Abstract
Although the function of protein kinase D1 (PKD) in cardiac cells has remained enigmatic, recent work has shown that PKD phosphorylates the nuclear regulators HDAC5/7 (histone deacetylase 5/7) and CREB, implicating this kinase in the development of dysfunction seen in heart failure. Additional studies have shown that PKD also phosphorylates multiple sarcomeric substrates to regulate myofilament function. Initial studies examined PKD through adenoviral vector expression of wild type PKD, constitutively active PKD (caPKD), or dominant negative PKD in cultured adult rat ventricular myocytes. Confocal immunofluorescent images of these cells reveal a predominant distribution of all PKD forms in a non-nuclear, Z-line localized, striated reticular pattern, suggesting the importance of PKD in Ca2+ signaling in heart. Consistent with an established role of PKD in targeting cardiac troponin I (cTnI), caPKD expression led to a marked decrease in contractile myofilament Ca2+ sensitivity with an unexpected electrical stimulus dependence to this response. This desensitization was accompanied by stimulus-dependent increases in cTnI phosphorylation in control and caPKD cells with a more pronounced effect in the latter. Electrical stimulation also provoked phosphorylation of regulatory site Ser916 on PKD. The functional importance of this phospho-Ser916 event is demonstrated in experiments with a phosphorylation-defective mutant, caPKD-S916A, which is functionally inactive and blocks stimulus-dependent increases in cTnI phosphorylation. Dominant negative PKD expression resulted in sensitization of the myofilaments to Ca2+ and blocked stimulus-dependent increases in cTnI phosphorylation. Taken together, these data reveal that localized PKD may play a role as a dynamic regulator of Ca2+ sensitivity of contraction in cardiac myocytes.
Keywords: Cardiac Muscle, Contractile Protein, Phosphorylation Enzymes, Protein Kinases, Signal Transduction, Calcium Signaling, Protein Kinase D
Introduction
There has been much interest in the cellular signaling cascades that contribute to the pathological cardiac remodeling seen in response to hypertension or acute myocardial infarction. Recently, protein kinase D1 (PKD)2 has emerged as a critical transducing enzyme that is activated in heart in response to stress (1–3) and has a potential role in contractile regulation, cardioprotection, heart development, and disease progression (2). Not only does expression of constitutively active PKD lead to chamber dilation and death in animal models, but it is up-regulated in the failing human heart as well (4). Further, mice with heart-specific deletion of PKD showed diminished hypertrophic and genetic responses to a variety of pathological stimuli (3).
Although the molecular sequelae of cardiac PKD activation are largely unknown, critical targets are being identified. The earliest studies revealed that the regulation of gene expression is due to PKD-dependent phosphorylation of class II histone deacetylases (1, 4, 5) and cAMP response element-binding protein (CREB) (6, 7). PKD-mediated pathways regulate cardiac myofilament function through phosphorylation of multiple sarcomeric proteins. Cardiac troponin I (cTnI) can be phosphorylated by PKD at canonical PKA sites, Ser22/Ser23, desensitizing the myofilaments to intracellular Ca2+ (8, 9). PKD accelerates cross-bridge cycling kinetics through a distinct mechanism, most likely a consequence of phosphorylation of cardiac myosin-binding protein C at a novel PKD site, Ser302 (10). PKD-dependent regulation of L-type Ca2+ channel activity and sarcoplasmic reticulum Ca2+ load have been reported, although specific phosphorylation reactions remain to be defined (11, 12). Therefore, in addition to its well established role in gene expression, these results broaden the set of biological functions of PKD to the regulation of the excitation-contraction coupling (EC coupling) cascade in heart as well.
The discovery of multiple PKD-dependent phosphorylation reactions in cardiac cells raises the issue of when such reactions are operable in normal and diseased heart. The overlap of PKD and PKA phosphorylation sites on multiple protein targets requires new studies to clarify when PKD signaling plays a critical functional role (6–10). For example, because the Ser22/Ser23 sites of cTnI are substrates for both PKD and PKA, it is not clear under what circumstances PKD-mediated phosphorylation of cTnI is an important functional event (8–10, 13–15). In failing hearts, β-adrenergic receptors are desensitized, resulting in attenuation of downstream cAMP/PKA cascades (16). This has led to the appealing view that PKD-dependent phosphorylation of cTnI is likely to be of greater functional significance in the failing myocardium (10, 15). New approaches are needed to define when PKD-mediated events are critical for cardiac myocyte function.
The diversity of functional substrates for PKD in cardiac myocytes requires a complex network of activation cascades. The activation of PKD following hypertrophic stimuli in neonatal cardiac ventricular myocytes includes well established PKC-dependent cascades (17–20). However, other studies reveal that PKC-independent activation mechanisms are also important in cardiac cells. When cultured adult ventricular myocytes were exposed to high glucose/palmitic acid, PKD was activated and recruited to the plasma membrane (21). PKC-independent mechanisms require additional factors, such as diacylglycerol (DAG), in neonatal myocytes (7). In this regard, a recent study in COS cells identified a role for agonist-evoked increases in DAG levels in Golgi, allowing this intracellular membrane to serve as a platform for signaling by PKD distinct from those at the plasma membrane (22). Electrical stimulation of isolated adult ventricular cardiac myocytes rapidly increased PKD activity by 50% (23) and suggests that electrical stimulation itself may play a critical role in regulating PKD at specific subcellular sites. Because activation pathways for PKD are cell-specific and will vary between neonatal and adult cardiac myocytes, it is important to define these functional cascades in normal and failing heart.
In the present study, the role of PKD in regulation of excitation-contraction coupling in adult cardiac myocytes was critically examined. A combination of molecular genetics, functional assays, and complementary molecular studies revealed an unexpected role for PKD in electrical stimulus-dependent dynamic regulation of myofilament Ca2+ sensitivity in cultured cardiac myocytes that may be operable in intact heart tissue. Molecular genetic studies revealed that phosphorylation of PKD at Ser916 is a critical step in this stimulus-dependent signaling cascade. Finally, through a characterization of the PKD probes developed in this study, new information on the non-nuclear subcellular distribution of PKD in adult rat ventricular mycoytes was obtained.
EXPERIMENTAL PROCEDURES
Construction of Recombinant Adenoviruses
Production of recombinant adenovirus (Ad5)-containing wild type PKD (wtPKD) cDNA, constitutively active PKD (caPKD) cDNA, or kinase-deficient dominant negative PKD (dnPKD) cDNA and the corresponding fluorescent fusion proteins with the red-shifted GFP mutant, Plum, was carried out according to methods described previously (24). Each of these recombinant adenoviral constructs contained an N-terminal hemagglutinin (HA) epitope tag to monitor heterologous PKD expression in cultured adult rat ventricular myocytes (ARVMs). The constitutively active mutant, caPKD, was generated by serine to glutamic acid phosphomimetic substitutions in the activation loop (S744E/S748E) of the catalytic domain (25). The kinase-deficient mutant, dnPKD, was generated by lysine to methionine (K618M) substitutions at the ATP binding site in the catalytic domain (26). The Plum plasmids were constructed by placing mPlum (27) at the N terminus of wtPKD and caPKD. Additionally, a fluorescent chimera Plum-caPKD mutant was constructed with the autophosphorylation site within the catalytic domain, Ser916, mutated to an unphosphorylatable alanine residue (Plum-caPKD-S916A (S916A)) (28).
Ventricular Myocyte Isolation, Cell Culturing, and Adenoviral Infection
Single ventricular myocytes were obtained from adult male hearts by a standard enzymatic technique as described previously (29). Briefly, hearts were excised from adult male Sprague-Dawley rats (225–250g) lethally anesthetized with 1 ml (i.p.) of Nembutal solution with a dose of 100 mg/kg. The aorta was cannulated for Langendorff perfusion with Hepes-buffered solution. Coronary arteries were perfused with digest solution containing type II collagenase (2.4 mg/ml; Worthington). Isolated ventricular myocytes were separated by mechanical dispersion and reintroduced to 1.8 mm Ca2+ stepwise. Acutely dissociated cells were resuspended in Hepes-buffered DMEM with 10% fetal bovine serum and added to 35-mm tissue culture dishes containing 25-mm glass coverslips coated with laminin (2.5–5 μg/cm2; Invitrogen). After 2 h in culture (37 °C, 5% CO2), plates were washed with DMEM and transiently infected with replication-deficient human adenovirus type 5 mutant (Ad-d1312, control adenovirus) or with recombinant adenoviral constructs described above to drive overexpression of wtPKD, caPKD, dnPKD, or Plum-tagged derivatives in cultured ARVMs. In order to achieve comparable heterologous PKD expression, we adjusted the concentration of virus used to infect cultured ARVMs according to the viral titer of each adenoviral construct. All subsequent experiments were carried out 18–24 h after infection. All animal protocols were approved by the Animal Use and Care Committee of the University of Maryland, School of Medicine.
Western Blotting
Whole-cell extracts of cultured ARVMs were prepared by homogenizing cells in a standard lysis buffer. When probing with phospho-specific antibodies, a phosphatase inhibitor mixture containing 1 mm EGTA, 1 mm EDTA, 50 mm NaF, 0.2 mm NaVO3, 10 mm sodium pyrophosphate, and 10 mm β-glycerol phosphate was added to the standard lysis buffer. Phospho-specific proteins were detected before and after electrical stimulation (three sets of a train of 20 depolarizing pulses at 1 Hz with a 1-min rest between each set) after treatment with 200 mm phorbol 12-myristate 13-acetate (PMA) for 15 min at 37 °C or after treatment with 1 μm isoproterenol for 5 min at 37 °C. For small molecule inhibitor experiments, cells were pretreated with 5 μm bisindolylmaleimide I (BIM I) or Gö 6976 for 45 min at 37 °C, followed by rapid harvesting before and after electrical stimulation or after treatment with 200 nM PMA as described above. Extracts (∼5 μg/lane) were resolved on 10% SDS-polyacrylamide gels, and proteins were transferred to BioTrace polyvinylidene difluoride (PVDF) membrane (VWR Scientific). Western blots were blocked with 5% milk or 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) with 0.1% Tween and incubated overnight at 4 °C with primary antibody. Primary antibodies used in Western blots in this study include mouse monoclonal anti-HA (1:3000; Covance), rabbit polyclonal anti-PKD (1:1000; Cell Signaling Technologies), rabbit polyclonal anti-phospho-PKD-Ser916 (P-Ser916) (1:1000; Cell Signaling Technologies), rabbit polyclonal anti-phospho-cTnI-Ser22/Ser23 (P-cTnI) (1:1000; Abcam), or rabbit polyclonal anti-phospho-PLB-Ser16 (1:1000; Abcam). The anti-P-cTnI antibody is designed to recognize cTnI phosphorylation at Ser22/23 in rats, which corresponds to Ser23/24 cTnI phosphorylation in mouse and human samples and is referred to in this way in other in vitro studies (30). The anti-phospho-PLB-Ser16 antibody is designed to recognize phospholamban (PLB) phosphorylation at the well established PKA site, Ser16. After incubation with the horseradish peroxidase-labeled secondary antibody, blots were developed using enhanced chemiluminescence (Pierce Supersignal). All signals were recorded using a darkroom imaging system for quantification and captured on film for representation. Volume quantitation was performed using ImageJ software to integrate the intensity of all pixels in a band excluding background. Equal protein loading was ensured by reprobing for anti-GAPDH-HRP (1:5000; Abcam) or for total protein with anti-cTnI-HRP (1:5000; Abcam). All experiments were performed in duplicate from multiple, independent culture preparations. Samples shown together in representative blots within the same panel of a figure were run and developed on the same gel and in separated lanes when indicated. Each band was normalized to total protein or loading control for a particular sample. Control, wtPKD, caPKD, and dnPKD values were further normalized to prestimulation levels when detecting phosphoproteins.
