors were reported to internalize effectively in an phosphorylation-independent fashion: δ-opioid receptors (62), leukotriene B4 receptors (63), and substance P receptor (64).
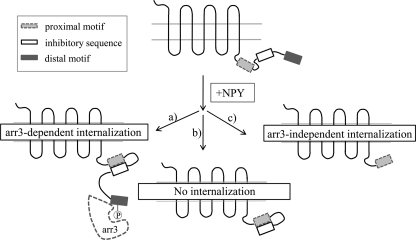
Because Δ369 does not internalize, and even Δ369_5Q does not bind arr-3, we hypothesized that the basic amino acid stretch 357FKAKKNLEVRKN368 located between the internalization motifs is an inhibitory element. Our data suggest that the distal motif is required to reverse the inhibition and allow arr-3 binding. Based on our data, we propose the following mechanism of hY2R internalization. First, receptor activation results in a conformational change and allows phosphorylation of the distal C-terminal motif by GRKs, which mediates the binding of arr-3, which connects the receptor with the internalization machinery of the cell. The inhibitory element, located just N-terminal to the distal motif, precludes arrestin interaction in the absence of phosphorylation and thereby prevents constitutive internalization. Phosphorylation of the distal motif functionally neutralizes the inhibitory segment, allowing arrestin binding to wild type receptor. When the distal motif is absent, the inhibitory element blocks the proximal motif and therefore completely prevents internalization (e.g. Δ369). If both the distal motif and the inhibitory sequence are eliminated, the receptor (e.g. Δ357) internalizes in an arr-3-independent manner (Fig. 9).
FIGURE 9.
Proposed mechanism underlying hY2R internalization. The hY2R C terminus consists of three regulatory sequence motifs: a proximal internalization motif (light gray bar), an inhibitory sequence (white bar with a black line), and a distal internalization motif (dark gray bar). Agonist stimulation induces a conformational change within the receptor, and depending on the exposure of distinct motifs multiple mechanisms operate: a) the distal motif is exposed to the cytoplasm to get phosphorylated, which then enables arr-3 binding and subsequent internalization (wild type receptor). b) When the receptor lacks the distal motif the inhibitory element blocks the proximal internalization motif, precluding internalization (Δ369). c) When the receptor lacks both the distal motif and the inhibitory sequence, the proximal internalization motif is accessible and acts as backup motif to promote phosphorylation- and arr-3-independent internalization (Δ357).
It is worth noting that multiple internalization mechanisms were proposed for the related Y1R subtype. Arr-3-dependent internalization was shown to be predominant, but arrestin-independent mechanisms have been implicated in the control of internalization (45). Muscarinic M2 receptor was also shown to bind arrestins, but internalize via an arrestin-independent pathway (65, 66). Interestingly, M2 (65, 66) and opioid (67) receptors also have inhibitory elements that act as “brakes” of internalization. In both cases the inhibition is normally relieved by the phosphorylation of these elements, but it can be artificially eliminated by their deletion.
Once the receptors have been internalized, they can either be targeted to recycling endosomes and traffic back to the cell surface or translocate to lysosomes for degradation (68, 69). Receptor down-regulation is related to the binding affinities of GPCRs for arrestins. GPCRs were classified into two groups. Class A receptors have a higher affinity for arr-3 and form transient GPCR-arrestin complexes, which dissociate near the cell surface (e.g. β2-adrenergic, μ-opioid, and dopamine D1A receptors). Class B receptors have equal affinities for arr-2 and arr-3, and the GPCR-arrestin complex is stable and remains intact during the internalization process (e.g. vasopressin V2, neurotensin 1, and angiotensin II type 1A receptors) (68, 70–72). Decreased or increased affinity of the mutant arrestin complex with the same β2-adrenergic receptor was shown to direct it to recycling or the degradation pathway, respectively (38, 73). We found that after ligand exposure, the redistributed arr-3 is confined to the cell periphery and does not traffic along with the activated hY2R to endocytic vesicles, suggesting that the hY2R belongs to class A. Class A GPCRs are known to be recycled rather than degraded. Indeed, the identified sequence within the proximal C-terminal tail is responsible for fast hY2R recycling in different cellular environments (Fig. 8 and supplemental Fig. S2). Our data indicate that early dissociation of hY2R/arr-3 complex at the plasma membrane is a prerequisite for rapid recycling, because the dissociation of arrestins from the receptor makes it accessible for phosphatases in early endosomes and consequently allows receptor recycling (74). Interestingly, the recycling motif in hY2R overlaps with the conserved motif (φ-H-(S/T)-(E/D)-V-(S/T)-X-T), which also contributes to arr-3-independent internalization.
