Abstract
Mammalian reoviruses replicate in a broad range of hosts, cells, and tissues. These viruses display strain-dependent variation in tropism for different types of cells in vivo and ex vivo. Early steps in the reovirus life cycle, attachment, entry, and disassembly, have been identified as pivotal points of virus-cell interaction that determine the fate of infection, either productive or abortive. However, in studies of the differential capacity of reovirus strains type 1 Lang and type 3 Dearing to replicate in Madin-Darby canine kidney (MDCK) cells, we found that replication efficiency is regulated at a late point in the viral life cycle following primary transcription and translation. Results of genetic studies using recombinant virus strains show that reovirus tropism for MDCK cells is primarily regulated by replication protein μ2 and further influenced by the viral RNA-dependent RNA polymerase protein, λ3, depending on the viral genetic background. Furthermore, μ2 residue 347 is a critical determinant of replication efficiency in MDCK cells. These findings indicate that components of the reovirus replication complex are mediators of cell-selective viral replication capacity at a post-entry step. Thus, reovirus cell tropism may be determined at early and late points in the viral replication program.
Keywords: Double-stranded RNA Viruses, Reovirus, Viral Genetics, Viral Polymerase, Viral Replication, Viral Tropism
Introduction
Viral tropism, defined by the range of hosts and tissues productively infected, creates natural biologic groupings among viruses that correlate with infection pathology, clinical disease expression, and epidemiology. Delineating the molecular basis of viral cell tropism is fundamental to the elucidation of disease mechanisms and the identification of viral and cellular targets for treatment and prevention of infection. Growing threats posed by zoonotic viral diseases with global pandemic potential (1) underscore the necessity for an enhanced understanding of unifying principles that influence viral tropism and host range. We are conducting studies using mammalian reoviruses to better understand the nature of virus-cell interactions that dictate unique tropism properties.
Mammalian orthoreoviruses (hereafter referred to as reoviruses) are an established model for studies of viral replication and pathogenesis (2–6). Reoviruses have contributed to the development of paradigms of viral disease based on discrete patterns of viral tropism for particular host cells and tissues. The reovirus virion is a nonenveloped, double layered, icosahedral particle consisting of an outer shell surrounding an inner core that contains 10 double-stranded (ds) RNA gene segments. The viral genome encodes eight structural and three nonstructural proteins. Viral RNA gene segments can be resolved into small, medium, and large size classes by SDS-PAGE, corresponding to σ, μ, and λ proteins, respectively. Four reovirus serotypes have been identified based on neutralization and hemagglutination studies (7–9). Strains type 1 Lang (T1) 3 and type 3 Dearing (T3) serve as prototypes for serotype 1 and 3 reoviruses, respectively. T1 and T3 display numerous phenotypic differences, and the genetic basis of several biologic polymorphisms has been defined using reassortant viruses derived from co-infections of T1 and T3 (6). Such analyses have provided key insights into the mechanisms of viral replication and disease.
The reovirus replication program (reviewed in Ref. 6) begins with attachment using proteinaceous or carbohydrate receptors followed by entry into host cells by receptor-mediated endocytosis. Proteolytic disassembly of the outer capsid within endosomes facilitates membrane penetration, leading to release of transcriptionally active cores into the cytoplasm. Full-length, positive-strand, capped, and nonpolyadenylated RNAs synthesized by core particles serve as templates for translation and synthesis of new genomic dsRNA. Secondary viral mRNAs transcribed from new dsRNA templates fuel subsequent rounds of protein synthesis. The source of these late transcripts is presumed to be subviral particles. Virion assembly is completed by the addition of outer capsid proteins to core particles. Reovirus replication and assembly are thought to occur within viral intracytoplasmic inclusions (10), which strictly depend on nonstructural proteins σNS and μNS and structural protein μ2 for proper formation and function in reovirus-infected cells (11, 12). Inclusions are detectable within 4 h post-infection, lack a delimiting membrane, contain viral proteins, dsRNA and virion particles at various stages of morphogenesis, and are composed of highly ordered arrays of mature virions at late times of infection (13–19).
The 83-kDa μ2 protein, encoded by the M1 gene segment, is essential for viral replication (20). The μ2 protein functions in viral transcription (21, 22) and virion particle assembly (22). As an integral component of viral inclusions, μ2 determines kinetics of inclusion formation (23) and inclusion morphology (24). Fluorescence microscopy performed on a variety of cell types has shown that T1 and T3 form filamentous and globular inclusions, respectively (11, 20, 23, 24), which is explained by a differential capacity of T1 and T3 μ2 proteins to bind microtubules and anchor inclusions to the cytoskeleton (24).
The μ2 protein is a component of the reovirus core, which contains ∼20 molecules of μ2 per virion particle (6, 25, 26). Although the precise position of μ2 is unknown, several lines of evidence from biochemical, structural, and genetic studies indicate that μ2 resides at the vertex base in close approximation to λ3, the viral RNA-dependent RNA polymerase (RdRp) (22, 27–31). Single-stranded RNA and dsRNA (32) are bound by μ2, which possesses nucleotide triphosphatase and RNA triphosphatase activities enhanced by the presence of λ3 (27, 28). Two regions of μ2 sequence bare similarity to the nucleotide-binding motifs of ATPases and are essential for triphosphatase action (27). Strain-specific differences in transcriptional efficiency of core particles are determined by μ2 (21), and reovirus strains containing temperature-sensitive lesions in μ2 protein display defects in replication and particle assembly at restrictive temperatures (22). Thus, μ2 is a probable subunit of the functional reovirus polymerase complex.
In addition to its central place in viral replication, μ2 also regulates reovirus cell tropism and virulence. Strain-dependent viral replication efficiency has been genetically linked to μ2 in studies using primary and transformed cells, including murine cardiac cells (33–36), bovine aortic endothelial cells (37), and Madin-Darby canine kidney (MDCK) cells (38). The capacity of reovirus to cause myocarditis in neonatal mice is regulated by μ2 and tied to the innate antiviral response. Reovirus induction of and sensitivity to type 1 interferon (IFN) in cardiac myocytes is coupled through μ2 to viral replication efficiency, organ pathology, and survival (34–36). The μ2 protein of myocarditic strains such as T1 antagonizes the IFN response by mediating nuclear sequestration of interferon regulatory factor-9 (39).