Immunofluorescence
Cultured ARVMs were washed with warm PBS (37 °C) and fixed in warm 4% paraformaldehyde (37 °C) for 10 min at room temperature followed by 100% ice-cold methanol for 5 min at −20 °C. Cells were rehydrated in PBS and then permeabilized in PBS containing 0.1% Triton X-100 for 15 min at room temperature. Cells were blocked in PBS containing 5% goat serum and 3% BSA for 1 h at room temperature and incubated in primary antibody overnight at 4 °C. Primary antibodies include mouse monoclonal anti-HA (1:50; Covance), rabbit anti-titin-Z (to the first two Ig domains; 3 μg/ml), and rabbit anti-obscurin Rho-GEF (Rho-GEF domain of obscurin is primarily found in structures at the level of the M-band; 3 μg/ml) (31). These rabbit polyclonal antibodies were a generous gift from Dr. Katia Kontrogianni-Konstantopoulos (University of Maryland School of Medicine, Baltimore, MD) and have been reported as established markers of the Z-line (31) and M-line (31), respectively. After washes with the above blocking solution, cells were incubated in secondary antibody (Alexa 488 and Alexa 633, Molecular Probes) for 2 h at room temperature and mounted using slow anti-fade kit (Molecular Probes). Images were collected on a Zeiss 510 confocal microscope (×63 oil objective, 1.4 numerical aperture (Zeiss), pinhole equals 1.0 Airy disc) using Carl Zeiss imaging software. All channels were collected on the same photomultiplier tube. Control adenovirus-treated cells were used as a negative control and allowed us to appropriate gain settings to obtain minimal background fluorescence (data not shown). Images were imported into CorelDraw for cropping and linear contrast adjustment. Three-dimensional reconstructions were created using a z-stack of 50–60 consecutive confocal scans obtained at intervals of 0.3 μm and assembled using Volocity software (version 4.4; Improvision). Pixel sizes for all images collected were less than 0.1 × 0.1 μm.
Cell Shortening and [Ca2+]i Transient Measurements
The IonOptix detection system was used to measure changes in sarcomere length twitch amplitude (percentage of resting length, %RL) and Ca2+ transients (F/F0) simultaneously during field stimulation of intact ARVMs. Cultured cells were washed with a modified Ringer solution and loaded with the fluorescent Ca2+ indicator Fluo-3/AM (4–5 μm; Molecular Probes) for 15 min prior to all experiments. Cells were placed in an experimental chamber, perfused with modified Ringer solution at 35 °C, and field-stimulated (Grass S48) at 1 Hz with 1-ms electrical pulses through platinum wires mounted in the bottom of the chamber. All measurements were collected on a Nikon Eclipse (TS100) inverted microscope equipped with an IonOptix photomultiplier tube for fluorescent detection and recorded using a selected region of interest within a single, isolated ventricular myocyte. Data of five consecutive steady-state contractions (cell shortening and fluorescence) were used to construct phase-plane plots of contractile shortening as a function of the Ca2+ transient to better assess Ca2+ sensitivity over a broad range of [Ca2+]i rather than at a single peak Ca2+ amplitude.
Data Analysis
All results are representative of duplicate experiments from at least three independent culture preparations and are reported as means ± S.E. where quantified (n is the number of rat hearts in the biochemical experiments or the number of myocytes in functional experiments). Unpaired Student's t test and one-way analysis of variance with Newman-Keuls test were applied when appropriate to determine statistical significance of the differences. p < 0.05 was considered statistically significant, except where noted.
RESULTS
Localization of PKD in Adult Rat Ventricular Myocytes and Translocation following PMA-induced Activation
We expressed a fluorescent chimera, Plum-wtPKD, in cultured ARVMs. Confocal images of live, non-contracting myocytes reveal a subcellular distribution with extensive non-nuclear PKD segregation, which includes perinuclear targeting and a striated pattern throughout the cell (Fig. 1A). When cells were stimulated with PMA, we observed a rapid (within minutes) translocation of Plum-wtPKD to the plasma membrane that was sustained after 10 min (Fig. 1B). Although acute PMA-evoked changes in Plum-wtPKD spatial distribution are difficult to reconcile with a purely nuclear function, Kim et al. (21) report a similar sarcolemmal translocation of endogenous PKD in cardiac myocytes in response to stimulation with high glucose/palmitic acid. PMA is often used as a pharmacological probe because it is a direct activator of PKD and phorbol ester-sensitive PKC isoforms and is a useful tool to assess effects of large, if not maximal, activation of PKD in cardiac myocytes (1, 15, 26, 32). Although these non-physiological translocation events must be interpreted with caution, they do highlight the critical importance of PKD activation state in specifying its subcellular distribution in ARVMs.
FIGURE 1.
Localization and movement of Plum-tagged wild type PKD (Plum-wtPKD) in myocytes activated with phorbol ester. ARVMs were infected with an adenovirus construct to drive expression of Plum-wtPKD and cultured for 24 h as described under “Experimental Procedures.” A, fluorescent confocal image of a single isolated ARVM that shows localization of Plum-wtPKD in the steady state. Scale bar, 10 μm. B, cells were stimulated with the phorbol ester derivative, PMA (2 nm), and a series of confocal images were acquired as indicated from the same cell after the addition of PMA.
Expression of wtPKD and caPKD in Cultured ARVMs
We constructed and characterized recombinant adenoviral vectors to drive expression of epitope-tagged (HA) wild-type (wt), or constitutively active (ca) PKD in cultured ARVMs (see mutation scheme in supplemental Fig. 1A). Western blot analyses developed with anti-HA reveal that adenovirus vectors promote the expression of wtPKD and caPKD in a dose-dependent manner in cultured ARVMs (supplemental Figs. 1B and 2A). In fact, it was possible to adjust viral titers to express equivalent levels of wtPKD and caPKD in 24-h cultures as demonstrated in Western blots with anti-HA (supplemental Fig. 1C) and with anti-PKD antibody (supplemental Fig. 1D).
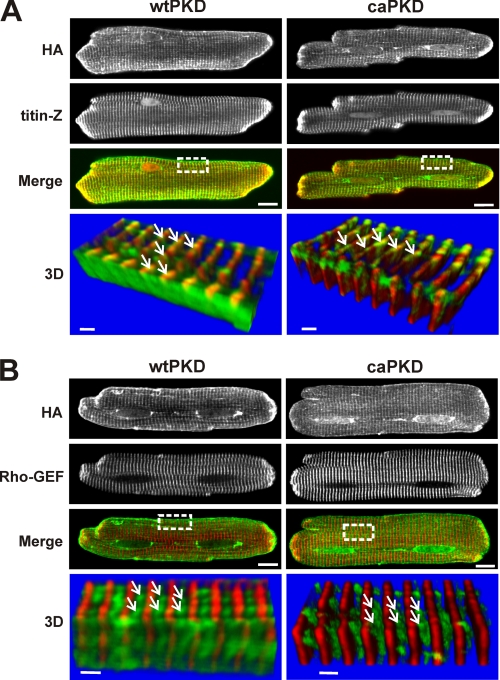
Because the activation state of PKD can dictate its subcellular distribution, we examined the localization of HA-tagged PKD constructs through immunofluorescent confocal microscopy of fixed cells (Fig. 2). The upper panels of Fig. 2A show that, overall, the distributions for wtPKD and caPKD are quite similar, with the majority of the protein imaged in a striated pattern comparable with the localization of Plum-wtPKD in live cells (Fig. 1A). The localization of these PKD striations was further explored with the use of established Z-line and M-line markers (31). When cells were co-stained with anti-titin-Z, an established Z-line marker (31), a pattern similar to that of PKD was observed (second row of images). The Z-line localization for both PKD forms is clearly illustrated in the merged images, where the yellow regions indicate co-localization (third row of Fig. 2A). To further confirm these results, a set of parallel studies exploited antibodies against the Rho-GEF domain of obscurin, an established M-line marker (31). As shown in the confocal images of cells co-stained with anti-obscurin Rho-GEF (Fig. 2B, second row of images), there is no overlap between HA-PKDs and the M-line region.
FIGURE 2.
Both wtPKD and caPKD localize to Z-line regions in cultured ARVMs. Myocytes were infected with HA-tagged wtPKD or caPKD adenoviruses for 24 h. Cells were prepared for double label confocal immunofluorescent analysis as described under “Experimental Procedures.” A, cells were labeled with anti-HA antibody (green) and Z-line marker, anti-titin-Z (red). The merged image shows colocalization in yellow. Scale bar, 10 μm. The lower panels (labeled 3D) are three-dimensional images constructed from consecutive Z-scans (0.3 μm apart) derived from the region marked in a white box in the merged image. The stack was rotated by about 200° so that the sarcolemma edge is closest to the viewer. Scale bars, 1.5 μm. The white arrows identify a punctate pattern of HA localization along the Z-line. B, confocal fluorescent images similar to A, except cells were co-labeled with the M-line marker, anti-obscurin Rho-GEF (red), along with the HA tag (green). The white arrows identify HA localization between M-lines with a punctate, lattice-like/reticular distribution pattern.
