Abstract
Spt6 is an essential transcription elongation factor and histone chaperone that binds the C-terminal repeat domain (CTD) of RNA polymerase II. We show here that Spt6 contains a tandem SH2 domain with a novel structure and CTD-binding mode. The tandem SH2 domain binds to a serine 2-phosphorylated CTD peptide in vitro, whereas its N-terminal SH2 subdomain, which we previously characterized, does not. CTD binding requires a positively charged crevice in the C-terminal SH2 subdomain, which lacks the canonical phospho-binding pocket of SH2 domains and had previously escaped detection. The tandem SH2 domain is apparently required for transcription elongation in vivo as its deletion in cells is lethal in the presence of 6-azauracil.
Keywords: Chromatin, Crystal Structure, Peptide Interactions, RNA Polymerase II, Transcription Elongation Factors
Introduction
Spt6 is an essential nuclear protein (1, 2) that is required for transcription elongation together with Spt4 and Spt5 (3, 4). Spt6 co-localizes with Spt5 on transcribed genes in Drosophila polytene chromosomes (5, 6). Spt6 stimulates the elongation rate of RNA polymerase II in vivo (7). There is strong evidence that Spt6 is a histone chaperone. In cells lacking functional Spt6, transcription can start from cryptic promoters within the coding regions of genes (8), and transcription reinitiation from regular promoters is facilitated (9). This suggests that Spt6 is required for the correct positioning of nucleosomes in transcribed regions. Consistently, Spt6 interacts with histone H3 and can deposit nucleosomes on DNA (10). Spt6 is further involved in mRNA 3′-end formation (11) and export (12).
The C-terminal region of mouse Spt6 binds the Ser-2-phosphorylated CTD,2 suggesting a mechanism for how Spt6 is recruited to elongating RNA polymerase II (12). The Spt6 C-terminal region was predicted to contain an SH2 domain. We recently reported the crystal structure of this SH2 domain (13), which is the only SH2 domain in the yeast genome (14). The structure contained residues 1251–1349 of Candida glabrata Spt6 and showed a fold that combined features of both SH2 domain subfamilies present in higher eukaryotes (15, 16), likely resembling a common evolutionary ancestor of all SH2 domains (13). The structure also revealed a shallow phospho-binding pocket that explained the unusual binding specificity for phospho-serine rather than phospho-tyrosine. Our previous work, however, also showed that the SH2 domain alone did not bind CTD phospho-peptides (13), although a more extended C-terminal region does (12).
To resolve this discrepancy, we investigated the structure and CTD binding of C-terminal fragments of Spt6 encompassing the SH2 domain and additional flanking regions. We found that a second SH2 domain is present directly C-terminal of the known SH2 domain. X-ray crystallography revealed that the two SH2 domains form an extended interface and are arranged in a previously unobserved manner into a rigid “tandem SH2 domain.” In contrast to the previously studied single SH2 domain, the tandem SH2 domain binds Ser-2-phosphorylated CTD peptides in vitro. The tandem SH2 domain is apparently also required for transcription elongation in vivo as shown by 6-azauracil (6-AU) phenotyping. Our results reveal a novel CTD-binding protein module and indicate that the structure and CTD binding function of the Spt6 C-terminal region is conserved in eukaryotes.
EXPERIMENTAL PROCEDURES
Preparation of the Spt6 Tandem SH2 Domain
DNA containing the tandem SH2 domain of C. glabrata Spt6 (residues 1250–1444) was amplified by polymerase chain reaction from genomic DNA and was cloned into expression vector pET28b(+) (Novagen) using restriction sites NdeI and NotI. This results in an open reading frame that encodes the tandem SH2 domain with an N-terminal hexahistidine tag with a thrombin cleavage site. The sequence was modified by the introduction of codons for methionine residues at the positions of residues Leu-1309, Leu-1317, and Leu-1411 by the rapid PCR site-directed mutagenesis (17). The resulting variant was used for selenomethionine labeling as described (18, 19). The tandem SH2 domain was expressed overnight at 18 °C in Escherichia coli BL21 CodonPlus(DE3)RIL strain cells (Stratagene). For selenomethionine labeling, we used E. coli strain B834(DE3)pLysS (Novagen). Cells were harvested, resuspended in 50 mm HEPES, pH 7.0, 1 m NaCl, 10 mm β-mercaptoethanol, 10 mm imidazole, and lysed by sonication. Cell lysates were subjected to affinity chromatography on a nickel-nitrilotriacetic acid column (Qiagen) followed by cleavage of the hexahistidine tag with thrombin and dialysis overnight at 4 °C. The tag and the undigested protein were removed on a second nickel-nitrilotriacetic acid column. DNA was removed by cation exchange chromatography (Mono S 10/100 GL, Amersham Biosciences). After gel filtration (Superose 12 10/300 GL, Amersham Biosciences), the sample was concentrated to 14.5 mg/ml and stored at 4 °C in 50 mm HEPES, pH 7.0, 100 mm NaCl, 5 mm DTT.