A previous study using T1 × T3 reassortant viruses showed that the M1 and λ3-encoding L1 genes segregate with differences in the replication efficiency of T1 and T3 in MDCK cells (38). This dichotomy presents a useful experimental platform to rigorously address the individual and cooperative roles of μ2 and λ3 in reovirus cell tropism and identify μ2-sensitive steps in the viral life cycle. Therefore, we conducted studies to genetically and biochemically characterize strain-specific differences in reovirus replication efficiency in MDCK cells. Our findings indicate the μ2 protein is the primary determinant of viral replication efficiency in these cells and that λ3 is a conditional co-regulator of μ2 function depending on the viral genetic background. Furthermore, the critical μ2-dependent step in MDCK cells occurs at a later point in the reovirus replication program subsequent to primary rounds of viral transcription and translation. These results enhance our understanding of the viral replication apparatus and the viral and cellular requirements crucial for its activity.
EXPERIMENTAL PROCEDURES
Cells and Viruses
L cells were grown in Joklik's modified Eagle's minimal essential medium (Lonza, Walkersville, MD) supplemented to contain 5% fetal calf serum (Cellgro, Manassas, VA), 2 mm l-glutamine (Invitrogen), 100 units of penicillin G/ml (Invitrogen), 100 μg of streptomycin/ml (Invitrogen), and 0.25 μg of amphotericin B/ml (Sigma). MDCK cells were grown in DMEM supplemented to contain 4.5 g/liter sodium pyruvate (Cellgro), 10% fetal calf serum, 2 mm l-glutamine, 100 units of penicillin G/ml, 100 μg of streptomycin/ml, and 0.25 μg of amphotericin B/ml. BHK-T7 cells were grown in DMEM (Invitrogen) supplemented to contain 5% fetal calf serum, 2 mm l-glutamine, 2% minimal essential medium amino acid solution (Invitrogen), and 1 mg/ml geneticin (Invitrogen). Strains T1, T3, T1-T3M1, T3-T1M1, and all other reassortant strains used in this study were recovered by reverse genetics as described previously (40, 41). Virus was purified from L cells by CsCl gradient centrifugation (42). Viral titers were determined by plaque assay using L cell monolayers as described previously (43). Attenuated vaccinia virus strain rDIs-T7pol expressing T7 RNA polymerase (44) was propagated in chicken embryo fibroblasts (40).
Construction of Mutant Viruses
T1 × T3 M1 chimeric cDNAs were generated by inserting the EcoRV-RsrII (Ch1), MfeI-RsrII (Ch2), PsiI-RsrII (Ch3), and NdeI-RsrII (Ch4) fragments of pT7-M1T1L (41) into the pT7-M1T3D vector (40). M1 point mutants were generated using PCR with mutagenic primers and the pT7-M1T1L and pT7-M1T3D plasmids. Vectors containing mutant M1 genes were substituted for WT M1 to recover recombinant viruses using reverse genetics. Plasmid sequences were determined to confirm fidelity of mutagenesis and cloning.
Quantification of Virus Infectivity
Monolayers of L cells or MDCK cells (∼5 × 105 cells) seeded in 24-well plates (Costar, Corning, NY) were adsorbed with virus at an m.o.i. of 2 pfu/cell. After 1 h of adsorption at room temperature, the viral inoculum was removed, cells were washed with PBS, and fresh medium was added. Cells were incubated at 37 °C for various intervals and removed to −80 °C. Viral titers in cell lysates were determined by plaque assay using L cells (43). Viral yield was calculated as the difference between log10 titer at 24 h and log10 titer at 0 h. Negative differences were assigned a value of zero. For the purpose of calculating fold differences in viral yields, non-log transformed data were used, and a yield of less than 1 was considered to be 1.
Immunofluorescence Detection of Reovirus Infection
L cells plated on untreated glass coverslips and MDCK cells plated on poly-l-lysine (Sigma)-treated glass coverslips in 24-well plates were adsorbed with virus at an m.o.i. of 20 pfu/cell. Following incubation at 37 °C for various intervals, cells were fixed and stained with rabbit μ2-specific (45) and guinea pig σNS-specific (46) antisera followed by Alexa 546-conjugated goat anti-rabbit secondary antibody (Invitrogen), Alexa 488-conjugated goat anti-guinea pig secondary antibody (Invitrogen), and ToPro3 (Invitrogen). Images were acquired using a Zeiss LSM 510 META inverted confocal microscope.
Analysis of Viral RNA Production Using Reverse Transcription-Quantitative PCR
Monolayers of L cells or MDCK cells (∼5 × 105 cells) seeded in 24-well plates were adsorbed with virus at an m.o.i. of 10 pfu/cell. After 1 h of adsorption at room temperature, the viral inoculum was removed; cells were washed with PBS, and fresh medium was added. Following incubation at 37 °C for various intervals, cultures were frozen at −80 °C, and total RNA was extracted from 140 μl of thawed lysate using the RNeasy mini kit (Qiagen, Valencia, CA). RNA was incubated at 95 °C for 3 min and immediately placed on ice. Reovirus S4 RNA in 10 μl of RNA extract was quantified using the SuperScript III Platinum One-Step qRT-PCR system (Invitrogen). PCRs were prepared according to the manufacturer's specifications with minor modifications. The S4-specific fluorogenic probe used was 5′-dFAM-AGCGCGCAAGAGGGATGGGA-BHQ-1–3′ (Biosearch Technologies, Novato, CA). Either forward (S4 83F, 5′-CGCTTTTGAAGGTCGTGTATCA-3′) or reverse (S4 153R, 5′-CTGGCTGTGCTGAGATTGTTTT-3′) primer corresponding to the viral S4 gene was used for reverse transcription performed at 50 °C for 15 min. Following 3 min of incubation at 95 °C, the second primer was added, and 40 cycles of quantitative PCR were performed at 95 °C for 15 s followed by 60 °C for 30 s. Total S4 RNA (combined dsRNA and mRNA) was quantified using the S4-specific reverse primer (complementary to the positive strand) in the reverse transcription step, and double-stranded S4 RNA was quantified using the S4-specific forward primer (complementary to the negative strand) in the reverse transcription step. Inclusion of only the reverse or forward primer in the reverse transcription step resulted in specific detection of the positive or negative strand of S4 RNA, respectively. Total S4 RNA is represented by the amount of positive-sense template, whereas dsRNA is equivalent to the amount of negative-sense template. S4 mRNA was calculated by subtracting the amount of dsRNA from total RNA. Standard curves relating Ct values to copies of positive- or negative-sense RNA template were generated using 10-fold dilutions of viral RNA extracted from purified virion particles (QIAamp viral RNA purification kit, Qiagen) and quantified by spectrophotometry. The amount of total and dsRNA in each sample was then extrapolated from standard curves generated with the reverse and forward primers, respectively. Standard curves generated using forward and reverse primers in the reverse transcription step were consistently similar, reflecting comparable amplification efficiencies of S4 RNA positive- and negative-sense strands.