Three-dimensional reconstructions provide more detail on the distribution of PKD constructs in this Z-line region of the myocyte. The lower panels labeled 3D in Fig. 2, A and B, display a series of z-stack confocal images taken from the boxed regions in the merged images above. In three-dimensional images of Fig. 2A, not only is the Z-line marker/PKD overlap more clearly defined, but both wtPKD and caPKD display an uneven, punctate distribution, as indicated by the white arrows. The three-dimensional reconstructions in the lower panels of Fig. 2B emphasize this conclusion and more clearly reveal a lattice-like/reticular pattern for both PKD forms. Although such a high resolution analysis of myocardial PKD has not been performed before, this novel distribution reported here for the first time is reminiscent of the three-dimensional spatial distribution of the cardiac T-tubular/junctional sarcoplasmic reticulum network reported in another imaging study with dextran-linked fluorescein in live cells (33). Although a minor fraction of wtPKD shows some decoration of the plasma membrane (see “Discussion”), overall the results of Figs. 1 and 2 reveal that both wtPKD and caPKD constructs distribute in subcellular patterns in myocardial cells indistinguishable at the level of light microscopic resolution.
PKD Regulation of Myofilament Ca2+ Sensitivity
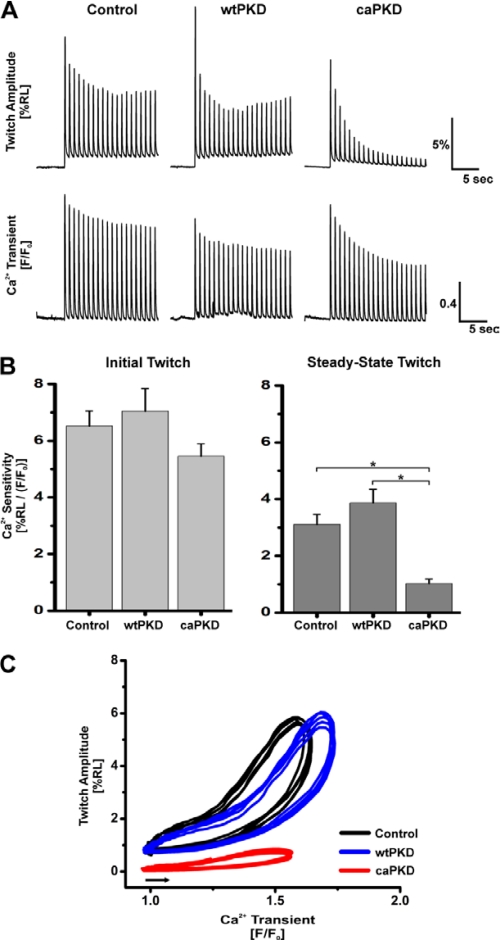
The intriguing subcellular distribution of PKD constructs seen in Fig. 2 suggests that PKD plays unappreciated roles in regulation of EC coupling. Effects of PKD on EC coupling have been examined previously in cardiac myocytes overexpressing wtPKD following endothelin-1 (ET1) stimulation (9). However, ET1 evokes multiple signaling cascades, including nPKCs, Ras·GTP, ERK1/2, Akt, RhoA, and Rac1 in cardiac cells (34). Thus, we developed a strategy where PKD activation mutants could be used to directly assess the impact of this kinase on EC coupling in heart cells in situ. To test the hypothesis that PKD is a critical regulator of the EC coupling cascade, we stimulated isolated Fluo-3-loaded ARVMs with a protocol of 20 depolarizing pulses at 1 Hz. Simultaneous measurements of cell shortening and [Ca2+]i transients allowed for broad assessment of EC coupling properties of ARVMs expressing wtPKD and caPKD compared with those infected with control adenovirus. Fig. 3A shows representative traces of cell shortening and [Ca2+]i transients during the stimulus protocol for ARVMs that were initially at rest. For control cells, there was a small decline in cell shortening and the magnitude of the [Ca2+]i transient that reached steady-state values by the end of the pulse protocol. Nearly identical responses were seen in wtPKD cells. However, EC coupling was markedly different in the caPKD cells. Although the magnitudes of the [Ca2+]i transients were comparable with those in control and wtPKD cells throughout the train of stimuli, there was a profound decrease in the twitch amplitude that progressed to only 1.5% resting length as the number of stimuli increased to 20. The summary data for values in the initial twitch and the steady state (20th twitch) are shown in Table 1. Note that there are no differences in the magnitude of the [Ca2+]i transients between all three groups of cells, initially or at steady state. Although the magnitudes of the twitches in control and wtPKD cells are not statistically different, there is a small decrease in the initial twitch of 18% in caPKD cells, which develops into a 71% decline (relative to control cells) in the steady state (Table 1). Taken together, under these conditions, there were no significant changes in [Ca2+]i transients when PKD constructs were expressed, whereas caPKD did evoke marked declines in contractility manifest in a stimulus-dependent manner.
FIGURE 3.
Analysis of sarcomere shortening and [Ca2+]i transients reveals a stimulus-dependent Ca2+ desensitization enhanced in caPKD cells. Myocytes were cultured for 24 h after infection with control, wtPKD, or caPKD adenoviruses as in Fig. 2. Cells were loaded with Fluo-3 and field-stimulated with a train of 20 pulses at 1 Hz. Cell shortening and Ca2+ transients were recorded as described under “Experimental Procedures.” A, representative traces of shortening (twitch amplitude as %RL) and Ca2+ transients (F/F0) measured simultaneously from individual cardiomyocytes. Upper panels, change in sarcomere length; lower panels, corresponding change in the [Ca2+]i transients measured simultaneously. B, summary data of Ca2+ sensitivity of contraction (assessed as ratio of peak %RLmax/(F/F0)max) in control adenovirus, wtPKD, and caPKD cells for first twitch (Initial Twitch) and final twitch (Steady-State Twitch) in pulse protocol. Data are means ± S.E. (error bars) from multiple culture preparations, n = 20–40 cells. *, statistical significance, p < 0.05. C, representative phase-plane plots of instantaneous cell shortening as a function of the Ca2+ transient during five consecutive steady-state twitch contractions for control (black), wtPKD (blue), and caPKD (red). Phase-plane loops proceed counterclockwise and start from points indicated by the arrow.
TABLE 1.
Summary of the effects of electrical stimulation on cell shortening and magnitude of [Ca2+]i transients
Maximal amplitude of cell shortening and of [Ca2+]i transients were derived as described in the legend to Fig. 3 and under “Experimental Procedures.” The results are reported as %RL for cell shortening and F/F0 for [Ca2+]i transients. The data are presented as the means ± S.E. for control (n = 22), wtPKD (n = 21), and caPKD (n = 40), where n equals the number of cells analyzed.
| Control |
wtPKD |
caPKD |
||||
|---|---|---|---|---|---|---|
| %RL | F/F0 | %RL | F/F0 | %RL | F/F0 | |
| Initial | 12.22 ± 0.78 | 2.00 ± 0.13 | 12.14 ± 1.17 | 1.90 ± 0.18 | 9.98 ± 0.61a | 2.03 ± 0.11 |
| Steady-state | 5.08 ± 0.59 | 1.65 ± 0.08 | 5.85 ± 0.73 | 1.62 ± 0.14 | 1.49 ± 0.18b,c | 1.52 ± 0.07 |
a Significantly different from initial control %RL values (p < 0.05).
b Significantly different from steady-state control %RL values (p < 0.05).
c Significantly different from steady-state wtPKD %RL values (p < 0.05).
These declines in twitch amplitude may be due to a decrease in myofilament Ca2+ sensitivity. Dynamic myofilament Ca2+ sensitivity can be quantified as the ratio of maximum cell shortening (%RL) to the magnitude of the Ca2+ transients (F/F0)max. As shown in Fig. 3B, Ca2+ sensitivity of the first contraction following rest in caPKD cells shows a trend toward decrease relative to control and wtPKD. However, these experiments did not reach statistical significance in this sampling set. In contrast, although control and wtPKD cells show comparable values of steady-state myofilament Ca2+ sensitivity, this value is reduced by 67% in cells expressing caPKD (Fig. 3B). The results shown in Fig. 3B report Ca2+ sensitivity at only one point in the twitch cycle, at the peak of cell shortening and the [Ca2+]i transient. To assess myofilament Ca2+ sensitivity over a broad range of [Ca2+]i, phase-plane plots of contractile shortening as a function of the [Ca2+]i transient were constructed. Data from five consecutive steady-state contractions were used to generate these plots, which are shown in Fig. 3C. The phase plot of dynamic Ca2+ sensitivity for caPKD cells (red) is markedly depressed throughout the entire range of [Ca2+]i in both contraction and relaxation phases. Consistent with a dynamic phosphorylation-dephosphorylation mechanism, once desensitization was established in caPKD cells, the effects were reversible. However, restitution of normal Ca2+ sensitivity was slow, requiring 5 min of rest for return to the control levels (data not shown).
To further verify the specificity of caPKD, functional properties of cells expressing much lower levels of caPKD were analyzed (supplemental Fig. 2). Importantly, marked stimulus-dependent Ca2+ desensitization was still observed in caPKD cells where caPKD is overexpressed only 2-fold relative to endogenous PKD levels (supplemental Fig. 2B).
The dynamic in situ studies in Fig. 3 are consistent with and extend results from previous studies where equilibrium myofilament Ca2+ sensitivity was assessed in skinned cardiac myocytes exposed to active PKD (recombinant PKD catalytic domain with the regulatory domain deleted) (8, 10). In those studies, a significant rightward shift of the equilibrium sarcomere length versus pCa relationship was seen. Also, the lack of a phenotype in wtPKD cells in Fig. 3 is consistent with a previous study where contractile shortening was identical in control and wtPKD cardiac myocytes following electrical stimulation (9). In that report, PKD-dependent changes in EC coupling were seen only following stimulation with ET1 in such wtPKD cells. These results suggest that electrical stimulation has a more subtle, localized effect on PKD signaling compared with G-protein-coupled receptor activation by ET1. Taken together, studies with caPKD have unmasked a novel stimulus-dependent regulation of dynamic Ca2+ sensitivity of contraction in situ.