Furthermore, only the variants that carry the functional motif are able to recycle as fast as wild type receptor. Mutations within this motif resulted in a dramatically reduced recycling capability or even completely prevented recycling. However, with respect to specific recycling motifs there is only little information available. All of the identified C-terminal motifs differ in length and sequence from each other and from the identified hY2R motif: β1-adrenergic (ESKV) and β2-adrenergic receptor (DSLL), μ-opioid (LENLEAE), and κ-opioid receptors (NKPV), thyrotropin stimulating hormone receptor (TVL), and CRTH2 receptor (DSEL) (57, 75). It appears that in addition to the sequence itself, the nature of the surrounding residues, as well as sterical and conformational requirements play a major role in receptor recycling.
In summary, the removal of ligand-activated GPCRs from the cell surface by internalization and subsequent down-regulation or recycling contributes to the tight regulation of GPCR responsiveness. Understanding this complex mechanism is particularly important for pharmaceutical targets, such as hY2R. Different therapeutic interventions are conceivable at the level of receptor internalization. Because multiple peptide hormone receptors are known to be overexpressed in several human cancer cells, ligand-induced internalization could serve as a means of anti-tumor therapy and diagnosis (76). The drug often needs to be shuttled into tumor cells, and ligand-induced receptor internalization could be used as a delivery route. Effective receptor internalization allowed the use of agonists as functional antagonists of the S1P1 receptor (5). A third therapeutic approach utilizing internalization was described for the prevention of HIV-1 infection: the administration of receptor-selective ligands leading to the removal of co-receptors (like CCR5 and/or CXCR4) from the cell surface reduces the probability of HIV-1 infection (60). Thus, the molecular mechanisms of GPCR internalization could be exploited in many ways for therapeutic purposes.
In conclusion, we demonstrated that hY2R recruits arr-3 to the plasma membrane. Moreover, we characterized two independent C-terminal internalization motifs in the hY2R: proximal and distal motifs that trigger arr-3-independent and -dependent internalization. We also showed for the first time that the hY2R C terminus contributes to receptor recycling, and identified an inhibitory sequence located in the central C-terminal tail. Thus, the hY2R is subject to a highly sophisticated trafficking regulation, which involves multiple molecular events. Our findings identified distinct structural determinants playing crucial roles in internalization and recycling processes.
Supplementary Material
Acknowledgments
We highly appreciate the excellent technical assistance of Kristin Löbner in cell culture and Janet Schwesinger in plasmid sequencing. We are grateful to Dr. Jonathan A. Javitch for expert advice on receptor-arrestin BRET and the plasmid encoding Venus, Dr. Nevin A. Lambert for plasmid encoding R. luciferase variant 8, Dr. Antonio De Blasi for the plasmid encoding human GRK2, and Dr. Carl Johnson for use of POLARstar Optima dual channel luminometer and fluorimeter microplate reader.
This work was supported, in whole or in part, by National Institutes of Health Grant GM081756 (to V. V. G.) and Deutsche Forschungsgemeinschaft (AGB-S) Grant SFB-610/TP-A1.
Dedicated to Horst Kunz on the occasion of his 70th birthday.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- GPCR
- G protein-coupled receptors
- hY2R
- human neuropeptide Y2 receptor
- ELISA
- enzyme-linked immunosorbent assay
- arr-2
- bovine arrestin-2 (β-arrestin and β-arrestin-1)
- arr-3
- bovine arrestin-3 (β-arrestin-2)
- GRK2
- G protein-coupled receptor kinase 2
- NPY
- neuropeptide Y
- PP
- pancreatic polypeptide
- PYY
- intestinal peptide YY
- p
- porcine
- [3H]pNPY
- propionylated [3H]porcine neuropeptide Y
- YR
- neuropeptide Y receptor
- EYFP
- enhanced yellow fluorescent protein
- BRET
- bioluminescence resonance energy transfer
- COS-7
- African green monkey (kidney) cells.