Analysis of Viral Protein Synthesis
Monolayers of L cells or MDCK cells (∼5 × 105 cells) seeded in 24-well plates were adsorbed with virus at an m.o.i. of 100 pfu/cell. After 1 h at room temperature, the inoculum was removed; fresh medium was added, and cells were incubated at 37 °C for various intervals. Cells were incubated with 50 mCi of [35S]methionine-cysteine (PerkinElmer Life Sciences) in methionine-free medium (MP Biomedicals, Solon, OH) for 1 h prior to harvest; the medium was removed, and 50 μl of lysis buffer (0.1 m NaCl, 1 mm EDTA, 10 mm Tris (pH 7.4), and 0.5% IGEPAL) was added. Cell lysates were centrifuged at 14,000 × g at 4 °C for 15 min, and 20 μl of the resultant supernatant was mixed with an equal volume of Laemmli sample buffer (Bio-Rad). Samples were incubated at 95 °C for 5 min and electrophoresed in a 10% SDS-polyacrylamide gel, which was dried onto filter paper and exposed to film (BioMaz MR film, Eastman Kodak Co.).
RESULTS
Reovirus Strain-specific Replication in MDCK Cells
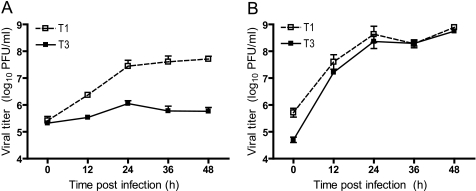
Previous studies revealed a strain-specific capacity of reovirus T1 and T3 to replicate in MDCK cells (38). Using classic reassortant analysis, this phenotype was genetically mapped to the λ3-encoding L1 and μ2-encoding M1 gene segments. To confirm that recombinant viruses recovered using reverse genetics (40, 41) recapitulate growth characteristics of native T1 and T3 in MDCK cells, recombinant strain T1 and recombinant strain T3 were used to infect MDCK cells at an m.o.i. of 2 pfu/cell, and viral titers in cell lysates were monitored over the course of infection. Consistent with previous findings, strain T1 achieved an ∼100-fold increase in viral titer over 48 h of growth (Fig. 1A). Conversely, strain T3 exhibited a minimal increase in titer during the course of infection. In contrast to growth in MDCK cells, both strains replicated with similar kinetics and produced equivalently high titers in L cells (Fig. 1B). These results demonstrate that differences in tropism for MDCK cells displayed by the native viruses are recapitulated by the recombinant strains.
FIGURE 1.
Growth of reovirus in cultured cells. MDCK cells (A) or L cells (B) were infected at an m.o.i. of 2 pfu/cell, and viral titers in cell lysates were determined at the time points shown by plaque assay using L cell monolayers. Results represent the mean of triplicate experiments. Error bars indicate S.D.
Genetic Analysis of Reovirus Replication in MDCK Cells
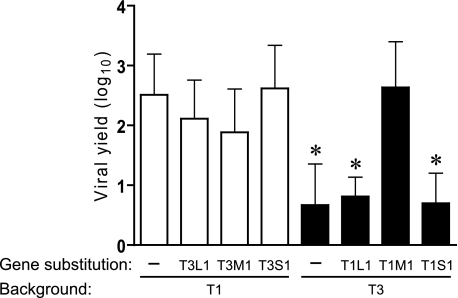
To more fully understand the contributions of individual reovirus genes to viral replication efficiency in MDCK cells, we generated monoreassortant viruses (containing nine gene segments from one strain, denoted first in the virus name, and one gene derived from a different strain, indicated second in the virus name) and assessed production of infectious progeny virions. L1, M1, and S1 monoreassortant viruses were generated in both T1 and T3 genetic backgrounds. L1 and M1 were reassorted because these genes were previously identified as determinants of reovirus tropism for MDCK cells (38). Monoreassortant viruses encoding reciprocal exchanges of the S1 gene, encoding viral attachment protein σ1 and nonstructural protein σ1s, were tested for growth in MDCK cells because receptor engagement is responsible for differences in the infectivity of type 1 and type 3 reoviruses for a variety of cells and tissues (6, 47–50). Parental and monoreassortant viruses were used to infect MDCK cells at an m.o.i. of 2 pfu/cell, followed by quantification of viral yields after 24 h of growth. Consistent with the earlier mapping study (38), the T1 M1 gene was sufficient to support growth of T3 to the level of T1, whereas the S1 gene was not associated with strain-specific differences in viral yield (Fig. 2). However, contrary to previous findings (38), an independent association of the L1 gene with viral replication was not observed using L1 monoreassortant viruses; T1-T3L1 and T3-T1L1 displayed high and low yields, respectively, mimicking their parental strains. Furthermore, the effect of M1 on viral replication was unidirectional; the T3 M1 gene segment alone did not diminish T1 yield. These results indicate that the μ2 protein regulates reovirus replication efficiency in MDCK cells, but the effects of μ2 on replication are subject to modulation by other viral determinants. We assessed potential cooperativity between the L1 and M1 genes in viral yield experiments using L1/M1 double reassortant viruses in the T1 (T1-T3L1M1) and T3 (T3-T1L1M1) backgrounds. Pairing T3 L1 and T3 M1 genes in the T1 background significantly reduced T1 viral yield in MDCK cells at 24 h post-infection (Fig. 3A). In contrast, growth of T3-T1M1 was unaffected by the strain origin of the L1 gene.
FIGURE 2.
Growth of monoreassortant viruses in MDCK cells. Cells were infected with L1, M1, or S1 monoreassortant viruses at an m.o.i. of 2 pfu/cell, and viral yields (relative to 0 h) at 24 h post-infection were determined by plaque assay using L cell monolayers. Results represent the mean of triplicate experiments. Error bars indicate S.D. *, p < 0.05 in comparison with strain T1 (Student's t test).
