Abstract
CC chemokine receptor 5 (CCR5), the major HIV coreceptor, is a G protein-coupled receptor (GPCR) involved in cell activation and migration in response to chemokines. Blockade of CCR5 is an effective anti-HIV strategy, and potent anti-HIV chemokine analogs such as PSC-RANTES have been developed. These inhibitors act by interfering with receptor trafficking, thereby inducing prolonged intracellular sequestration of CCR5. Like many GPCRs, CCR5 is desensitized following agonist activation. The initial steps in this process are well understood, but later stages, including where CCR5 is sequestered during desensitization, and how anti-HIV chemokine analogs intervene to achieve prolonged sequestration, have yet to be elucidated in detail. In this study we demonstrate that CCR5 cycles to and from the cell surface via the endosome recycling compartment and the trans-Golgi network during desensitization, accumulating in the trans-Golgi network following internalization by both PSC-RANTES and CCL5, the native ligand from which it was derived. In addition, we show that unlike CCR5 sequestered by CCL5, CCR5 sequestered by PSC-RANTES cannot be induced to return to the cell surface by addition of the small molecule CCR5 inhibitor, TAK-779, and that association of PSC-RANTES with CCR5 is more durable than that of native CCL5 during desensitization. Our findings reconcile the previously conflicting descriptions of the location of sequestered CCR5 during desensitization, as well as providing more general insights into potential trafficking routes for endocytosed GPCRs and further elucidation of the unusual inhibitory mechanism of chemokine analogs with potent anti-HIV activity.
Keywords: Antiviral Agents, Chemokines, Endocytosis, G Protein-coupled Receptor, HIV, Intracellular Trafficking, Receptor Desensitization, Receptor Recycling, PSC-RANTES, Trans-Golgi Network
Introduction
Receptor down-modulation, i.e. reduction of cell surface receptor concentration, is a component of the well characterized desensitization/resensitization process that is seen across the G protein-coupled receptor (GPCR)2 superfamily (1, 2). Following endocytosis, GPCRs are either routed for degradation or recycled back to the cell surface in a resensitized form (2). Many details of this sorting process are currently unknown, however, and its study represents an emerging field in GPCR biology (1).
The chemokine receptor CCR5 is a GPCR expressed predominantly on leukocyte subsets, with a principal physiological role in the recruitment of effector cells to sites of inflammation (3). CCR5 also acts as an HIV coreceptor and plays a key role both in HIV transmission from person to person and in disease progression once infection has occurred (3). As a consequence, CCR5 is a key target for anti-HIV strategies directed toward both prevention and therapy (4).
The physiological ligands of CCR5, CCL5, CCL3, and CCL4, have intrinsic anti-HIV activity (5), resulting from both steric blockade of coreceptor interaction with the HIV envelope glycoprotein complex (6) and induction of CCR5 down-modulation (7). N-terminally modified analogs of CCR5 ligands, including AOP-RANTES (8) and PSC-RANTES (9), exhibit much higher anti-HIV activity than the physiological ligands. Although it is clear that the enhanced potency of these analogs is due to induction of stronger and more prolonged CCR5 down-modulation (9, 10), the details of this unusual inhibitory mechanism have yet to be fully elucidated.
The early steps of CCR5 desensitization, which have been quite extensively characterized (for review, see Ref. 11), involve processes common to many GPCRs: C-terminal phosphorylation, association with β-arrestin proteins, and clathrin-dependent endocytosis. The subsequent steps are less clearly understood, however. First, although it has been established that CCR5 accumulates in a perinuclear region during desensitization, several different locations for the site of accumulation have been proposed: the early endosome (12), the endosomal recycling compartment (ERC) (10, 13–15), and the Golgi apparatus (16, 17). Second, it has been demonstrated that during desensitization the pool of sequestered CCR5 receptors remains accessible to the cell surface by continually cycling to and from the plasma membrane (13, 15), but little is currently known about how the receptor resensitization process is triggered. Finally, although some progress has been made (10, 13, 18), the mechanism by which anti-HIV chemokine analogs interfere with the desensitization process to achieve prolonged receptor down-modulation has yet to be elucidated in detail.