Crystal Structure Determination
The purified native and selenomethionine-labeled Spt6 tandem SH2 domain was crystallized by vapor diffusion using 22–24% PEG 8000, 50 mm MES, pH 6.5–7, 0.2 m sodium acetate as reservoir solution. Complete diffraction data at 1.7 and 1.6 Å resolution were obtained from crystals of the selenomethionine-labeled protein and unlabeled protein, respectively, and were processed by XDS and scaled by XSCALE (20). The structure was solved by multiwavelength anomalous diffraction using SOLVE, and density modification was performed with RESOLVE (21). The model was automatically built by the program ARP/wARP (22) with some manual rebuilding using Coot (23) and refined with program PHENIX (24). We added water molecules initially with Coot when a peak height greater than 3.0 σ was obtained in Fourier difference maps. We added more water molecules manually and further refined the model with PHENIX using bulk solvent scaling and individual isotropic B-factors. Refinement statistics are shown in Table 1.
TABLE 1.
X-ray diffraction data and refinement statistics for the Spt6 tandem SH2 domain
r.m.s., root mean square.
| Crystal | Se-Met | Native | ||
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | ||
| Unit cell dimensions (Å) | 109.2, 57.5, 85.1 | 111.2, 57.0, 88.0 | ||
| Peak | Inflection | Remote | ||
| Wavelength (Å) | 0.97958 | 0.97964 | 0.90784 | 0.91881 |
| Resolution (Å) | 71.1-1.70 | 54.9-1.72 | 54.9-1.72 | 40-1.6 |
| (1.74-1.70) | (1.74-1.72) | (1.76-1.72) | (1.64-1.60) | |
| Rsym (%) | 3.4 (51.2) | 2.9 (53.6) | 3.1 (55.0) | 3.2 (40.5) |
| I/σI | 14.4 (2.1) | 15.2 (2.0) | 15.2 (2.0) | 13.9 (2.5) |
| Completeness (%) | 95.7 (97.9) | 95.7 (97.6) | 95.6 (97.2) | 95.0 (95.6) |
| Redundancy | 2.43 (2.41) | 2.43 (2.41) | 2.45 (2.49) | 2.12 (2.17) |
| Refinement | ||||
| Resolution (Å) | 35-1.6 | |||
| Unique reflections | 56,064 | |||
| Rwork/Rfree (%) | 19.8/24.0 | |||
| No. of atoms | ||||
| Protein | 3351 | |||
| Water | 484 | |||
| Ligand/Ion | 28 | |||
| B-factors (Å2) | ||||
| Protein | 25.8 | |||
| Water | 37.8 | |||
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | |||
| Bond angles (°) | 0.994 | |||
Phosphopeptide Interaction Assay
For fluorescence anisotropy measurements, we used a HORIBA Jobin-Yvon Fluoromax 3 spectrofluorometer. The CTD phospho-peptides were synthesized by PANATecs GmbH (Tübingen), had the sequences P1 = SYpSPTSPSYpSPTSPS (pS, phospho-serine), P2 = SYpSPTSPSYSPTSPS, and P3 = SYSPTSPSYpSPTSPS, P4 = SYSPTSPSYSPTSPS, and were N-terminally labeled with fluorescein aminocaproic acid. The phospho-peptide (500 nm) was dissolved in 20 mm HEPES, pH 8.0, 10 mm NaCl, 5 mm DTT and was titrated at 20 °C with pure tandem SH2 domain or its mutant variants R1281K, K1434R, or R1281K/K1434R, and the fluorescence anisotropy was measured after 2 min when equilibrium was reached. Data were analyzed by non-linear least squared regression against A = A0 + ΔA × 0.5 × (b−(b2 − P × L)½) with b = P + L + KD (KD, apparent dissociation constant; P, protein concentration; L, ligand concentration).