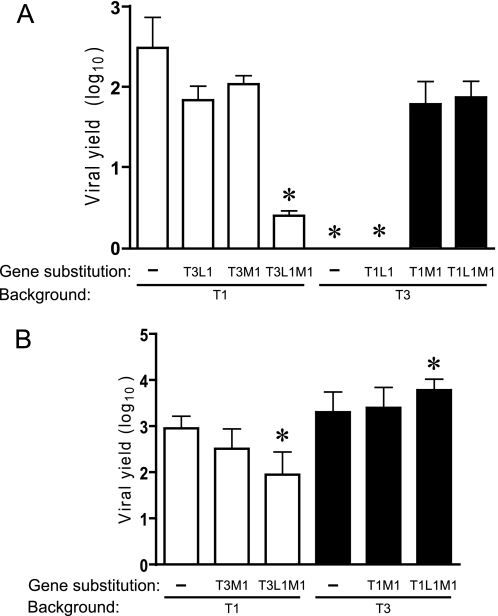
FIGURE 3.
Yields of L1/M1 reassortant viruses in cultured cells. MDCK cells (A) or L cells (B) were infected at an m.o.i. of 2 pfu/cell, and viral yields (relative to 0 h) were determined at 24 h post-infection by plaque assay using L cell monolayers. Results represent the mean of triplicate experiments. Error bars indicate S.D. *, p < 0.05 in comparison with strain T1 (Student's t test).
Yields of M1 monoreassortant and L1/M1 double-reassortant viruses were also determined using L cells to control for potential constitutive effects of the T1 and T3 L1 and M1 genes on viral replication. In these experiments, all strains produced high yields in L cells (Fig. 3B). Reassortant strain T1-T3L1M1 grew less efficiently in L cells compared with the T1 parental strain. However, the reduction in viral yield was 10-fold compared with a 100-fold difference in MDCK cells. The yield of T3-T1L1M1 exceeded that of T3 by ∼3-fold, but this difference was modest compared with the 70-fold increase in MDCK cells. Taken together, results of experiments using M1 and L1 reassortant viruses indicate that μ2 is a key determinant of reovirus tropism for MDCK cells and, additionally, provide evidence for co-regulation of viral replication by polymerase protein λ3, conditioned on the viral genetic background.
Identification of Sequences in μ2 That Mediate Reovirus Tropism for MDCK Cells
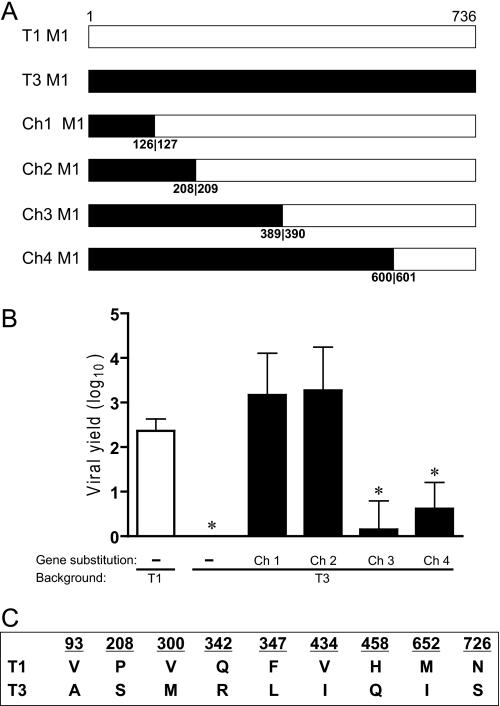
To identify μ2 sequence features of reovirus replication efficiency in MDCK cells, we generated a panel of T1 × T3 μ2-chimeric viruses in the T3 genetic background (Fig. 4A). Chimera 1 and chimera 2, containing T3 μ2-derived sequences 1–126 and 1–208, respectively, produced high viral yields similar to T1, whereas chimera 3 and chimera 4, containing T3 μ2-derived sequences 1–389 and 1–600, respectively, produced low yields similar to T3 (Fig. 4B). Relative replication efficiencies of the four chimeric viruses suggest that μ2 sequences regulating reovirus replication in MDCK cells are bounded by amino acid residues 209 and 389.
FIGURE 4.
Identification of a μ2 domain that mediates reovirus tropism for MDCK cells. A, schematic of T1 × T3 chimeric (Ch) M1 gene segments. Chimeric junctions were introduced at the nucleotide position corresponding to the amino acid numbers shown. Viruses containing M1 chimeric gene segments were generated in a T3 strain background. B, MDCK cells were infected with M1 chimeric viruses at an m.o.i. of 2 pfu/cell, and viral yields (relative to 0 h) were determined at 24 h post-infection by plaque assay using L cell monolayers. Results represent the mean of triplicate experiments. Error bars indicate S.D. *, p < 0.05 in comparison with strain T1 (Student's t test). C, T1 and T3 μ2 proteins differ at nine amino acid positions.
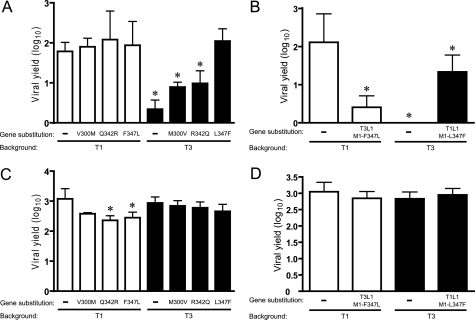
Three sequence polymorphisms with respect to T1 and T3 (300, 342, and 347) are present within the μ2 region that controls viral growth efficiency in MDCK cells (Fig. 4C). The relative contribution of these amino acids to viral growth was assessed by reciprocal exchanges between the μ2 proteins of T1 and T3 using reverse genetics. Moderately increased yields (∼4-fold) resulted from replacement of T3 μ2 amino acids with the corresponding T1 μ2 residues at positions 300 and 342. In contrast, substitution of Leu-347 in T3 μ2 with Phe (T3-M1L347F), which is found at this position in T1 μ2, enabled T3 to grow equivalently to T1 (∼100-fold yield). The yield of T1-M1F347L, isogenic to T1 except for Leu at μ2 amino acid position 347, approximated that of T1 (Fig. 5A). Yields of μ2 point-mutant viruses T1-M1Q342R and T1-M1F347L, which contain T3 residues at μ2 amino acid positions 342 and 347, were modestly reduced in L cells, relative to parental strain T1 (Fig. 5C). However, the other μ2 point-mutant strains and the parental viruses all exhibited equivalently high yields in L cells, and no influence of the L1 gene was detected (Fig. 5, C and D). Therefore, constitutive replication defects are unlikely to account for disparities in yields of μ2-mutant viruses when propagated in MDCK cells.
FIGURE 5.