REFERENCES
- 1.Millar R. P., Newton C. L. (2010) Mol. Endocrinol. 24, 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rana B. K., Shiina T., Insel P. A. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 593–624 [DOI] [PubMed] [Google Scholar]
- 3.Spiegel A. M. (1996) Annu. Rev. Physiol. 58, 143–170 [DOI] [PubMed] [Google Scholar]
- 4.Schöneberg T., Schulz A., Biebermann H., Hermsdorf T., Römpler H., Sangkuhl K. (2004) Pharmacol. Ther. 104, 173–206 [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann V. (2009) Br. J. Pharmacol. 158, 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calebiro D., Nikolaev V. O., Persani L., Lohse M. J. (2010) Trends Pharmacol. Sci. 31, 221–228 [DOI] [PubMed] [Google Scholar]
- 7.Kenakin T., Miller L. J. (2010) Pharmacol. Rev. 62, 265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrele C., Beck-Sickinger A. G. (2000) J. Pept. Sci. 6, 97–122 [DOI] [PubMed] [Google Scholar]
- 9.Herzog H. (2003) Eur. J. Pharmacol. 480, 21–29 [DOI] [PubMed] [Google Scholar]
- 10.Parker E., Van Heek M., Stamford A. (2002) Eur. J. Pharmacol. 440, 173–187 [DOI] [PubMed] [Google Scholar]
- 11.Dumont Y., Fournier A., St-Pierre S., Schwartz T. W., Quirion R. (1990) Eur. J. Pharmacol. 191, 501–503 [DOI] [PubMed] [Google Scholar]
- 12.Widdowson P. S. (1993) Brain Res. 631, 27–38 [DOI] [PubMed] [Google Scholar]
- 13.Lindner D., Stichel J., Beck-Sickinger A. G. (2008) Nutrition 24, 907–917 [DOI] [PubMed] [Google Scholar]
- 14.Sheikh S. P., O'Hare M. M., Tortora O., Schwartz T. W. (1989) J. Biol. Chem. 264, 6648–6654 [PubMed] [Google Scholar]
- 15.Smith-White M. A., Hardy T. A., Brock J. A., Potter E. K. (2001) Br. J. Pharmacol. 132, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flood J. F., Morley J. E. (1989) Peptides 10, 963–966 [DOI] [PubMed] [Google Scholar]
- 17.El Bahh B., Balosso S., Hamilton T., Herzog H., Beck-Sickinger A. G., Sperk G., Gehlert D. R., Vezzani A., Colmers W. F. (2005) Eur. J. Neurosci. 22, 1417–1430 [DOI] [PubMed] [Google Scholar]
- 18.Karra E., Chandarana K., Batterham R. L. (2009) J. Physiol. 587, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Körner M., Reubi J. C. (2007) Peptides 28, 419–425 [DOI] [PubMed] [Google Scholar]
- 20.Reubi J. C., Gugger M., Waser B., Schaer J. C. (2001) Cancer Res. 61, 4636–4641 [PubMed] [Google Scholar]
- 21.Böhme I., Stichel J., Walther C., Mörl K., Beck-Sickinger A. G. (2008) Cell. Signal. 20, 1740–1749 [DOI] [PubMed] [Google Scholar]
- 22.Beck-Sickinger A. G., Wieland H. A., Wittneben H., Willim K. D., Rudolf K., Jung G. (1994) Eur. J. Biochem. 225, 947–958 [DOI] [PubMed] [Google Scholar]
- 23.Dinger M. C., Bader J. E., Kobor A. D., Kretzschmar A. K., Beck-Sickinger A. G. (2003) J. Biol. Chem. 278, 10562–10571 [DOI] [PubMed] [Google Scholar]
- 24.Merten N., Lindner D., Rabe N., Römpler H., Mörl K., Schöneberg T., Beck-Sickinger A. G. (2007) J. Biol. Chem. 282, 7543–7551 [DOI] [PubMed] [Google Scholar]
- 25.Lindner D., Walther C., Tennemann A., Beck-Sickinger A. G. (2009) Cell. Signal. 21, 61–68 [DOI] [PubMed] [Google Scholar]
- 26.Celver J., Vishnivetskiy S. A., Chavkin C., Gurevich V. V. (2002) J. Biol. Chem. 277, 9043–9048 [DOI] [PubMed] [Google Scholar]