Yeast Strains
Saccharomyces cerevisiae strains containing a C-terminal tandem affinity purification-tagged version of Spt6 and the same strain lacking amino acid residues 1250–1451 of Spt6 were as described (13).
RESULTS
Spt6 Contains a Tandem SH2 Domain
When we examined the Spt6 amino acid sequences flanking the known SH2 domain, we noted additional predicted secondary structure elements C-terminal to the known SH2 domain. In particular, the arrangement of predicted α-helices and β-strands suggested that a second SH2 domain could be present that had escaped detection by standard domain predication procedures. To investigate this, we prepared a recombinant variant of the Spt6 C-terminal region that spanned the known SH2 domain and the predicted SH2 domain (see “Experimental Procedures”). Soluble protein was purified and crystallized. The x-ray structure was determined by multiwavelength anomalous diffraction and refined at 1.6 Å resolution (Table 1).
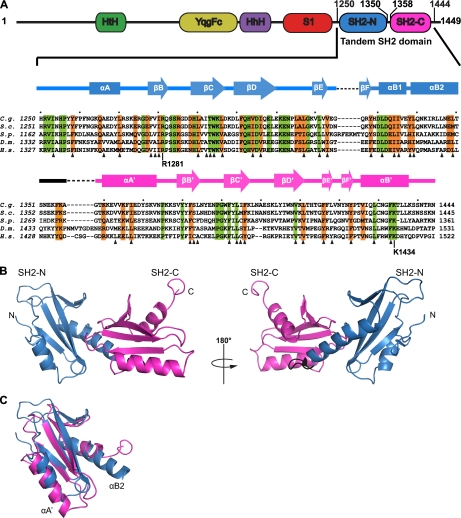
The structure revealed the previously reported SH2 domain (13) and additionally a second SH2 domain (Fig. 1). The two domains, designated hereafter subdomains SH2-N and SH2-C, intimately pack against each other. Therefore, we refer to this structure as tandem SH2 domain. The C-terminal helix of subdomain SH2-N is composed of two parts (αB1 and αB2). The αB2 part extends into subdomain SH2-C and makes extensive hydrophobic contacts with it, thereby stabilizing the tandem structure. Superposition of the two subdomains revealed that both have a similar structure, although the N- and C-terminal helices in SH2-C were longer and shorter, respectively, than in SH2-N (Fig. 1C). Because hydrophobic core residues are conserved from yeast to human (Fig. 1A, triangles), the tandem SH2 domain fold is conserved among eukaryotes.
FIGURE 1.
Structure of the Spt6 tandem SH2 domain. A, domain architecture of Spt6 (41) and alignment of the tandem SH2 domain amino acid sequence with corresponding regions in other eukaryotic Spt6 sequences (C. g., C. glabrata; S. c., S. cerevisiae; S. p., Schizosaccharomyces pombe; D. m., Drosophila melanogaster; H. s., Homo sapiens). Secondary structure elements are indicated above the sequences (cylinders, α-helices; arrows, β-strands). SH2-N is colored in blue, whereas SH2-C is in magenta. Invariant residues are in green, whereas conserved residues are in orange. Residues in the hydrophobic core are marked with triangles. HtH, helix-turn-helix domain, binds to double-stranded DNA; YqgFc, predicted to be a resolvase or ribonuclease, but in Spt6, catalytic residues are exchanged, thus probably not active; HhH, triple-helix-domain, binding to double-stranded DNA; S1, RNA-binding domain (13, 41). The sequence alignment of Spt6 tandem SH2 domains was obtained with ClustalW (42). B, ribbon representation of the Spt6 tandem SH2 domain structure. Subdomains are colored according to A. The left and right views are related by a 180° rotation around the vertical axis. C, superposition of SH2-N and SH2-C; structures were aligned by DALI-Lite (43), resulting in a root mean square deviation of 2.1 Å for 83 residues. The subdomains are colored according to A.