Stimulus-dependent Phosphorylation of cTnI in ARVMs
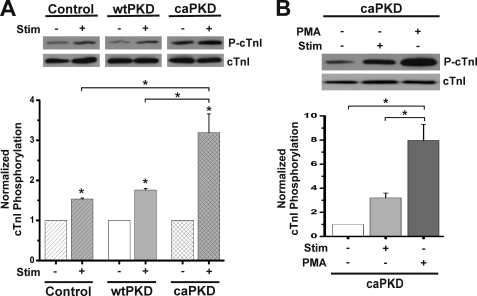
The results from Fig. 3 and Table 1 suggest that stimulus-dependent phosphorylation of cTnI at PKD sites Ser22 and Ser23 should be seen in caPKD cells. To test this hypothesis, ARVMs treated with control adenovirus or those expressing wtPKD or caPKD were rapidly harvested before and after electrical stimulation. Western blots were developed with a phospho-specific cTnI antibody (P-cTnI) to detect levels of cTnI phosphorylation at established PKD sites, Ser22/Ser23 (8–10). As shown in Fig. 4A, there is a 3-fold increase in cTnI phosphorylation following electrical stimulation of caPKD cells. Control and wtPKD cells both show a small but significant increase in cTnI phosphorylation following electrical stimulation, which is in accord with the small Ca2+ desensitization that is seen in these cells following the pulse protocol (Fig. 3 and Table 1). In order to calibrate the magnitude of the stimulus-evoked cTnI phosphorylation and thus the likely functional impact seen in stimulated cells, PMA was exploited in parallel experiments. PMA is a direct activator of PKD and PKCs (1, 15, 26, 32) and results in large, if not maximal, phosphorylation of cTnI in cardiac cells (35–37). As expected, PMA provoked a large response with an 8-fold increase in phospho-cTnI (Fig. 4B). Although the stimulus-dependent responses were 20–40% of the PMA signal, it is important to note that even modest increases in cTnI phosphorylation can lead to significant Ca2+ desensitization in cardiac cells (38). Thus, the results in Fig. 4 are in accord with stimulus-dependent functional results reported in Fig. 3 and Table 1.
FIGURE 4.
Electrical stimulation of control and caPKD myocytes increases cTnI phosphorylation. Cultured myocytes were subjected to 1-Hz field stimulation or PMA treatment (200 nm, 15 min) as described under “Experimental Procedures.” Cells were rapidly harvested, and protein extracts were analyzed by Western blot analyses. Blots were probed with anti-P-cTnI to detect phosphorylation at Ser22/23 and anti-cTnI to detect total cTnI protein levels. A, representative blot of cTnI phosphorylation before stimulation (−) and after electrical stimulation (+) as indicated. Control, wtPKD, and caPKD samples shown were run and developed on the same gel in separated lanes. The lower histogram shows summary data from multiple preparations of cTnI phosphorylation normalized to cTnI total protein (means ± S.E. (error bars), n = 3). Control, wtPKD, and caPKD values were further normalized to prestimulation levels. B, representative blot of cTnI phosphorylation in caPKD cells before and after electrical stimulation or after PMA treatment as indicated. The lower histogram shows summary data for these experiments (means ± S.E., n = 3) with caPKD values normalized to prestimulation levels. *, statistical significance, p < 0.01 versus prestimulation levels for control, wtPKD, and caPKD or differences as indicated with brackets.
Stimulus-dependent Phosphorylation of cTnI Is Linked to PKD
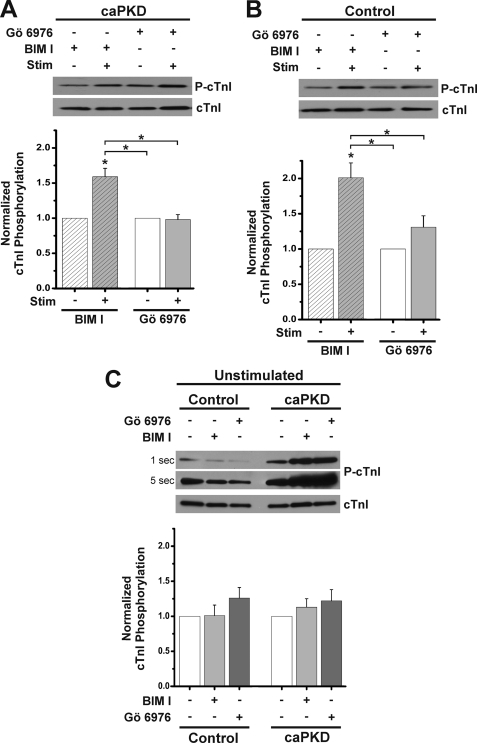
A complementary pharmacological approach with small molecule kinase inhibitors was used to further test the notion that PKD mediates stimulus-dependent phosphorylation of cTnI. Accordingly, we exploited a PKD inhibitor, Gö 6976, that does not inhibit nPKC isoforms (4, 30, 39), along with the PKC inhibitor BIM I (40, 41), which does not directly inhibit PKD (19, 26, 39, 42). As shown in Fig. 5A, stimulus-dependent cTnI phosphorylation was still evident in caPKD cells following BIM I pretreatment at doses known to inhibit cardiac PKCs (32, 40, 41). In contrast, Gö 6976 pretreatment at doses known to inhibit PKD in cardiac cells (7, 30) resulted in complete ablation of the stimulus-dependent increase in cTnI phosphorylation in caPKD cells (Fig. 5A). These results are consistent with in vitro cTnI Ser23/24 phosphorylation experiments (30). If this PKD cascade is operable in normal myocytes, then a similar pharmacological pattern should be seen in control ARVMs. Indeed, Fig. 5B confirms this notion with a 2-fold increase in stimulus-dependent cTnI phosphorylation in BIM I-treated control cells but a more effective inhibition of the stimulus-dependent increase in Gö 6976-treated control cells.
FIGURE 5.
Small molecule PKD inhibitor blocks stimulus-dependent cTnI phosphorylation. Cultured myocytes were treated with 5 μm BIM I or Gö6976 for 45 min prior to electrical stimulation as described in the legend to Fig. 4. Western blots from cell extracts were probed with anti-phospho-cTnI antibody to detect levels of P-cTnI and anti-cTnI to detect total cTnI protein level within each sample. A, upper panel shows a representative blot of cTnI phosphorylation in caPKD cells before and after stimulation in presence of inhibitors as indicated. The histogram below displays summary data with cTnI phosphorylation normalized to total cTnI protein (means ± S.E. (error bars), n = 4). BIM I and Gö 6976 values were further normalized to prestimulation levels. B, representative blot of cTnI phosphorylation in an experiment parallel to that in A except that control myocytes were examined. A summary histogram shows the means ± S.E. (n = 4). BIM I and Gö 6976 values were further normalized to prestimulation levels. *, statistical significance for caPKD or control, p < 0.01 versus prestimulation levels for BIM I or Gö 6976 or differences as indicated with brackets. C, representative blot of cTnI phosphorylation in unstimulated control and caPKD cells in the absence (−) or presence (+) of inhibitor as noted. Samples shown together were run and developed on the same gel. Exposure times for a representative blot are given on the left side of the blot. Summary data are shown below (means ± S.E., n = 4) with control and caPKD values further normalized to unstimulated (without inhibitor) levels.
In order to further validate the experiments with small molecule inhibitors, the effects of these agents on basal cTnI phosphorylation were quantified. Accordingly, extracts were prepared from unstimulated control and caPKD cells following incubations in the absence or presence of inhibitor. The levels of phospho-cTnI were lower in control cells compared with caPKD cells, as reflected in longer exposure times in Fig. 5C, and were consistent with Fig. 4A. Furthermore, small molecule kinase inhibitors had no influence on basal cTnI phosphorylation. As shown in Fig. 5C, the histogram reveals no significant differences in cTnI phosphorylation in control or caPKD cells following incubation with PKC and PKD kinase inhibitors. Although results with small molecule kinase inhibitors on their own should be interpreted with caution, the results of Fig. 5 provide more corroborative evidence linking PKD to the regulation of stimulus-dependent cTnI phosphorylation in caPKD cells. They also support the view that a stimulus-PKD cascade is a critical determinant of Ca2+ sensitivity in normal cardiac cells as well.
It is possible that PKA is activated in caPKD cells, which might explain some of these results. To assess activation of PKA, the phosphorylation state of PLB at the well established PKA site, Ser16, was examined (see supplemental Fig. 3). Importantly, PKA phosphorylation of PLB was minimal and identical in control and caPKD cells. Additionally, phospho-PLB did not increase following electrical stimulation in either control or caPKD cells (supplemental Fig. 3). Thus, PKA is not likely to play a prominent role in stimulus-dependent signaling in caPKD cells.
Stimulus-dependent Phosphorylation of Ser916 Regulatory Site within the Catalytic Domain of PKD
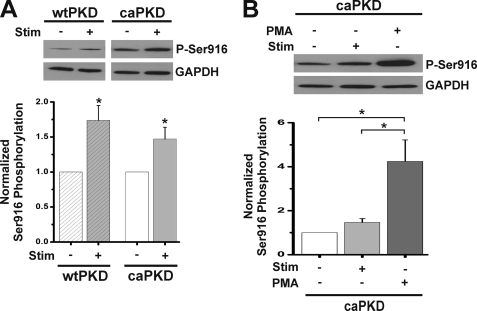
The results reported above provide compelling evidence that electrical stimulation evokes changes in the activation state of PKD. It was surprising to observe that even for caPKD myocardial cells, additional activation events are required for Ca2+ desensitization. It is well recognized that phosphorylation of PKD at Ser916, if not a direct readout for kinase activation, is a sensitive surrogate for stimulus-dependent, post-translational modification cascades impacting on PKD (2, 23, 30, 43). Multiple Western blot experiments with control ARVMs revealed that endogenous PKD levels are too low to detect phospho-Ser916 before or after stimulation (data not shown). Accordingly, caPKD cells were exploited in in situ experiments because they displayed marked functional and molecular responses to electrical stimulation (Figs. 3 and 4), and they express the caPKD construct that harbors the Ser916 available for potential phosphorylation events (see supplemental Fig. 1A). As shown in Fig. 6A, there is a 50% increase in phospho-Ser916 following electrical stimulation of caPKD cells and a 70% increase in wtPKD cells. Note that in this analysis, wtPKD and caPKD values are normalized to their prestimulation levels. As shown in blots in Fig. 6A, the intensity of caPKD bands before and after electrical stimulation are both markedly greater than those for stimulated wtPKD. In fact, direct quantitative comparison reveals that phospho-Ser916 levels are 2.78 ± 0.32-fold greater in unstimulated caPKD cells compared with stimulated wtPKD cells. To calibrate the magnitude of stimulus-evoked Ser916 phosphorylation, PMA was exploited in parallel experiments to elicit a maximal response of Ser916 phosphorylation, an approach reported previously (23). Fig. 6B shows phospho-Ser916 in electrically stimulated compared with PMA-stimulated caPKD cells. As expected for the robust activator of PKCs and PKD (see Fig. 1B), PMA had a more potent effect on Ser916 phosphorylation than electrical stimulation. Taken together, these data provide in situ evidence that electrical stimulation leads to post-translational modification of caPKD required to provoke Ca2+ desensitization in ARVMs.