- 27.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 28.Loening A. M., Fenn T. D., Wu A. M., Gambhir S. S. (2006) Protein Eng. Des. Sel. 19, 391–400 [DOI] [PubMed] [Google Scholar]
- 29.Höfliger M. M., Castejón G. L., Kiess W., Beck Sickinger A. G. (2003) J. Recept. Signal Transduct. Res. 23, 351–360 [DOI] [PubMed] [Google Scholar]
- 30.Böhme I., Mörl K., Bamming D., Meyer C., Beck-Sickinger A. G. (2007) Peptides 28, 226–234 [DOI] [PubMed] [Google Scholar]
- 31.Haack M., Beck-Sickinger A. G. (2009) Chem. Biol. Drug. Des. 73, 573–583 [DOI] [PubMed] [Google Scholar]
- 32.Kocan M., Pfleger K. D. (2009) Methods Mol. Biol. 552, 305–317 [DOI] [PubMed] [Google Scholar]
- 33.Kuravi S., Lan T. H., Barik A., Lambert N. A. (2010) Biophys. J. 98, 2391–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namkung Y., Dipace C., Javitch J. A., Sibley D. R. (2009) J. Biol. Chem. 284, 15038–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namkung Y., Dipace C., Urizar E., Javitch J. A., Sibley D. R. (2009) J. Biol. Chem. 284, 34103–34115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schöneberg T., Liu J., Wess J. (1995) J. Biol. Chem. 270, 18000–18006 [DOI] [PubMed] [Google Scholar]
- 37.Lindner D., van Dieck J., Merten N., Mörl K., Günther R., Hofmann H. J., Beck-Sickinger A. G. (2008) Biochemistry 47, 5905–5914 [DOI] [PubMed] [Google Scholar]
- 38.Pan L., Gurevich E. V., Gurevich V. V. (2003) J. Biol. Chem. 278, 11623–11632 [DOI] [PubMed] [Google Scholar]
- 39.Berglund M. M., Schober D. A., Statnick M. A., McDonald P. H., Gehlert D. R. (2003) J. Pharmacol. Exp. Ther. 306, 147–156 [DOI] [PubMed] [Google Scholar]
- 40.Gicquiaux H., Lecat S., Gaire M., Dieterlen A., Mély Y., Takeda K., Bucher B., Galzi J. L. (2002) J. Biol. Chem. 277, 6645–6655 [DOI] [PubMed] [Google Scholar]
- 41.Ouedraogo M., Lecat S., Rochdi M. D., Hachet-Haas M., Matthes H., Gicquiaux H., Verrier S., Gaire M., Glasser N., Mély Y., Takeda K., Bouvier M., Galzi J. L., Bucher B. (2008) Traffic 9, 305–324 [DOI] [PubMed] [Google Scholar]
- 42.Parker S. L., Kane J. K., Parker M. S., Berglund M. M., Lundell I. A., Li M. D. (2001) Eur. J. Biochem. 268, 877–886 [DOI] [PubMed] [Google Scholar]
- 43.Kilpatrick L. E., Briddon S. J., Hill S. J., Holliday N. D. (2010) Br. J. Pharmacol. 160, 892–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holliday N. D., Lam C. W., Tough I. R., Cox H. M. (2005) Mol. Pharmacol. 67, 655–664 [DOI] [PubMed] [Google Scholar]
- 46.Kule C. E., Karoor V., Day J. N., Thomas W. G., Baker K. M., Dinh D., Acker K. A., Booz G. W. (2004) Regul. Pept. 120, 141–148 [DOI] [PubMed] [Google Scholar]
- 47.Reiner S., Ziegler N., Leon C., Lorenz K., von Hayn K., Gachet C., Lohse M. J., Hoffmann C. (2009) Mol. Pharmacol. 76, 1162–1171 [DOI] [PubMed] [Google Scholar]
- 48.Mundell S. J., Matharu A. L., Nisar S., Palmer T. M., Benovic J. L., Kelly E. (2010) Br. J. Pharmacol. 159, 518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones B. W., Song G. J., Greuber E. K., Hinkle P. M. (2007) J. Biol. Chem. 282, 12893–12906 [DOI] [PubMed] [Google Scholar]
- 50.Potter R. M., Maestas D. C., Cimino D. F., Prossnitz E. R. (2006) J. Immunol. 176, 5418–5425 [DOI] [PubMed] [Google Scholar]