Unique Structure of the Tandem SH2 Domain
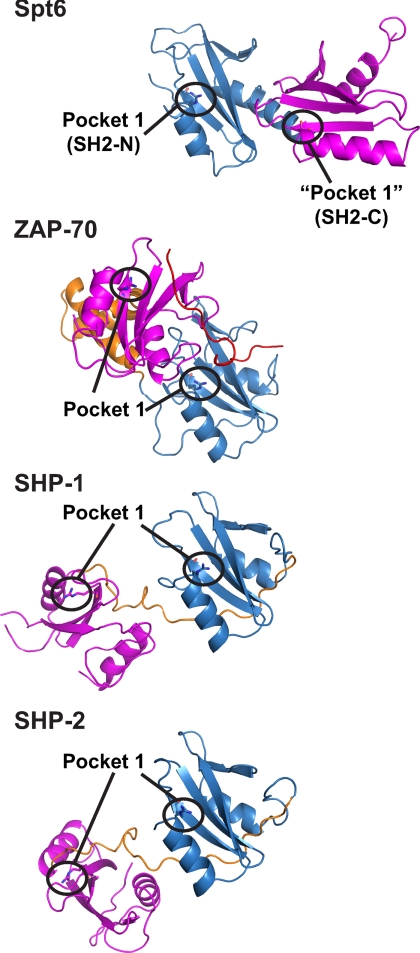
An arrangement of two subsequent SH2 domains was previously observed in signaling proteins present in higher eukaryotes. Structures of four proteins with two adjacent SH2 domains were published from the related tyrosine kinases ZAP-70 (25, 26) and Syk (27) and from the related tyrosine phosphatases SHP-2 (28, 29) and SHP-1 (30). A comparison revealed that these structures are all strikingly different from the Spt6 tandem SH2 domain and showed different arrangements of the two SH2 domains (Fig. 2). In addition, the two SH2 domains are always linked by an insertion of varying structure that does not exist in the Spt6 tandem SH2 domain. Although the SH2 domains pack against each other in ZAP-70 and Syk, they do not in the SHP phosphatases and are instead positioned via additional domains present in these proteins.
FIGURE 2.
Comparison with other SH2 domain tandem arrangements. The SH2 domains of ZAP-70 (Protein Data Bank (PDB) code 2oq1), SHP-1 (PDB code 2b3o), and SHP-2 (PDB code 2shp) are shown, with their N-terminal and C-terminal SH2 domains in blue and magenta, respectively. Additional structural elements between both SH2 domains are represented in orange, and a bound phospho-peptide in ZAP-70 is in red. The N-terminal SH2 domains in these proteins were superimposed onto Spt6 subdomain SH2-N.
The Tandem SH2 Domain Binds to the Ser-2-phosphorylated CTD in Vitro
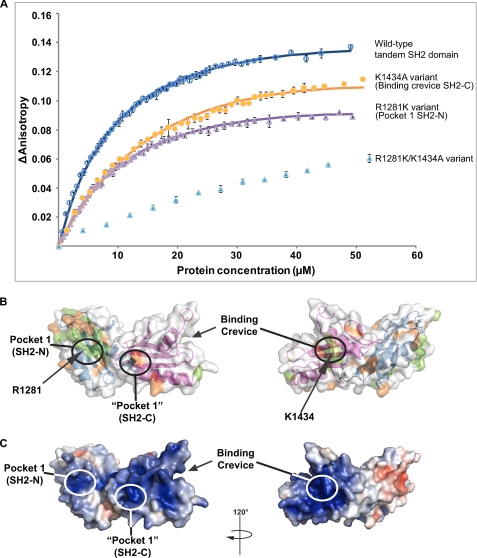
We used fluorescence anisotropy to investigate whether the tandem SH2 domain binds to CTD peptides with two heptapeptide repeats (see “Experimental Procedures”). We used four peptides. One peptide was phosphorylated on both Ser-2 residues (P1), whereas another one was unphosphorylated (P4). One peptide was phosphorylated only on the N-terminal Ser-2 residue (P2), and one was phosphorylated only at the C-terminal Ser-2 residue (P3). Titration equilibrium measurements revealed binding curves indicative for a specific protein-peptide interaction for the Ser-2-phosphorylated peptides P1, P2, and P3, whereas the unphosphorylated peptide P4 did not bind (Fig. 3A and Table 2). Regression analysis led to a dissociation constant of 10 μm for P1, and the binding affinity dropped 3–5-fold when only a single phosphorylated Ser-2 was present. Isolated SH2-N did not show specific binding to the same phospho-peptide (13). Thus, the tandem SH2 domain, but not SH2-N alone, binds to Ser-2-phosphorylated CTD peptides in vitro, and phosphorylation of two residues enhances binding.