FIGURE 6.
Electrical stimulation increases Ser916 phosphorylation in wtPKD and caPKD. Cultured myocytes were electrically stimulated or treated with PMA (200 nm, 15 min) as described under “Experimental Procedures.” Cell extracts were analyzed by Western blot methods and were probed with anti-phospho-Ser916 and then with anti-GAPDH as a loading control. A, representative blot of Ser916 phosphorylation before (−) and after (+) electrical stimulation of wtPKD and caPKD cells as indicated. Samples shown were run and developed on the same gel in separated lanes. The histogram below shows summary data of Ser916 phosphorylation normalized to loading control (means ± S.E. (error bars), n = 3). wtPKD and caPKD values were further normalized to prestimulation levels. B, representative blot of Ser916 phosphorylation in caPKD cells before and after electrical stimulation or treatment with PMA as indicated. *, statistical significance, p < 0.05 versus prestimulation levels for wtPKD and caPKD or differences as indicated with brackets.
Effects of Phosphorylation-deficient caPKD-S916A Mutant Expression on Myofilament Ca2+ Desensitization
Additional experiments were designed to directly test the notion that phosphorylation of Ser916 is a critical step in the electrical depolarization-PKD-Ca2+ desensitization cascade. Accordingly, a new chimera was generated, caPKD that harbors a phosphorylation-defective site at Ser916, Plum-caPKD-S916A (S916A). Cultured ARVMs were infected with adenoviral constructs that drove the expression of equal levels of either HA-caPKD, Plum-caPKD, or S916A (data not shown). Because Ser916 resides at the C terminus in a PDZ-binding domain (44), potential aberrant targeting of Plum-caPKD-S916A could complicate interpretations of these experiments. Thus, confocal imaging analyses were used to compare the targeting of S916A expression in ARVMs to Plum-caPKD (supplemental Fig. 4) and HA-caPKD (data not shown) (Fig. 2). Importantly, double labeling and three-dimensional reconstructions for Plum-caPKD and S916A (supplemental Fig. 4) are consistent with those from HA-tagged caPKD and wtPKD (Fig. 2), which show that the striated pattern displays a punctate, lattice-like distribution at the Z-line and not the M-line. Thus, detailed imaging analysis confirmed that the S916A mutant distributed in a manner nearly identical to other PKD constructs in ARVMs.
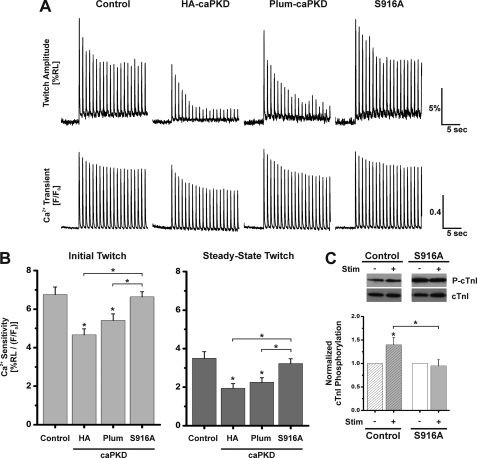
Another set of control experiments examined the possibility that the addition of the Plum sequence might disrupt PKD function. Fig. 7A shows representative traces of cell shortening and [Ca2+]i transients recorded during a train of depolarizing stimuli from cells that were initially at rest. Note that cell shortening was depressed in a similar manner in HA- and Plum-caPKD cells as steady-state values were achieved (Fig. 7A and summary data in Table 2). The Ca2+ sensitivity analysis in Fig. 7B confirms that the Plum-caPKD maintains electrically induced Ca2+ desensitization in cardiac cells. Whereas the Ca2+ sensitivities of the initial twitch of HA- and Plum-tagged caPKD cells in Fig. 7B decreased 30 and 20% relative to control, respectively, there is a similar trend toward decrease in caPKD relative to control in Fig. 3B. However, those experiments did not reach statistical significance in that sampling set.
FIGURE 7.
Cells expressing caPKD-S916A mutant display a marked increase in cell shortening. Measurements of sarcomere length and [Ca2+]i transients in field-stimulated ARVMs expressing various forms of caPKD were performed as described under “Experimental Procedures” and in the legend to Fig. 3. A, representative traces of cell shortening (twitch amplitude, %RL) and Ca2+ transients (F/F0) measured simultaneously from individual cardiomyocytes. Upper panels show the change in sarcomere length during 20-s stimulation at 1 Hz. Lower panels show the change in the [Ca2+]i transient magnitude in Fluo-3-loaded cells. B, summary data of Ca2+ sensitivity of contraction, expressed as the ratio %RLmax/(F/F0)max, for control adenovirus, HA-caPKD, Plum-caPKD, and S916A (Plum-caPKD-S916A) myocytes as indicated. The results are displayed for first twitch (Initial Twitch) and values at the end of a train of 20 pulses (Steady-State Twitch). Data are means ± S.E. (error bars) from multiple culture preparations, n = 49–87 cells. *, statistical significance, p < 0.05 versus control or differences as indicated with brackets. C, representative blot of cTnI phosphorylation before stimulation (−) and after electrical stimulation (+) as indicated. Cultured myocytes were subjected to 1-Hz field stimulation as described under “Experimental Procedures.” Cells were rapidly harvested, and protein extracts were analyzed by Western blot analyses. Blots were probed with anti-P-cTnI to detect phosphorylation at Ser22/23 and anti-cTnI to detect total cTnI protein levels. Control and S916A samples shown were run and developed on the same gel in separated lanes. The lower histogram shows summary data of cTnI phosphorylation normalized to cTnI total protein (means ± S.E., n = 4). Control and S916A values were further normalized to prestimulation levels. *, statistical significance, p < 0.05 versus prestimulation levels for control or differences as indicated with brackets.
TABLE 2.
Summary of the effects of electrical stimulation on cell shortening and magnitude of [Ca2+]i transients in PKD mutant-expressing cells
Maximal amplitude of cell shortening and of [Ca2+]i transients were derived as described in the legend to Fig. 3 and “Experimental Procedures.” The results are reported as %RL for cell shortening and F/F0 for [Ca2+]i transients. The data are derived from experiments illustrated in Fig. 7 and presented as the means ± S.E. for control (n = 49), HA-caPKD (n = 49), Plum-caPKD (n = 54), and Plum-caPKD-S916A (n = 87), where n equals the number of cells analyzed.
| Control |
HA-caPKD |
Plum-caPKD |
Plum-caPKD-S916A |
|||||
|---|---|---|---|---|---|---|---|---|
| %RL | F/F0 | %RL | F/F0 | %RL | F/F0 | %RL | F/F0 | |
| Initial | 11.14 ± 0.47 | 1.75 ± 0.06 | 8.52 ± 0.51a,b | 1.93 ± 0.07 | 9.81 ± 0.51 | 1.92 ± 0.06 | 11.14 ± 0.40 | 1.74 ± 0.04 |
| Steady-state | 4.97 ± 0.44 | 1.48 ± 0.04 | 3.04 ± 0.40c,d | 1.59 ± 0.06 | 3.62 ± 0.38c,d | 1.61 ± 0.05 | 5.04 ± 0.44 | 1.51 ± 0.04 |
a Significantly different from initial control %RL values (p < 0.05).
b Significantly different from initial Plum-caPKD-S916A %RL values (p < 0.05).
c Significantly different from steady-state control %RL values (p < 0.05).
d Significantly different from steady-state Plum-caPKD-S916A %RL values (p < 0.05).
In contrast to HA- and Plum-caPKD cells, with S916A cells, there was little impact of the pulse protocol on shortening as steady-state values were achieved (Fig. 7A). In fact, cell shortening in the S916A mutant was comparable with levels seen in control cells (Fig. 7A and Table 2). There was no significant difference in the magnitude of the [Ca2+]i transients between control, HA-caPKD, Plum-caPKD, and S916A cells (Table 2) that might account for the differences in cell shortening. Fig. 7B shows the analysis of Ca2+ sensitivity. As expected, contractile Ca2+ sensitivity is reduced in HA-caPKD cells as well as in Plum-caPKD cells compared with control (Fig. 7B). In contrast, there was no difference in Ca2+ sensitivity between control and S916A cells in either the first twitch or the steady-state twitch values.
These results imply that there is a lack of stimulus-dependent cTnI phosphorylation in S916A cells. To assess this notion, cells were rapidly harvested following electrical stimulation, and extracts were probed for cTnI phosphorylation. As shown in Fig. 7C, in contrast to the increases in stimulus-dependent cTnI phosphorylation seen in control cells, the intensity of the phospho-cTnI bands was unchanged by electrical stimulation in S916A cells. It is noted that S916A expression alone resulted in a 2.9-fold increase in basal cTnI phosphorylation relative to control cells (Fig. 7C), and this is comparable with the 2.7-fold increase seen in HA-tagged caPKD cells relative to control (e.g. see Fig. 4B). The mechanism for this response is not clear, but the inhibitor studies in Fig. 5 suggest this may be an indirect effect not strictly dependent on PKD or PKC activity. Taken together, these in situ results reveal that Ser916 phosphorylation plays a critical role in linking electrical stimulation and PKD signaling to myofilament Ca2+ sensitivity.
Increased Myofilament Ca2+ Sensitivity and Loss of Stimulus-dependent cTnI Phosphorylation in ARVMs Expressing Dominant Negative PKD
If PKD is a critical regulator of Ca2+ sensitivity in myocytes, then expression of dominant negative constructs should have an impact on stimulus-dependent contraction. The HA-tagged kinase-dead PKD mutant (dnPKD) was expressed (see mutation scheme in supplemental Fig. 1A), and Western blot analyses revealed that this adenoviral approach achieves equal expression of dnPKD compared with wtPKD and caPKD in cultured ARVMs (supplemental Fig. 1, C and D). Note that long exposure (2 h) Western blots with anti-PKD (supplemental Fig. 1D) demonstrate marked overexpression levels of dnPKD relative to endogenous protein, a condition critical for functional dnPKD experiments.