Identification of residues in μ2 that mediate reovirus tropism for MDCK cells. MDCK cells (A) or L cells (C) were infected with μ2 point-mutant viruses at an m.o.i. of 2 pfu/cell, and viral yields (relative to 0 h) were determined at 24 h post-infection by plaque assay using L cell monolayers. MDCK cells (B) or L cells (D) were infected at an m.o.i. of 2 pfu/cell with L1 monoreassortant viruses containing point mutations at residue 347 of μ2, and viral yields (relative to 0 h) were determined at 24 h post-infection by plaque assay using L cell monolayers. Results represent the mean of triplicate experiments. Error bars indicate S.D. *, p < 0.05 in comparison with strain T1 (Student's t test).
Consistent with the cooperative relationship of WT λ3 and μ2 proteins in viral growth (Fig. 3), we observed a functional interaction between λ3 and mutant μ2 governed by the specific viral genetic environment; yield of T1-M1F347L in MDCK cells was reduced relative to WT parental strain T1 when the mutant μ2 protein was accompanied by the T3 (strain T1-T3L1-M1F347L), but not T1 (strain T1-M1F347L), λ3 protein (Fig. 5, A and B).
Interestingly, the T1 λ3 protein acted synergistically with the μ2 L347F mutation to moderately suppress (∼13-fold reduction in yield) the growth of T3 (T3-T1L1-M1L347F) in MDCK cells (Fig. 5B), which further substantiates the functional linkage of λ3 and μ2 in reovirus tropism for these cells.
These results indicate that a sequence polymorphism at amino acid position 347 is the primary determinant of μ2-mediated reovirus replication efficiency in MDCK cells. Furthermore, the λ3 protein acts as a modifier of tropic phenotypes manifested by sequence variation at this position in μ2.
Viral Inclusion Formation in Reovirus-infected Cells
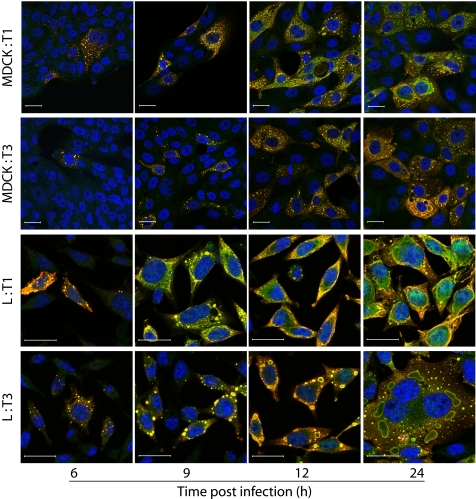
As an initial approach to delineating the μ2-sensitive step in viral replication in MDCK cells, we imaged viral inclusions over a time course of infection using confocal immunofluorescence microscopy. Cells were infected with T1 or T3, fixed at various intervals, and stained with anti-μ2 and anti-σNS antisera. In both T1- and T3-infected MDCK cells, inclusions initially appeared as small, punctate structures that gradually enlarged and assumed strain-dependent filamentous and globular features characteristic of T1 and T3, respectively, in L cells (Fig. 6) (23, 24). However, T1 inclusions in MDCK cells also displayed globular structures intermingled with filamentous forms, resulting in a more intermediate phenotype. Although the progress of inclusion formation for both T1 and T3 was slightly delayed in MDCK cells in comparison with L cells, inclusion development was comparable between MDCK cells and L cells by 12 h post-infection. Thus, kinetics of T3 inclusion maturation in MDCK cells was not overtly impaired. Typical inclusion maturation and abundant expression of σNS, a nonstructural protein, in T3-infected MDCK cells indicate that viral attachment, entry, and early rounds of transcription and translation proceed normally.
FIGURE 6.
Reovirus inclusion formation. MDCK or L cells were infected with T1 or T3 at an m.o.i. of 20 pfu/cell, fixed at the time points shown, stained with anti-μ2 (green) and anti-σNS (red) antibodies and ToPro3 (blue, nuclear), and imaged using confocal microscopy. Scale bars, 20 μm.
Quantification of Reovirus RNA in Infected Cells
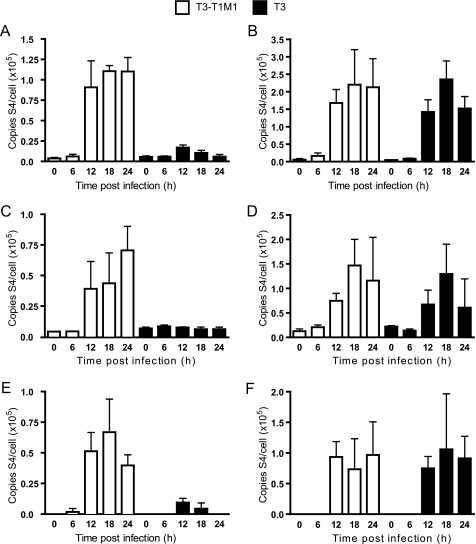
Although the presence of viral inclusions in T3-infected MDCK cells confirms the occurrence of viral transcription and translation, these findings cannot distinguish possible temporal defects occurring during primary and secondary rounds of gene expression. To monitor production of viral RNA over the course of infection, we used an RT-qPCR approach to specifically quantify positive and negative sense S4 gene RNA, which encodes viral structural protein σ3. Standard curves relating total viral genomic RNA to Ct were used for calculations of S4 total, double-stranded, and mRNA in RNA extracts from virus-infected cells. MDCK cells and L cells were infected at an m.o.i. of 10 pfu/cell with either T3-T1M1 or T3. Strain T3-T1M1 was used in place of T1 because the T3 and T3-T1M1 S4 alleles are identical. Growth of T3-T1M1 in MDCK cells recapitulated that of T1 (Fig. 2). At 12 h post-infection and the following time points, high levels of total S4 RNA were detected in T3-T1M1-infected MDCK cells compared with the minimal amount of RNA produced in T3-infected cells (Fig. 7A). Conversely, in L cells, significant levels of total RNA were present in both T3-T1M1- and T3-infected cells at 12 h post-infection and beyond (Fig. 7B). Similar patterns of S4 dsRNA production by T3-T1M1 and T3 were observed (Fig. 7, C and D). Significant levels of S4 mRNA were detected in MDCK cells at 12, 18, and 24 h post-infection with strain T3-T1M1 (Fig. 7E), whereas little S4 mRNA was detected in T3-infected cells at 12 and 18 h post-infection (Fig. 7E). The amount of S4 mRNA in T3-infected cells at 24 h post-infection was below the limit of detection (<10 copies of S4 RNA (data not shown)). These results indicate roughly equivalent efficiencies of early viral RNA synthesis in T3-T1M1- and T3-infected MDCK cells. However, late RNA synthesis is markedly diminished in T3-infected cells in comparison with that in T3-T1M1-infected cells, consistent with attenuated dsRNA production by T3 in MDCK cells.