FIGURE 3.
The tandem SH2 domain binds the RNA polymerase II CTD in vitro. A, fluorescence anisotropy titration of the CTD-derived phospho-peptide P1 (SYpSPTSPSYpSPTSPS; pS, phospho-serine) with different Spt6 tandem SH2 domain variants (open circles, wild-type tandem SH2 domain; open triangles, variant R1281K; filled circles, variant K1434A; filled triangles, variant R1281K/K1434A). The titration with the variant R1281K/K1434A could not be fitted with a simple binding model. The solid lines represent non-linear least square fits. B, Spt6 tandem SH2 domain surface conservation. Invariant and conserved residues are colored according to Fig. 1A. Indicated with circles are pocket 1 in SH2-N, the region that would correspond to pocket 1 in SH2-C, and the CTD-binding crevice in SH2-C. Residues Arg-1281 and Lys-1434 are represented as sticks and are labeled. The left and right views are related by a 120° rotation around the vertical axis. C, surface charge distribution. The domain surface is color-coded according to the electrostatic potential from negative (red) to positive (blue). Electrostatic surface potentials were calculated with APBS. The left and right views are related by a 120° rotation around the vertical axis.
TABLE 2.
Dissociation constants (mm) obtained by fluorescence anisotropy
NF, not fitted, i.e. the data could not be fitted with a nonlinear least-squared regression, indicating that there is no significant binding. ND, not determined.
| CTD peptide P1 | CTD peptide P2 | CTD peptide P3 | CTD peptide P4 | |
|---|---|---|---|---|
| Tandem SH2 variant | ||||
| WT | 10.4 ± 0.2 | 34.7 ± 2.2 | 46.0 ± 2.3 | NF |
| R1281K | 13.9 ± 0.6 | 40.7 ± 1.1 | NF | ND |
| K1434A | 25.7 ± 0.7 | NF | NF | ND |
| R1281K/K1434A | NF | NF | NF | NF |
CTD Binding Requires a Conserved Non-canonical Surface Crevice
The structure revealed that the canonical phospho-binding pocket in SH2-N remains accessible in the tandem domain (Fig. 3B, Pocket 1). The region in SH2-C corresponding to the canonical pocket 1, however, lacks the conserved arginine found in all other SH2 domains, and instead the side chain of Tyr-1393 occupies the space of the canonical pocket. However, a conserved surface crevice was observed on the opposite side of SH2-C (Fig. 3B). This crevice is positively charged and is flanked by the invariant residue Lys-1434 that could contribute to phospho-peptide binding (Figs. 1A and 3, B and C).
To investigate whether SH2-N pocket 1 and/or the SH2-C crevice are required for phospho-CTD binding, we prepared three mutant variants of the tandem SH2 domain and tested them for binding the doubly Ser-2-phosphorylated peptide P1 (Fig. 3A). Mutation of residue Arg-1281 in pocket 1 of SH2-N to lysine led to a 1.4-fold increase of the dissociation constant. Mutation of residue Lys-1434 in the crevice of SH2-C to alanine caused a 2.5-fold increase of the dissociation constant. When both point mutations were combined into a double mutant variant of the tandem SH2 domain, binding affinity dropped strongly. Binding to the phospho-CTD peptide did not reach saturation and could not be fitted using a simple binding model. This non-saturation behavior shows that the double mutant only interacts non-specifically with the phospho-peptide. Thus, both patches contribute to the binding affinity, and the CTD peptide apparently extends over the tandem domain surface from pocket 1 in SH2-N via a positively charged surface to the non-canonical crevice in SH2-C that strongly contributes to the binding affinity (Fig. 3).