- 51.Tobin A. B., Butcher A. J., Kong K. C. (2008) Trends Pharmacol. Sci. 29, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas W. G., Motel T. J., Kule C. E., Karoor V., Baker K. M. (1998) Mol. Endocrinol. 12, 1513–1524 [DOI] [PubMed] [Google Scholar]
- 53.Bouvier M., Hausdorff W. P., De Blasi A., O'Dowd B. F., Kobilka B. K., Caron M. G., Lefkowitz R. J. (1988) Nature 333, 370–373 [DOI] [PubMed] [Google Scholar]
- 54.Karoor V., Wang L., Wang H. Y., Malbon C. C. (1998) J. Biol. Chem. 273, 33035–33041 [DOI] [PubMed] [Google Scholar]
- 55.Kara E., Crépieux P., Gauthier C., Martinat N., Piketty V., Guillou F., Reiter E. (2006) Mol. Endocrinol. 20, 3014–3026 [DOI] [PubMed] [Google Scholar]
- 56.Liu Q., Dewi D. A., Liu W., Bee M. S., Schonbrunn A. (2008) Mol. Pharmacol. 73, 292–304 [DOI] [PubMed] [Google Scholar]
- 57.Roy S. J., Parent A., Gallant M. A., de Brum-Fernandes A. J., Stanková J., Parent J. L. (2010) Eur. J. Pharmacol. 630, 10–18 [DOI] [PubMed] [Google Scholar]
- 58.Lee K. B., Ptasienski J. A., Bunemann M., Hosey M. M. (2000) J. Biol. Chem. 275, 35767–35777 [DOI] [PubMed] [Google Scholar]
- 59.Onorato J. J., Palczewski K., Regan J. W., Caron M. G., Lefkowitz R. J., Benovic J. L. (1991) Biochemistry 30, 5118–5125 [DOI] [PubMed] [Google Scholar]
- 60.Kenakin T. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 349–379 [DOI] [PubMed] [Google Scholar]
- 61.Prossnitz E. R., Kim C. M., Benovic J. L., Ye R. D. (1995) J. Biol. Chem. 270, 1130–1137 [DOI] [PubMed] [Google Scholar]
- 62.Zhang X., Wang F., Chen X., Li J., Xiang B., Zhang Y. Q., Li B. M., Ma L. (2005) J. Neurochem. 95, 169–178 [DOI] [PubMed] [Google Scholar]
- 63.Jala V. R., Shao W. H., Haribabu B. (2005) J. Biol. Chem. 280, 4880–4887 [DOI] [PubMed] [Google Scholar]
- 64.Richardson M. D., Balius A. M., Yamaguchi K., Freilich E. R., Barak L. S., Kwatra M. M. (2003) J. Neurochem. 84, 854–863 [DOI] [PubMed] [Google Scholar]
- 65.Pals-Rylaarsdam R., Gurevich V. V., Lee K. B., Ptasienski J. A., Benovic J. L., Hosey M. M. (1997) J. Biol. Chem. 272, 23682–23689 [DOI] [PubMed] [Google Scholar]
- 66.Lee K. B., Ptasienski J. A., Pals-Rylaarsdam R., Gurevich V. V., Hosey M. M. (2000) J. Biol. Chem. 275, 9284–9289 [DOI] [PubMed] [Google Scholar]
- 67.Whistler J. L., Tsao P., von Zastrow M. (2001) J. Biol. Chem. 276, 34331–34338 [DOI] [PubMed] [Google Scholar]
- 68.Moore C. A., Milano S. K., Benovic J. L. (2007) Annu. Rev. Physiol. 69, 451–482 [DOI] [PubMed] [Google Scholar]
- 69.Drake M. T., Shenoy S. K., Lefkowitz R. J. (2006) Circ. Res. 99, 570–582 [DOI] [PubMed] [Google Scholar]
- 70.Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (1999) J. Biol. Chem. 274, 32248–32257 [DOI] [PubMed] [Google Scholar]
- 71.Oakley R. H., Laporte S. A., Holt J. A., Caron M. G., Barak L. S. (2000) J. Biol. Chem. 275, 17201–17210 [DOI] [PubMed] [Google Scholar]
- 72.Pierce K. L., Lefkowitz R. J. (2001) Nat. Rev. Neurosci. 2, 727–733 [DOI] [PubMed] [Google Scholar]
- 73.Shenoy S. K., Lefkowitz R. J. (2003) J. Biol. Chem. 278, 14498–14506 [DOI] [PubMed] [Google Scholar]
- 74.Krueger K. M., Daaka Y., Pitcher J. A., Lefkowitz R. J. (1997) J. Biol. Chem. 272, 5–8 [DOI] [PubMed] [Google Scholar]
- 75.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 76.Reubi J. C. (2003) Endocr. Rev. 24, 389–427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.