FIGURE 7.
Analysis of viral RNA synthesis. Cells were infected with T3-T1M1 or T3 at an m.o.i. of 10 pfu/cell, and total RNA was extracted from cell lysates at the indicated time points. RT-qPCR of total S4 RNA in MDCK cells (A) and L cells (B) was performed using an S4-specific reverse primer (complementary to the (+)-strand) in the reverse transcription step, followed by qPCR. A standard curve relating Ct to the amount of S4 RNA isolated from purified virion particles was used to determine S4 (+)-strand RNA copy number. RT-PCR of dsRNA in infected MDCK cells (C) and L cells (D) was performed using an S4-specific forward primer (complementary to the (−)-strand) in the reverse transcription step, followed by qPCR. A standard curve relating Ct to the amount of S4 RNA isolated from purified virion particles was used to determine S4 (−)-strand RNA copy number. Amounts of viral mRNA in MDCK cells (E) and L cells (F) are expressed as the difference between total RNA and dsRNA at each time point shown. Results represent the mean of triplicate experiments. Error bars indicate S.D.
Protein Synthesis in Reovirus-infected Cells
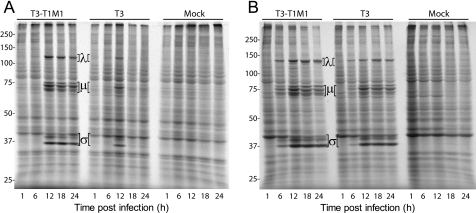
We next assessed viral protein synthesis in infected MDCK cells and L cells. Strains T1 and T3 differ in the capacity to inhibit host cell protein synthesis, a phenotype unrelated to the M1 gene segment (51, 52). Therefore, T3-T1M1 was used instead of T1 to remove this variable. MDCK cells and L cells were infected at an m.o.i. of 100 pfu/cell with either T3-T1M1 or T3. One hour prior to harvest, cells were incubated with 50 mCi [35S]methionine-cysteine, and solubilized whole-cell proteins were resolved using SDS-PAGE, followed by autoradiography. New viral protein synthesis was observed at 12, 18, and 24 h post-infection in T3-T1M1-infected MDCK cells, yet only at 12 h post-infection in T3-infected cells (Fig. 8A). These data generally mirror T3-T1M1 and T3 RNA production in MDCK cells. In contrast, the kinetics and magnitude of new viral protein synthesis by T3-T1M1 and T3 did not significantly differ in infected L cells (Fig. 8B). Thus, early viral protein synthesis was roughly equivalent between T3-T1M1 and T3 in MDCK cells, but late protein synthesis by T3 was undetectable, consistent with diminished mRNA production at later time points.
FIGURE 8.
Analysis of viral protein synthesis. MDCK cells (A) or L cells (B) were infected at an m.o.i. of 100 pfu/cell with reovirus strain T3-T1M1 or T3 and incubated for 1 h with [35S]methionine/cysteine at the indicated times post-infection. Radiolabeled polypeptides were resolved by SDS-PAGE and visualized using autoradiography. Positions of viral λ, μ, and σ proteins are indicated. Molecular mass standards (in kDa) are shown.
DISCUSSION
The purpose of this study was to define post-receptor control mechanisms of reovirus cell tropism. Our primary findings emerging from this study are the following: 1) μ2 controls efficiency of reovirus growth in MDCK cells; 2) residue 347 is the primary determinant of μ2-mediated viral replication potential in these cells; 3) polymerase protein λ3 is a co-regulator of viral replication in a strain-dependent manner; and 4) μ2 protein appears to regulate a later step in the reovirus replication program subsequent to primary rounds of viral transcription and translation. These findings suggest a unique structural or functional interaction between μ2 and λ3 that facilitates productive viral replication in a manner responsive to specific viral and host-cell environments.
Initial studies of reovirus replication in MDCK cells revealed a genetic association of the λ3 and μ2 proteins with viral replication efficiency using classical reassortant analysis (38). A reverse genetics system for reovirus has permitted us to generate viruses containing specific combinations of T1 and T3 gene segments and clearly define the relative contributions of μ2 and λ3 to this phenotype (Figs. 2 and 3). T1 μ2 is associated with efficient reovirus replication in MDCK cells irrespective of the viral genetic background. The influence of T3 μ2 on viral replication is more complex; its quantitative effect on production of infectious progeny requires the co-association of T3 λ3. The μ2 protein is thought to be a cofactor of the viral RdRp (6). However, there is limited insight into interactions between μ2 and λ3. Biochemical data show that the nucleotide triphosphatase and RNA triphosphatase activities of μ2 are enhanced in the presence of λ3, and the two proteins interact in immunoprecipitation assays (27). Furthermore, the L1 and M1 genes display nonrandom segregation in T1 × T3 reassortant viruses (53), suggesting co-evolution of λ3 and μ2 to optimize concerted roles in viral replication. Data presented in this study showing the unidirectional μ2-mediated regulation of replication efficiency in MDCK cells and viral background-dependent co-regulation by λ3 suggest a unique structural or functional interaction between μ2 and λ3 in specific viral and cellular contexts. Our findings provide additional genetic evidence linking the function of these two proteins and implicate the role of the polymerase complex in determining viral cell tropism.
Using recombinant viruses containing chimeric and point-mutant μ2 proteins, we were able to systematically define sequence determinants of reovirus replication efficiency in MDCK cells. Our results indicate that residue 347 is primarily responsible (Fig. 5A). The influence of this polymorphic position on viral replication potential is modulated by λ3 in a manner consistent with the effect of λ3 on full-length μ2 (Fig. 5B). A crystal structure of μ2 has not been reported; however, a number of functional sequence determinants have been defined (20, 22, 24, 27, 28). It is possible that critical residues identified in this study are part of a larger domain of μ2 that controls reovirus tropism for different types of cells. For example, a temperature-sensitive strain of reovirus (tsH11.2) containing mutations at residues 399 and 414 of μ2 displays defective dsRNA synthesis and particle assembly at restrictive temperatures. The proximity of these residues to amino acid position 347 in primary sequence raises the possibility that all three are part of a functional unit responsive to the host cell environment. Residue 347 is also near a predicted leucine-rich nuclear export signal spanning residues 328–335 (NetNES 1.1 (54)). Because μ2 distributes to both the cytoplasm and nucleus (Fig. 6) (20, 24) and contains two predicted nuclear localization signals, one of which has been shown to be important for viral replication (20), it is plausible that a polymorphism at position 347 could deleteriously shift the nucleocytoplasmic balance of μ2.