We next investigated how the domain mutations influence binding of CTD peptides with single Ser-2 phosphorylations. The mutant tandem SH2 domain variants generally did not bind to single phosphorylated CTD peptides anymore, except for variant R1281K, which bound the single phosphorylated CTD peptide P2 with almost the same affinity as the wild-type domain. This indicated that the N-terminal phosphorylated Ser-2 residue binds to the conserved surface crevice on SH2-C. However, confirmation of this proposal and additional details of the interaction await a structure of the tandem domain in complex with a CTD peptide. Modeling indicates that a single CTD repeat cannot span the distance between the binding sites on the two SH2 subdomains, consistent with multiple CTD repeats contributing in the interaction.
Deletion of the Tandem SH2 Domain Causes an Elongation Defect in Vivo
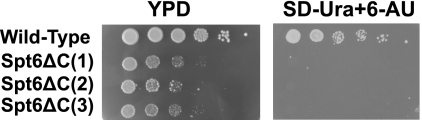
Deletion of the C-terminal region in Spt6 that corresponds to the tandem SH2 domain in yeast causes a slow growth phenotype (13, 31). To investigate whether this phenotype goes along with a transcription elongation defect, we tested the sensitivity of this deletion strain to 6-AU. Sensitivity to this nucleotide-depleting drug is observed in many elongation factor mutants (32), including mutants of the Spt6 gene (4, 33). We observed that the deletion strain failed to grow on 6-AU-containing (50 μg/ml) medium (Fig. 4). This shows that the Spt6 tandem SH2 domain is essential for cell viability under conditions when transcription elongation is impaired.
FIGURE 4.
The SH2 tandem domain is apparently involved in transcription elongation in vivo. Cultures of wild-type and Spt6 tandem domain deletion yeast strains (13, 31) were grown in YPD medium at 30 °C overnight and diluted to an optical density at 600 nm of 5 with fresh medium. The same amount of cells was spotted on plates in 10-fold serial dilutions. Plates were incubated for 5 days at 30 °C and inspected daily. 6-AU was added to SD-Ura plates at a concentration of 50 μg/ml.
DISCUSSION
Here we report that the C-terminal region of the essential transcription elongation factor Spt6 contains a tandem SH2 domain that combines the previously described SH2 domain (subdomain SH2-N) with a second, subsequent SH2 domain (subdomain SH2-C) into a single domain of novel structure. We show that the tandem SH2 domain, in contrast to SH2-N alone, binds to Ser-2-phosphorylated CTD peptides in vitro. A single phosphorylated Ser-2 residue is required and sufficient for binding, but the presence of a second phosphorylated Ser-2 residue strongly enhances binding, consistent with published data (34–36). The tandem domain is also required for cell growth in the presence of the nucleotide-depleting drug 6-AU in vivo, indicating its importance for transcription elongation.
We further show that the tandem SH2 domain of Spt6 is unique in its structure and mode of CTD binding. Two adjacent SH2 domains are found in several proteins in higher eukaryotes but are arranged in a different way in known three-dimensional structures (Fig. 2). Also, the tandem domain contains a canonical phospho-binding pocket only in its subdomain SH2-N, but not in its subdomain SH2-C, which lacks the invariant arginine residue found in all other SH2 domains.
Consistent with observed structural differences, the Spt6 tandem SH2 domain has a unique phospho-peptide binding mode. Two surface patches are involved in CTD binding, the canonical phospho-binding pocket in subdomain SH2-N and a non-canonical basic crevice in subdomain SH2-C that comprises an invariant lysine residue that contributes to binding. Mutating either one of these two conserved basic residues on the two CTD-binding surface patches resulted in a weaker interaction. It is likely that the SH2-C crevice interacts with a phospho-serine residue because a CTD peptide with a single phosphorylated Ser-2 residue binds equally well to wild-type tandem SH2 domain and a variant in which the SH2-N pocket is mutated. These results are consistent with an extended peptide-binding surface that reaches from the SH2-N pocket to the SH2-C crevice. Structural features that may provide specificity for CTD binding are, however, difficult to identify in the absence of a complex crystal structure.