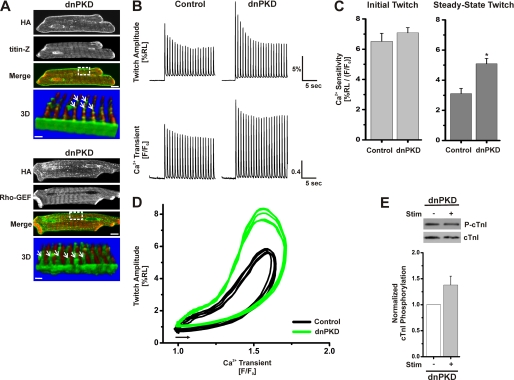
Because it has been reported that the dnPKD distributes very differently from PKD in neurons (44), confocal imaging analyses were used to examine the targeting of dnPKD expression in ARVMs. The upper panels of Fig. 8A show a striated distribution for dnPKD, which is comparable with the localization of Plum-wtPKD (Fig. 1A) and the distribution of HA-tagged wtPKD and caPKD (Fig. 2A). Importantly, double labeling and three-dimensional reconstruction images in Fig. 8A reveal a networked pattern identical to those of caPKD and wtPKD (Fig. 2). Thus, although dnPKD displays a very different distribution in neuronal cells (44), no differences were detected in ARVMs by high resolution confocal imaging.
FIGURE 8.
Impact of localized dnPKD expression in cardiac myocytes on myofilament Ca2+ sensitivity and stimulus-dependent changes in cTnI phosphorylation. Myocytes were cultured for 24 h after infection with dnPKD adenovirus as described under “Experimental Procedures” and in the legend to Fig. 2 to express dnPKD at levels comparable with those for caPKD and wtPKD (see supplemental material). A, confocal images of fixed cells prepared for double label immunofluorescent analysis using anti-HA antibody (green) with anti-titin-Z antibody (red) in the upper panel or with anti-obscurin Rho-GEF antibody (red) in the lower panel. Scale bar, 10 μm. The bottom panel (labeled 3D) shows a three-dimensional image constructed from consecutive Z-scans (0.3 μm apart) of the boxed region in the merged image and rotated by about 200° (so that the sarcolemma edge is close to the viewer). Scale bar, 1.5 μm. The white arrows identify HA localization along the Z-line with a punctate, lattice-like/reticular distribution pattern. Control data in B–D are identical to control data presented in Fig. 3 so that direct comparisons of data sets collected on the same day could be made. B, representative traces of cell shortening (twitch amplitude, %RL) and Ca2+ transients (F/F0) measured simultaneously from individual cardiomyocytes treated with control adenovirus or those expressing dnPKD. The upper traces show the change in sarcomere length during 20-s stimulation at 1 Hz. The lower traces display the corresponding change in the [Ca2+]i transient magnitude in these Fluo-3-loaded cells. C, summary data of Ca2+ sensitivity of contraction for control adenovirus and dnPKD myocytes. Data are means ± S.E. from multiple culture preparations, n = 20–40 cells. *, statistical significance, p < 0.05 versus control. D, representative phase-plane plots of instantaneous cell shortening as a function of the Ca2+ transient during five consecutive steady-state twitch contractions for control (black) and dnPKD (green). Phase-plane loops proceed counterclockwise and start from points indicated by the arrow. E, representative blot of cTnI phosphorylation before (−) and after (+) electrical stimulation for dnPKD myocytes. Western blots were probed with anti-phospho-cTnI antibody to detect levels of P-cTnI and anti-cTnI antibody to detect total cTnI protein level within each sample. The histogram below presents summary data of cTnI phosphorylation normalized to cTnI total protein. dnPKD values were further normalized to prestimulation level. Data are means ± S.E. (error bars) from multiple culture preparations, n = 3. Statistical significance, p < 0.01, was achieved in caPKD cells when comparing prestimulation levels with stimulation levels (data not shown) but not in dnPKD cells.
A detailed functional analysis of dnPKD cells focused on cell shortening and [Ca2+]i transients. Fig. 8B displays representative traces of cell shortening and [Ca2+]i transients recorded during a train of depolarizing stimuli from control or dnPKD cells that were initially at rest. Recall that, relative to control cells, the steady-state contractions were reduced in caPKD cells with no significant difference in [Ca2+]i transient magnitude (Figs. 3A and 7A). In contrast, the steady-state contractions were actually increased in dnPKD cells relative to control cells (Fig. 8B). The summary data in Fig. 8C show that the peak myofilament Ca2+ sensitivity in dnPKD cells following 20 pulses was 64% greater than that of control cells. This is contrasted with the opposite 67% decrease seen in caPKD cells relative to control cells (Fig. 3B). To better assess Ca2+ sensitivity over a dynamic range of [Ca2+]i, phase-plane plots of contractile shortening as a function of the [Ca2+]i transient were constructed as described in Fig. 3C. The representative phase-plane plots in Fig. 8D are consistent with the summary data (Fig. 8C) and show a marked increase in Ca2+ sensitivity throughout a major portion of contraction and relaxation phases relative to control. If endogenous PKD mediates stimulus-dependent cTnI phosphorylation, then overexpression of dnPKD should block this signaling event. Fig. 8E shows that, in contrast to the increases in stimulus-dependent cTnI phosphorylation in control and caPKD cells (data not shown but refer to Fig. 4A), there is a modest, non-significant increase in phospho-cTnI in dnPKD. These detailed in situ results provide important new insight into the dynamic regulation of Ca2+ sensitivity of contraction in cardiomyocytes.
DISCUSSION
Recent findings from several groups have shown that PKD is a key component in myocardial cell signaling that underlies cardiac hypertrophy and heart failure (1, 3, 4). PKD-mediated phosphorylation of class II histone deacetylases (1, 4, 5, 19) and CREB (6, 7) as well as PKD-mediated phosphorylation of multiple sarcomeric substrates (8–10) are likely reactions in these signaling cascades. Recent reports provide evidence that PKD can regulate cardiac L-type Ca2+ channels and sarcoplasmic reticulum Ca2+ load in intact cardiac myocytes as well (11, 12). Although the precise phosphorylation sites for these changes in Ca2+ signaling are not yet defined, these results reveal a broader scope of functions for this kinase in the regulation of excitation-contraction coupling in cardiac myocytes. Given that PKD is up-regulated in failing human heart and its inhibition is proposed as a promising therapeutic strategy for heart disease (2, 4, 19), it is critical to identify the hierarchy of functional targets for PKD throughout the cardiac myocyte. Through a combination of molecular genetics, functional assays, and complementary in situ molecular studies, we define a novel role for PKD in stimulus-dependent dynamic regulation of myofilament Ca2+ sensitivity in cultured ventricular cardiac myocytes. Molecular genetic studies reveal that phosphorylation of PKD at Ser916 is a critical step in the stimulus-dependent signaling cascade. Finally, through a characterization of the PKD probes developed in this study, new information on the subcellular distribution of PKD in adult ventricular cardiac myocytes is obtained. Through expression of wild type PKD as well as activation mutants, an important finding was that PKD distributes in a novel non-nuclear reticular pattern in adult myocytes that is not highly dependent on the activation state of the enzyme.
Role of PKD in Stimulus-dependent Regulation of Contractile Ca2+ Sensitivity
Previous studies show that PKD can reduce myofilament Ca2+ sensitivity through phosphorylation of Ser22/23 on cTnI in ARVMs (8–10). Because these interesting previous functional studies were performed in skinned myocytes with activated forms of PKD added exogenously, it is not known when this signaling reaction actually occurs in normal cardiac myocytes. Multiple groups (8–10) provide a plausible notion that PKD phosphorylation of cTnI may become prominent in failing heart cells where PKD expression and activity is increased (1–4). In the process of examining regulation of excitation-contraction coupling in situ with a gain of function PKD mutant, we found a profound Ca2+ desensitization of the twitch in caPKD cells. Although this contractile phenotype is consistent with these previous studies in skinned cells, it was unexpected to observe a remarkable stimulus dependence of this Ca2+ desensitization response most manifest in caPKD cells. Several lines of evidence support the conclusion that PKD mediates this response. Ca2+ desensitization in caPKD cells was accompanied by stimulus-dependent phosphorylation of a known PKD substrate, cTnI, at Ser22/23 (8–10). Although these serines are also substrates for PKA (45) and PKC (36), data presented here argue strongly against the involvement of either kinase in this process. The stimulus-dependent phosphorylation of cTnI was blocked by a known PKD inhibitor, Gö 6976, which does not inhibit novel PKCs. Further, a PKC inhibitor, BIM I, was not an efficient blocker of cTnI phosphorylation in caPKD cells. Finally, expression of a kinase-dead PKD mutant, dnPKD, blocked both the functional Ca2+ desensitization and phosphorylation of cTnI. Consistent with a dominant negative pattern of action, there was no stimulus dependence of effects in dnPKD cells, where contractions were more sensitive to [Ca2+]i throughout the train of stimuli, even on the first stimulus following rest. Further, the lack of phosphorylation of phospholamban at Ser16 reveals low levels of PKA activity in these caPKD cells. Taken together, these results are difficult to reconcile with a prominent role of PKA or PKC in this novel, stimulus-dependent regulation of cell shortening but rather illuminate a role for PKD in this signaling cascade in cultured cells.
A collection of related experiments reveals that this novel PKD contractile regulation is not only seen in caPKD cells but is operative in control myocytes as well. First, there was a stimulus-dependent Ca2+ desensitization response in control cells, a 52% decrease in Ca2+ sensitivity from the first to the steady-state twitch. Second, control cells also showed a small but significant increase in cTnI phosphorylation at Ser22/23 following stimulation that was sensitive to Gö 6976. This increase was comparable with that seen in wtPKD cells but significantly less than that seen in caPKD cells. It is important to note that the magnitude of changes in cTnI phosphorylation, although rather small, is physiologically relevant because it has been reported that small changes in cTnI phosphorylation can result in a marked decrease of cardiac contractile function (38, 46). For example, Belin et al. (38) report that only an 11% increase in the fraction of phosphorylated cTnI in control rat myocytes compared with those from heart failure models resulted in a 35% decrease in developed force. Finally, the ability of dnPKD expression to block the stimulus-dependent increases seen in control cells is best explained through the action of endogenous PKD in these responses. These in situ experiments provide new evidence implicating PKD as a regulator of cardiac myofilament Ca2+ sensitivity. Thus, stimulus-dependent PKD must be considered as part of the hierarchy of kinases, including PKA and PKCs, that regulate cTnI phosphorylation and Ca2+ sensitivity of contraction of heart tissue.