The three-dimensional structure of the μ2 protein is unknown; therefore, it is not possible to rule out an interaction of residue 347 with other structural or functional domains distantly located in the primary sequence, such as the nucleotide-binding motifs located at residues 414–420 and 445–450 (27). These sequences are required for nucleotide triphosphatase and RNA triphosphatase activities of μ2 (27), and we found them necessary for viral replication using an RNAi trans-complementation system (20). Therefore, changes at μ2 residue 347 plausibly could alter the function of these domains, with a resultant impact on viral replication efficiency in MDCK cells.
Although negative-stand RNA synthesis and particle assembly have been ascribed to viral inclusions, the discrete series of steps that occur between entry of transcriptionally active core particles into the cytoplasm and the appearance of mature progeny particles within viral inclusions remain poorly defined. The absence of detectable viral RNA and protein synthesis at late times of infection in T3-infected MDCK cells (Figs. 7 and 8) despite normal-appearing inclusions (Fig. 6) indicates that the impasse occurs at one or more of these undefined steps. Several potential but nonmutually exclusive mechanisms can be proposed to explain the replication block in T3-infected MDCK cells, including the following: 1) mislocalization of RNA; 2) decreased RNA stability; 3) malfunction of the viral replicase during positive- or negative-strand synthesis; and 4) defective genome packaging or particle assembly. Each of these possibilities is currently under investigation. Notably, by confocal immunofluorescence microscopy, core protein λ2 (which serves as the viral RNA-capping enzyme) exhibits the expected pattern of localization to inclusions in T3- and T3-T1M1-infected MDCK cells, using infected L cells as a reference for λ2 distribution in permissive cells (data not shown). These results suggest that recruitment of virion core proteins occurs normally in T3-infected MDCK cells and the defect in viral replication is not ascribable to mislocalization of replicase components.
The capacity of MDCK cells to discriminate reovirus strains T1 and T3 in a μ2- and λ3-dependent manner at a later point in the viral life cycle indicates the existence of a tightly controlled post-entry replication checkpoint responsive to structural or functional features of the viral polymerase complex. Conceivably, the regulatory mechanism could be positive or negative, i.e. active stimulation of T1 replication or suppression of T3 replication. A passive mechanism also is possible in which MDCK cells fail to provide a function essential for T3, but not T1, replication. The mediator of replication permissivity might be as simple as a single protein or as intricate as a biochemical pathway.
RdRp complexes of diverse RNA viruses may function in viral adaptation to different intracellular environments (55–57). In the influenza A paradigm, host-range restriction involves an active inhibitory process that protects against cross-species transmission of nonadapted viruses. It is provocative that reovirus strains T1 and T3 sharply contrast in their capacities to grow in canine cells, whereas both strains productively infect cells derived from several other species, including mouse (L (Fig. 1)), human (HeLa and 293T (11, 58)), and primate (Vero and CV-1 (59, 60)), raising the possibility that μ2 is a host-range determinant of reovirus infection. However, this hypothesis awaits formal testing.
The reovirus μ2 protein is a pleiotropic mediator of reovirus replication operating at the intersection of virus multiplication, cellular regulation of infection, and host disease. Based on this study, the capacity of μ2 to mediate viral tropism at the post-entry level can be added to the list of μ2 functions in the reovirus replication program. Further investigation of μ2 interactions with the viral and cellular metabolic machineries will provide sharper insights into mechanisms of cellular permissivity to reovirus infection and create a framework for conceptualizing and testing general models of viral pathogenesis, epidemiology, and adaptation.
Acknowledgments
Acquisition and analysis of confocal imaging data were performed in part through the use of the Vanderbilt University Medical Center Imaging Shared Resource.
This work was supported, in whole or in part, by National Institutes of Health Grants K08 AI62862 (to J. D. C.) and R01 AI32539 (to T. S. D.) from the United States Public Health Service. This work was supported by the Vanderbilt Department of Pathology (to L. S. O.), a fellowship from the Naito Foundation (to T. K.), and the Elizabeth B. Lamb Center for Pediatric Research.
- T1
- type 1 Lang
- T3
- type 3 Dearing
- MDCK
- Madin Darby canine kidney
- L cells
- murine 929 L cells
- dsRNA
- double-stranded RNA
- RdRp
- RNA-dependent RNA polymerase
- pfu
- plaque-forming unit
- m.o.i.
- multiplicity of infection
- qPCR
- quantitative PCR.