Canonical SH2 domains bind phosphorylated target peptides via two pockets, one that recognizes the phosphorylated tyrosine side chain (pocket 1) and one that contacts residues C-terminal of the phospho-tyrosine (pocket 2), generally at register +3. Pocket 2 generally contributes to the specificity of the interaction. In Spt6 SH2-N, pocket 1 is present and used for peptide binding, but the previously described pocket 2, which was proposed to play a similar role as in other SH2 domains (13), is occluded in the tandem SH2 domain structure (Fig. 3B). In SH2-C, neither pocket 1 nor pocket 2 is observed, but instead a conserved crevice on the opposite face contributes to peptide binding.
Despite all these differences between the Spt6 tandem SH2 domain and other arrangements of two SH2 domains, there is a functional analogy. Consistent with our observations, individual ZAP-70 SH2 domains do not bind the target phospho-peptide but do so when in a tandem arrangement (37). More generally, the phospho-peptide binding specificity of single SH2 domains is increased by the simultaneous binding of the peptide to an additional domain (38). Tandem SH2 domains therefore may generally increase target specificity due to cooperative peptide binding as suggested (39).
Recent data indicate that the tandem SH2 domain is required for retaining Spt6 at the end of transcribed genes, rather than recruiting it to genes. We have recently shown by high resolution genome-wide chromatin immunoprecipitation that Spt6 is recruited during an early transition in the 5′-region of genes near the transcription start site before high levels of Ser-2 phosphorylation are reached (40). Deletion of the genomic region encoding the tandem SH2 domain lowered the level of recruitment, in particular at the 3′-region around the polyadenylation site, where levels of Ser-2 phosphorylation are high (40). These results indicated that Spt6 is recruited near the transcription start site with the use of a CTD Ser-2 phosphorylation-independent mechanism in vivo, but full recruitment and retaining of Spt6 at the polyadenylation site require the CTD-binding tandem SH2 domain.
Acknowledgments
We thank Dirk Kostrewa and other members of the Cramer laboratory for help. Part of this work was performed at the Swiss Light Source (SLS) at the Paul Scherrer Institute (Villigen, Switzerland). We thank the crystallization facility at the Max Planck Institute (MPI) for Biochemistry, Martinsried, Germany.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the Transregio 5, the Nanosystems Initiative Munich (NIM) the Elitenetzwerk Bayern, the Fonds der Chemischen Industrie, and the Jung Stiftung.
The atomic coordinates and structure factors (code 3PJP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CTD
- C-terminal repeat domain
- 6-AU
- 6-azauracil.
REFERENCES
- 1.Clark-Adams C. D., Winston F. (1987) Mol. Cell. Biol. 7, 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swanson M. S., Carlson M., Winston F. (1990) Mol. Cell. Biol. 10, 4935–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson M. S., Winston F. (1992) Genetics 132, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartzog G. A., Wada T., Handa H., Winston F. (1998) Genes Dev. 12, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrulis E. D., Guzmán E., Döring P., Werner J., Lis J. T. (2000) Genes Dev. 14, 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan C. D., Morris J. R., Wu C., Winston F. (2000) Genes Dev. 14, 2623–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardehali M. B., Yao J., Adelman K., Fuda N. J., Petesch S. J., Webb W. W., Lis J. T. (2009) EMBO J. 28, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan C. D., Laprade L., Winston F. (2003) Science 301, 1096–1099 [DOI] [PubMed] [Google Scholar]