Ser916 Phosphorylation Is an Important Step in the PKD Activation Cascade
In general, it is well recognized that receptor activation of PKD includes a complex cascade of sequential auto- and transphosphorylation events (7, 30, 43). The stimulus-dependent behavior for the “constitutively active” PKD construct used here may appear counterintuitive because this PKD mutant has conserved phosphomimetic S744E/S748E substitutions in the activation loop, known substrates for nPKCs. However, although caPKD can be recovered in the resting state as a fully active, Ser916-phosphorylated form in many cells that possess high endogenous PKC/PKD activity (47, 48), more recent reports reveal that this is not the case in other cell types, such as lymphocytes (25) and cardiac myocytes (7). The apparent low resting state activity of caPKD may reflect insufficient levels of DAG cofactor in quiescent cardiac myocytes, as speculated by Ozgen et al. (7) (see Fig. 9).
FIGURE 9.
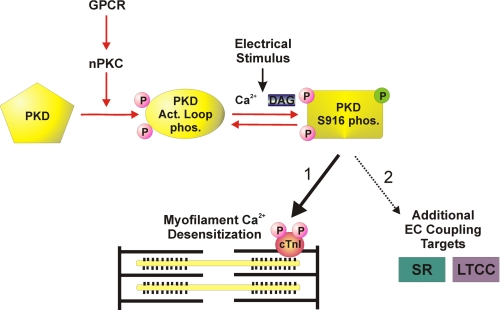
Schematic diagram emphasizing a localized PKD signaling cascade that contributes to the dynamic regulation of Ca2+ sensitivity of contraction in cardiac myocytes. The diagram depicts a multistep model for stimulus-dependent PKD activation, emphasizing an nPKC-dependent pathway to achieve PKD activation loop phosphorylations (oval labeled Act. Loop phos.), which is mimicked by our caPKD construct. An electrical stimulus is required to achieve further activation of this kinase. This step is reversible and most likely reflects the stimulus-dependent autophosphorylation of Ser916 (rectangle labeled PKD S916 phos.) within the catalytic domain of PKD and possibly at other sites within the regulatory domain of PKD (30). Although phosphorylation of Ser916 may or may not be a direct readout of PKD activation state, it is required for the subsequent stimulus-dependent phosphorylation of cTnI at Ser22/23 and development of myofilament Ca2+ desensitization (pathway 1). Pathway 2 demonstrates additional EC coupling targets of PKD signaling that are consistent with the spatial distribution of PKD and functional analyses on sarcoplasmic reticulum (SR) Ca2+ load and L-type calcium channel (LTCC) current (ICa) (11, 12).
The phosphorylation of Ser916 has been associated with several important functions, which may be cell-specific. Although phosphorylation of Ser916 has been linked to the PKD activation pathway, it may or may not be a direct readout of PKD activation state (2, 23, 30, 43). Further, this residue is found in a PDZ binding motif and is associated with a phosphorylation-dependent scaffolding and subcellular localization function in neuronal cells (44). Thus, we suspected that, regardless of its function, phosphorylation of Ser916 is critical in the stimulus-dependent PKD cascade and explains the stimulus-dependent behavior of caPKD cells. This view is supported by results showing a 50% increase of phospho-Ser916 in caPKD cells following electrical stimulation. As a direct test of this conclusion, cells were infected with a phosphorylation defective mutant, caPKD-S916A. Such cells responded normally to electrical stimulation with none of the enhanced desensitization or cTnI phosphorylation seen in their caPKD counterparts. Because Ser916 phosphorylation is important in scaffolding and targeting in neuronal cells (44), a series of high resolution confocal imaging studies examined subcellular targeting of this mutant in cardiac myocytes. Because this localization was identical to that seen for caPKD in cardiac myocytes, aberrant targeting in the S916A mutant is not a likely explanation for the lack of responses in such cells. Taken together these results underscore the importance of Ser916 phosphorylation in the PKD cascade reported here.
Although the precise downstream events remain to be defined, these in situ results are consistent with a recent report that, in vitro, caPKD autophosphorylation can proceed at multiple sites following Ser916 phosphorylation (30). Such phosphorylation events could mediate changes in activity and/or subcellular targeting. Regardless, these results highlight the critical importance of Ser916 phosphorylation in the control of PKD signaling in stimulated myocytes as depicted in the schematic diagram in Fig. 9. It will be important to determine if the other PKD targets in excitation-contraction coupling that we have seen, the sarcoplasmic reticulum and L-type Ca2+ channel (11, 12), are also under stimulus-dependent control.
Spatial Segregation of PKD in Adult Cardiac Myocytes
Information on the localization of cardiac PKD is derived primarily from studies with neonatal myocytes. These include identification of localized PKD scaffolding to AKAPs (49, 50), targeting to Z-discs (41), and nuclear targeting and subsequent phosphorylation of class II histone deacetylases (1, 4, 5, 19) and CREB (6, 7). Because the subcellular architecture of cultured neonatal cells is unique and quite distinct from the central features seen in adult myocytes, it is not possible to extrapolate these interesting previous findings to those in the adult heart. It is important to note that low levels of PKD expression in adult myocytes limit precise confocal imaging of endogenous protein in that setting. However, the characterization of the epitope-tagged PKD mutant probes developed here afforded the opportunity to assess, for the first time, subcellular targeting of activation mutants in adult myocytes. Regardless of the activation state, all forms of PKD used here, including wild type, constitutively active, kinase-dead, and Plum-tagged wild type and constitutively active, distributed in a predominant striated pattern associated with Z-lines, which is reminiscent of Z-disc targeting in cultured neonatal myocytes (41). This contrasts with the distinctly different distribution of kinase-dead mutants compared with wild-type PKD in neurons (44). More precise three-dimensional analyses revealed a punctate, reticular pattern for all PKDs, indicating that localization is not strictly limited to the Z-disc but rather recalls a distinctive network indicative of the T-tubular/junctional sarcoplasmic reticulum found in adult cardiac myocytes in other imaging studies using dextran-linked fluorescein in live cells (33). Although other studies have shown targeting of PKD to perinuclear vesicular structures in unstimulated cells, consistent with localization to the Golgi compartment (51–55), to our knowledge, this is the first study to show localization of PKD at reticular structures in ARVMs. This novel spatial distribution suggests that PKD plays unappreciated roles in regulation of excitation-contraction coupling and is also consistent with PKD regulation of the sarcoplasmic reticulum and L-type Ca2+ channel we have reported (11, 12).
The z-stack confocal images also showed that the activation state of PKD was important in specifying a relatively minor localization at the plasma membrane. In particular, minor fractions of wtPKD and dnPKD, but not caPKD, were seen at the sarcolemmal membrane when all three forms were expressed to the same level in cardiac myocytes. These results are in accord with previous studies where both wild-type and kinase-dead PKD translocate to plasma membranes in activated lymphocytes, fibroblasts, and epithelial cells (56, 57). In addition to this plasma membrane translocation of fluorescently tagged PKD following stimulation of various cell types, it appears that a small fraction of wtPKD and kinase-dead PKD forms reside at distinct sites on the plasma membrane in these quiescent cells as well (55–59). For example, Spitaler et al. report that a kinase-dead PKD mutant showed partial accumulation at the plasma membrane and perinuclear vesicular structures in unstimulated T cells (55). It has been proposed from studies with fibroblasts and epithelial cells that the second cysteine-rich domain within the regulatory domain of PKD, and not the catalytic domain, is necessary and sufficient to mediate the translocation of this kinase to the plasma membrane (57). Additional evidence is consistent with a lack of plasma membrane targeting for caPKD reported here. In studies in activated lymphocytes, fibroblasts, and epithelial cells, phosphorylation of Ser744 and Ser748, mimicked by the caPKD mutant here, destabilized PKD-plasma membrane interactions (57). Accordingly, Rey et al. (57) concluded that in response to GPCR activation, PKC-dependent phosphorylation of Ser744 and Ser748 are key events necessary for the rapid reverse translocation of GFP-PKD from the plasma membrane. Taken together, the distinct pattern of plasma membrane localization of PKD activation mutants reported here for cardiac cells is in accord with proposed mechanisms in several cell types.
This study reveals that PKD can dynamically regulate the normal contraction-relaxation process in cardiac mycoytes (Fig. 9). Given that PKD activity is increased in failing hearts, it will be important to determine the role of increased signaling at these newly appreciated cellular locales in the development of heart failure. Additionally, because PKD inhibition is proposed as a therapeutic strategy for heart disease (2, 4, 19), this approach has the potential to produce marked negative effects on excitation-contraction coupling in addition to the desired effects on nuclear targets and gene expression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL070709 (to T. B. R.), T32-GM-008181 (to M. H. G. and A. Z.), T32-AR-07592 (to M. H. G.), MARC U*STAR T34-08663 (to R. D. W.), and T32-HL-072751 (to A. Z.). This work was also supported by an Economic Development Corporation of Maryland stem cell grant (to T. B. R. and W. J. L.) and the Leducq Foundation (to W. J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- PKD
- protein kinase D1
- ARVM
- adult rat ventricular myocyte
- wtPKD
- wild type PKD
- caPKD
- constitutively active PKD
- dnPKD
- dominant negative PKD
- CREB
- cAMP-response element-binding protein
- cTnI
- cardiac troponin I
- EC coupling
- excitation-contraction coupling
- DAG
- diacylglycerol
- PMA
- phorbol 12-myristate 13-acetate
- BIM I
- bisindolylmaleimide I
- %RL
- percentage of resting length
- nPKC
- novel protein kinase C
- P-cTnI
- phospho-specific cardiac troponin I at Ser22/23
- P-Ser916
- phospho-specific protein kinase D at Ser916
- S916A
- Plum-caPKD-S916A
- PKA
- protein kinase A
- PLB
- phospholamban
- ET1
- endothelin-1.