REFERENCES
- 1.Taubenberger J. K., Kash J. C. (2010) Cell Host Microbe 7, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guglielmi K. M., Johnson E. M., Stehle T., Dermody T. S. (2006) Curr. Top. Microbiol. Immunol. 309, 1–38 [DOI] [PubMed] [Google Scholar]
- 3.Clarke P., Debiasi R. L., Meintzer S. M., Robinson B. A., Tyler K. L. (2005) Apoptosis 10, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrest J. C., Dermody T. S. (2003) J. Virol. 77, 9109–9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherry B. (2002) Viral Immunol. 15, 17–28 [DOI] [PubMed] [Google Scholar]
- 6.Schiff L. A., Nibert M. L., Tyler K. L. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., pp. 1853–1915, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 7.Rosen L. (1962) Ann. N. Y. Acad. Sci. 101, 461–465 [DOI] [PubMed] [Google Scholar]
- 8.Sabin A. B. (1959) Science 130, 1387–1389 [DOI] [PubMed] [Google Scholar]
- 9.Attoui H., Biagini P., Stirling J., Mertens P. P., Cantaloube J. F., Meyer A., de Micco P., de Lamballerie X. (2001) Biochem. Biophys. Res. Commun. 287, 583–588 [DOI] [PubMed] [Google Scholar]
- 10.Fields B. N. (1971) Virology 46, 142–148 [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T., Chappell J. D., Danthi P., Dermody T. S. (2006) J. Virol. 80, 9053–9063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold M. M., Murray K. E., Nibert M. L. (2008) Virology 375, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomatos P. J., Tamm I., Dales S., Franklin R. M. (1962) Virology 17, 441–454 [DOI] [PubMed] [Google Scholar]
- 14.Spendlove R. S., Lennette E. H., Knight C. O., Chin J. N. (1963) J. Immunol. 90, 548–553 [PubMed] [Google Scholar]
- 15.Dales S., Gomatos P. J., Hsu K. C. (1965) Virology 25, 193–211 [DOI] [PubMed] [Google Scholar]
- 16.Silverstein S. C., Schur P. H. (1970) Virology 41, 564–566 [DOI] [PubMed] [Google Scholar]
- 17.Anderson N., Doane F. W. (1966) J. Pathol. Bacteriol. 92, 433–439 [DOI] [PubMed] [Google Scholar]
- 18.Sharpe A. H., Chen L. B., Fields B. N. (1982) Virology 120, 399–411 [DOI] [PubMed] [Google Scholar]
- 19.Silverstein S. C., Dales S. (1968) J. Cell Biol. 36, 197–230 [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T., Ooms L. S., Chappell J. D., Dermody T. S. (2009) J. Virol. 83, 2892–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin B., Whyatt R. M., Perera F. P., Randall M. C., Cooper T. B., Santella R. M. (1995) Free Radic. Biol. Med. 18, 1023–1032 [DOI] [PubMed] [Google Scholar]
- 22.Coombs K. M. (1996) J. Virol. 70, 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mbisa J. L., Becker M. M., Zou S., Dermody T. S., Brown E. G. (2000) Virology 272, 16–26 [DOI] [PubMed] [Google Scholar]
- 24.Parker J. S., Broering T. J., Kim J., Higgins D. E., Nibert M. L. (2002) J. Virol. 76, 4483–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coombs K. M. (1998) Virology 243, 218–228 [DOI] [PubMed] [Google Scholar]
- 26.Coombs K. M. (1998) Curr. Top. Microbiol. Immunol. 233, 69–107 [DOI] [PubMed] [Google Scholar]
- 27.Kim J., Parker J. S., Murray K. E., Nibert M. L. (2004) J. Biol. Chem. 279, 4394–4403 [DOI] [PubMed] [Google Scholar]
- 28.Noble S., Nibert M. L. (1997) J. Virol. 71, 7728–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinisch K. M., Nibert M. L., Harrison S. C. (2000) Nature 404, 960–967 [DOI] [PubMed] [Google Scholar]
- 30.Yin P., Cheang M., Coombs K. M. (1996) J. Virol. 70, 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Walker S. B., Chipman P. R., Nibert M. L., Baker T. S. (2003) Nat. Struct. Biol. 10, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brentano L., Noah D. L., Brown E. G., Sherry B. (1998) Virology 72, 8354–8357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matoba Y., Sherry B., Fields B. N., Smith T. W. (1991) J. Clin. Invest. 87, 1628–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherry B., Schoen F. J., Wenske E., Fields B. N. (1989) J. Virol. 63, 4840–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherry B., Blum M. A. (1994) J. Virol. 68, 8461–8465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherry B. (1998) Curr. Top. Microbiol. Immunol. 233, 51–66 [DOI] [PubMed] [Google Scholar]
- 37.Matoba Y., Colucci W. S., Fields B. N., Smith T. W. (1993) J. Clin. Invest. 92, 2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodgers S. E., Barton E. S., Oberhaus S. M., Pike B., Gibson C. A., Tyler K. L., Dermody T. S. (1997) J. Virol. 71, 2540–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zurney J., Kobayashi T., Holm G. H., Dermody T. S., Sherry B. (2009) J. Virol. 83, 2178–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi T., Antar A. A., Boehme K. W., Danthi P., Eby E. A., Guglielmi K. M., Holm G. H., Johnson E. M., Maginnis M. S., Naik S., Skelton W. B., Wetzel J. D., Wilson G. J., Chappell J. D., Dermody T. S. (2007) Cell Host Microbe 1, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi T., Ooms L. S., Ikizler M., Chappell J. D., Dermody T. S. (2010) Virology 398, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furlong D. B., Nibert M. L., Fields B. N. (1988) J. Virol. 62, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin H. W., 4th., Bassel-Duby R., Fields B. N., Tyler K. L. (1988) J. Virol. 62, 4594–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishii K., Ueda Y., Matsuo K., Matsuura Y., Kitamura T., Kato K., Izumi Y., Someya K., Ohsu T., Honda M., Miyamura T. (2002) Virology 302, 433–444 [DOI] [PubMed] [Google Scholar]
- 45.Zou S., Brown E. G. (1996) Virology 217, 42–48 [DOI] [PubMed] [Google Scholar]
- 46.Becker M. M., Goral M. I., Hazelton P. R., Baer G. S., Rodgers S. E., Brown E. G., Coombs K. M., Dermody T. S. (2001) J. Virol. 75, 1459–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyler K. L., McPhee D. A., Fields B. N. (1986) Science 233, 770–774 [DOI] [PubMed] [Google Scholar]
- 48.Kaye K. M., Spriggs D. R., Bassel-Duby R., Fields B. N., Tyler K. L. (1986) J. Virol. 59, 90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiner H. L., Drayna D., Averill D. R., Jr., Fields B. N. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5744–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dermody T. S., Nibert M. L., Bassel-Duby R., Fields B. N. (1990) J. Virol. 64, 5173–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zweerink H. J., Joklik W. K. (1970) Virology 41, 501–518 [DOI] [PubMed] [Google Scholar]
- 52.Sharpe A. H., Fields B. N. (1982) Virology 122, 381–391 [DOI] [PubMed] [Google Scholar]
- 53.Nibert M. L., Margraf R. L., Coombs K. M. (1996) J. Virol. 70, 7295–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 55.Neumann G., Kawaoka Y. (2006) Emerg. Infect. Dis. 12, 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehle A., Doudna J. A. (2008) Cell Host Microbe 4, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumann G., Noda T., Kawaoka Y. (2009) Nature 459, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell J. A., Schelling P., Wetzel J. D., Johnson E. M., Forrest J. C., Wilson G. A., Aurrand-Lions M., Imhof B. A., Stehle T., Dermody T. S. (2005) J. Virol. 79, 7967–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berry J. M., Barnabé N., Coombs K. M., Butler M. (1999) Biotechnol. Bioeng. 62, 12–19 [DOI] [PubMed] [Google Scholar]
- 60.Carvalho J., Arnold M. M., Nibert M. L. (2007) Virology 364, 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]