- 9.Adkins M. W., Tyler J. K. (2006) Mol. Cell 21, 405–416 [DOI] [PubMed] [Google Scholar]
- 10.Bortvin A., Winston F. (1996) Science 272, 1473–1476 [DOI] [PubMed] [Google Scholar]
- 11.Kaplan C. D., Holland M. J., Winston F. (2005) J. Biol. Chem. 280, 913–922 [DOI] [PubMed] [Google Scholar]
- 12.Yoh S. M., Cho H., Pickle L., Evans R. M., Jones K. A. (2007) Genes Dev. 21, 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dengl S., Mayer A., Sun M., Cramer P. (2009) J. Mol. Biol. 389, 211–225 [DOI] [PubMed] [Google Scholar]
- 14.Maclennan A. J., Shaw G. (1993) Trends Biochem. Sci. 18, 464–465 [DOI] [PubMed] [Google Scholar]
- 15.Koch C. A., Anderson D., Moran M. F., Ellis C., Pawson T. (1991) Science 252, 668–674 [DOI] [PubMed] [Google Scholar]
- 16.Pawson T., Gish G. D., Nash P. (2001) Trends Cell Biol. 11, 504–511 [DOI] [PubMed] [Google Scholar]
- 17.Weiner M. P., Costa G. L. (1994) PCR Methods Appl. 4, S131–S136 [DOI] [PubMed] [Google Scholar]
- 18.Meinhart A., Blobel J., Cramer P. (2003) J. Biol. Chem. 278, 48267–48274 [DOI] [PubMed] [Google Scholar]
- 19.Budisa N., Steipe B., Demange P., Eckerskorn C., Kellermann J., Huber R. (1995) Eur. J. Biochem. 230, 788–796 [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 21.Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 22.Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deindl S., Kadlecek T. A., Brdicka T., Cao X., Weiss A., Kuriyan J. (2007) Cell 129, 735–746 [DOI] [PubMed] [Google Scholar]
- 26.Hatada M. H., Lu X., Laird E. R., Green J., Morgenstern J. P., Lou M., Marr C. S., Phillips T. B., Ram M. K., Theriault K., Zoller M. J., Karas J. L. (1995) Nature 377, 32–38 [DOI] [PubMed] [Google Scholar]
- 27.Fütterer K., Wong J., Grucza R. A., Chan A. C., Waksman G. (1998) J. Mol. Biol. 281, 523–537 [DOI] [PubMed] [Google Scholar]
- 28.Eck M. J., Pluskey S., Trüb T., Harrison S. C., Shoelson S. E. (1996) Nature 379, 277–280 [DOI] [PubMed] [Google Scholar]
- 29.Hof P., Pluskey S., Dhe-Paganon S., Eck M. J., Shoelson S. E. (1998) Cell 92, 441–450 [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Liu L., He D., Song X., Liang X., Zhao Z. J., Zhou G. W. (2003) J. Biol. Chem. 278, 6516–6520 [DOI] [PubMed] [Google Scholar]
- 31.Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., Mellor J. (2008) Mol. Cell. Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Exinger F., Lacroute F. (1992) Curr. Genet. 22, 9–11 [DOI] [PubMed] [Google Scholar]
- 33.Bucheli M. E., Buratowski S. (2005) EMBO J. 24, 2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P., Kenney J. M., Stiller J. W., Greenleaf A. L. (2010) Mol. Biol. Evol. 27, 2628–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stiller J. W., Cook M. S. (2004) Eukaryotic Cell 3, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P., Greenleaf A. L., Stiller J. W. (2008) Mol. Biol. Evol. 25, 719–727 [DOI] [PubMed] [Google Scholar]
- 37.Wange R. L., Malek S. N., Desiderio S., Samelson L. E. (1993) J. Biol. Chem. 268, 19797–19801 [PubMed] [Google Scholar]
- 38.Machida K., Mayer B. J. (2005) Biochim. Biophys. Acta 1747, 1–25 [DOI] [PubMed] [Google Scholar]
- 39.Ottinger E. A., Botfield M. C., Shoelson S. E. (1998) J. Biol. Chem. 273, 729–735 [DOI] [PubMed] [Google Scholar]
- 40.Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010) Nat. Struct. Mol. Biol. 17, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 41.Johnson S. J., Close D., Robinson H., Vallet-Gely I., Dove S. L., Hill C. P. (2008) J. Mol. Biol. 377, 1460–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labarga A., Valentin F., Anderson M., Lopez R. (2007) Nucleic Acids Res. 35, W6–W11 [DOI] [PMC free article] [PubMed] [Google Scholar]