REFERENCES
- 1.Harrison B. C., Kim M. S., van Rooij E., Plato C. F., Papst P. J., Vega R. B., McAnally J. A., Richardson J. A., Bassel-Duby R., Olson E. N., McKinsey T. A. (2006) Mol. Cell. Biol. 26, 3875–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avkiran M., Rowland A. J., Cuello F., Haworth R. S. (2008) Circ. Res. 102, 157–163 [DOI] [PubMed] [Google Scholar]
- 3.Fielitz J., Kim M. S., Shelton J. M., Qi X., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossuyt J., Helmstadter K., Wu X., Clements-Jewery H., Haworth R. S., Avkiran M., Martin J. L., Pogwizd S. M., Bers D. M. (2008) Circ. Res. 102, 695–702 [DOI] [PubMed] [Google Scholar]
- 5.McKinsey T. A. (2007) Cardiovasc. Res. 73, 667–677 [DOI] [PubMed] [Google Scholar]
- 6.Johannessen M., Delghandi M. P., Rykx A., Dragset M., Vandenheede J. R., Van Lint J., Moens U. (2007) J. Biol. Chem. 282, 14777–14787 [DOI] [PubMed] [Google Scholar]
- 7.Ozgen N., Obreztchikova M., Guo J., Elouardighi H., Dorn G. W., 2nd, Wilson B. A., Steinberg S. F. (2008) J. Biol. Chem. 283, 17009–17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haworth R. S., Cuello F., Herron T. J., Franzen G., Kentish J. C., Gautel M., Avkiran M. (2004) Circ. Res. 95, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 9.Cuello F., Bardswell S. C., Haworth R. S., Yin X., Lutz S., Wieland T., Mayr M., Kentish J. C., Avkiran M. (2007) Circ. Res. 100, 864–873 [DOI] [PubMed] [Google Scholar]
- 10.Bardswell S. C., Cuello F., Rowland A. J., Sadayappan S., Robbins J., Gautel M., Walker J. W., Kentish J. C., Avkiran M. (2010) J. Biol. Chem. 285, 5674–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodall M. H., Teos L. Y., Goldblum R. R., Ziman A., duBell W. H., Lederer W. J., Randall W., Rogers T. B. (2009) Biophys. J. 96, 622a–623a [Google Scholar]
- 12.Goodall M. H., Goldblum R. R., Ziman A., Lederer W. J., Rogers T. B. (2009) J. Mol. Cell Cardiol. 46, S29 [Google Scholar]
- 13.Kentish J. C., McCloskey D. T., Layland J., Palmer S., Leiden J. M., Martin A. F., Solaro R. J. (2001) Circ. Res. 88, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 14.Solaro R. J. (2008) J. Biol. Chem. 283, 26829–26833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haworth R. S., Roberts N. A., Cuello F., Avkiran M. (2007) J. Mol. Cell Cardiol. 43, 686–695 [DOI] [PubMed] [Google Scholar]
- 16.Lohse M. J., Engelhardt S., Eschenhagen T. (2003) Circ. Res. 93, 896–906 [DOI] [PubMed] [Google Scholar]
- 17.Backs J., Olson E. N. (2006) Circ. Res. 98, 15–24 [DOI] [PubMed] [Google Scholar]
- 18.Rozengurt E., Rey O., Waldron R. T. (2005) J. Biol. Chem. 280, 13205–13208 [DOI] [PubMed] [Google Scholar]
- 19.Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Mol. Cell. Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X., Zhang T., Bossuyt J., Li X., McKinsey T. A., Dedman J. R., Olson E. N., Chen J., Brown J. H., Bers D. M. (2006) J. Clin. Invest. 116, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M. S., Wang F., Puthanveetil P., Kewalramani G., Hosseini-Beheshti E., Ng N., Wang Y., Kumar U., Innis S., Proud C. G., Abrahani A., Rodrigues B. (2008) Circ. Res. 103, 252–260 [DOI] [PubMed] [Google Scholar]
- 22.Kunkel M. T., Newton A. C. (2010) J. Biol. Chem. 285, 22748–22752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luiken J. J., Vertommen D., Coort S. L., Habets D. D., El Hasnaoui M., Pelsers M. M., Viollet B., Bonen A., Hue L., Rider M. H., Glatz J. F. (2008) Cell. Signal. 20, 543–556 [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Cseresnyés Z., Randall W. R., Schneider M. F. (2001) J. Cell Biol. 155, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood C. D., Marklund U., Cantrell D. A. (2005) J. Biol. Chem. 280, 6245–6251 [DOI] [PubMed] [Google Scholar]
- 26.Zugaza J. L., Sinnett-Smith J., Van Lint J., Rozengurt E. (1996) EMBO J. 15, 6220–6230 [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Jackson W. C., Steinbach P. A., Tsien R. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16745–16749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews S. A., Rozengurt E., Cantrell D. (1999) J. Biol. Chem. 274, 26543–26549 [DOI] [PubMed] [Google Scholar]
- 29.DuBell W. H., Lederer W. J., Rogers T. B. (2000) Am. J. Physiol. Heart Circ. Physiol 278, H886–H897 [DOI] [PubMed] [Google Scholar]
- 30.Rybin V. O., Guo J., Steinberg S. F. (2009) J. Biol. Chem. 284, 2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman A. L., Kontrogianni-Konstantopoulos A., Hirsch S. S., Geisler S. B., Gonzalez-Serratos H., Russell M. W., Bloch R. J. (2007) FEBS Lett. 581, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haworth R. S., Goss M. W., Rozengurt E., Avkiran M. (2000) J. Mol. Cell Cardiol. 32, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 33.Soeller C., Cannell M. B. (1999) Circ. Res. 84, 266–275 [DOI] [PubMed] [Google Scholar]
- 34.Sugden P. H. (2003) J. Mol. Cell Cardiol. 35, 871–886 [DOI] [PubMed] [Google Scholar]
- 35.Pi Y., Zhang D., Kemnitz K. R., Wang H., Walker J. W. (2003) J. Physiol. 552, 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumandea M. P., Rybin V. O., Hinken A. C., Wang C., Kobayashi T., Harleton E., Sievert G., Balke C. W., Feinmark S. J., Solaro R. J., Steinberg S. F. (2008) J. Biol. Chem. 283, 22680–22689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avner B. S., Hinken A. C., Yuan C., Solaro R. J. (2010) Am. J. Physiol. Heart Circ. Physiol. 299, H723–H730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belin R. J., Sumandea M. P., Kobayashi T., Walker L. A., Rundell V. L., Urboniene D., Yuzhakova M., Ruch S. H., Geenen D. L., Solaro R. J., de Tombe P. P. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H2344–H2353 [DOI] [PubMed] [Google Scholar]
- 39.Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. (1996) FEBS Lett. 392, 77–80 [DOI] [PubMed] [Google Scholar]
- 40.Rybin V. O., Sabri A., Short J., Braz J. C., Molkentin J. D., Steinberg S. F. (2003) J. Biol. Chem. 278, 14555–14564 [DOI] [PubMed] [Google Scholar]
- 41.Iwata M., Maturana A., Hoshijima M., Tatematsu K., Okajima T., Vandenheede J. R., Van Lint J., Tanizawa K., Kuroda S. (2005) Biochem. Biophys. Res. Commun. 327, 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zugaza J. L., Waldron R. T., Sinnett-Smith J., Rozengurt E. (1997) J. Biol. Chem. 272, 23952–23960 [DOI] [PubMed] [Google Scholar]
- 43.Sinnett-Smith J., Jacamo R., Kui R., Wang Y. M., Young S. H., Rey O., Waldron R. T., Rozengurt E. (2009) J. Biol. Chem. 284, 13434–13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sánchez-Ruiloba L., Cabrera-Poch N., Rodríguez-Martínez M., López-Menéndez C., Jean-Mairet R. M., Higuero A. M., Iglesias T. (2006) J. Biol. Chem. 281, 18888–18900 [DOI] [PubMed] [Google Scholar]
- 45.Metzger J. M., Westfall M. V. (2004) Circ. Res. 94, 146–158 [DOI] [PubMed] [Google Scholar]
- 46.Kirk J. A., MacGowan G. A., Evans C., Smith S. H., Warren C. M., Mamidi R., Chandra M., Stewart A. F., Solaro R. J., Shroff S. G. (2009) Circ. Res. 105, 1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iglesias T., Waldron R. T., Rozengurt E. (1998) J. Biol. Chem. 273, 27662–27667 [DOI] [PubMed] [Google Scholar]
- 48.Storz P., Döppler H., Toker A. (2004) Mol. Cell. Biol. 24, 2614–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carnegie G. K., Smith F. D., McConnachie G., Langeberg L. K., Scott J. D. (2004) Mol. Cell 15, 889–899 [DOI] [PubMed] [Google Scholar]
- 50.Carnegie G. K., Soughayer J., Smith F. D., Pedroja B. S., Zhang F., Diviani D., Bristow M. R., Kunkel M. T., Newton A. C., Langeberg L. K., Scott J. D. (2008) Mol. Cell 32, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prestle J., Pfizenmaier K., Brenner J., Johannes F. J. (1996) J. Cell Biol. 134, 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rey O., Rozengurt E. (2001) Biochem. Biophys. Res. Commun. 287, 21–26 [DOI] [PubMed] [Google Scholar]
- 53.Liljedahl M., Maeda Y., Colanzi A., Ayala I., Van Lint J., Malhotra V. (2001) Cell 104, 409–420 [DOI] [PubMed] [Google Scholar]
- 54.Jamora C., Yamanouye N., Van Lint J., Laudenslager J., Vandenheede J. R., Faulkner D. J., Malhotra V. (1999) Cell 98, 59–68 [DOI] [PubMed] [Google Scholar]
- 55.Spitaler M., Emslie E., Wood C. D., Cantrell D. (2006) Immunity 24, 535–546 [DOI] [PubMed] [Google Scholar]
- 56.Matthews S. A., Iglesias T., Rozengurt E., Cantrell D. (2000) EMBO J. 19, 2935–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rey O., Young S. H., Cantrell D., Rozengurt E. (2001) J. Biol. Chem. 276, 32616–32626 [DOI] [PubMed] [Google Scholar]
- 58.Oancea E., Bezzerides V. J., Greka A., Clapham D. E. (2003) Dev. Cell 4, 561–574 [DOI] [PubMed] [Google Scholar]
- 59.Liu Y., Contreras M., Shen T., Randall W. R., Schneider M. F. (2009) J. Physiol. 587, 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.