In this study we used both native chemokine ligands and anti-HIV chemokine analogs to study CCR5 trafficking in response to ligands. Through a series of experiments using Chinese hamster ovary (CHO)-CCR5 cell lines and primary T lymphocytes, we present data that help explain the differences seen in previous work on the location of sequestered CCR5 during desensitization, as well as refining the existing mechanistic model to explain physiological CCR5 trafficking during desensitization and how it is disrupted by anti-HIV chemokine analogs.
EXPERIMENTAL PROCEDURES
Antibodies
Primary antibodies used in this study are described in supplemental Table 1. For immunofluorescence experiments, secondary antibodies (all used at 1:100 dilution) were Alexa Fluor 488-conjugated donkey anti-mouse and donkey anti-goat antibodies (both from Molecular Probes); and rhodamine (TRITC)-conjugated AffiniPure donkey anti-mouse IgG antibody, donkey anti-rabbit IgG, and donkey anti-sheep IgG antibodies (all from Jackson Immunoresearch Laboratories).
Chemokines
Chemokines (CCL5 and CCL4) and chemokine analogs (AOP-RANTES, PSC-RANTES) used in this study were prepared by total chemical synthesis as described previously (9).
Cy5-labeled Chemokines
Aminooxyacetyl-functionalized Cy5 was prepared as already described (19). Chemokines were prepared as above except that the C terminus was elongated with a K(S)G peptide to yield, by very mild periodate oxidation after folding of the protein, a glyoxylyl functionality for site-specific attachment by oximation of the aminooxyfluorophore. The presence of an N-terminal serine in native CCL5 necessitated the use of a transient protecting group, 2-(methylsulfonyl)ethyl-carbonate, which was (i) appended to the N-terminal fragment during synthesis by acylation of the t-butoxycarbonyl-deprotected fragment with the succinimide derivative of 2-(methylsulfonyl)ethyl-carbonate and (ii) cleaved after periodate oxidation of the folded protein as described previously (20). In both cases, the oxidized protein was reacted overnight with a 4-fold excess of aminooxyacetyl-Lys(Cy5) in a 0.1 m sodium acetate buffer containing 40% acetonitrile, pH 4.6. The Cy5-chemokines were characterized for authenticity by MALDI-TOF and shown to have indistinguishable activity from the parent chemokines in an HIV envelope-dependent cell fusion assay (9) (data not shown).
Reagents
Nocodazole, cycloheximide, and saponin were purchased from Sigma; dextran-rhodamine and rhodamine-transferrin (Tf) were obtained from Molecular Probes.
Cell Lines
The CHO-CCR5 cells used in this study have been described previously (9, 21). Cell lines were cultured at 37 °C with 5% CO2 in F12 nutrient mixture supplemented with 5% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin. All cell culture reagents were obtained from Invitrogen.
Preparation of Primary T Lymphocytes
Peripheral blood mononuclear cells were isolated from buffy coat preparations (supplied by the blood transfusion center, Geneva Hospital) according to the institutional guidelines of the ethical committee of the University of Geneva. Buffy coat preparations diluted 1:2 with phosphate-buffered saline (PBS) were gently laid onto 15-ml Ficoll-Hypaque (Amersham Biosciences) and centrifuged at 650 × g for 20 min at 20 °C. The mononuclear cell layer was extracted and washed with PBS and PBS-EDTA (1 mm) to remove platelets. Peripheral blood mononuclear cells were then activated by a 3-day exposure to phytohemagglutinin (5 μg/ml; Sigma) and cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). Nonadherent cells were resuspended in fresh medium supplemented with 20 ng/ml recombinant human interleukin-2 (IL-2) (Peprotech). Activated primary T lymphocytes were used for CCR5 experiments after 12–14 days of activation, a time chosen to maximize baseline CCR5 expression. Cell preparations were routinely quality controlled by flow cytometry analysis (CD3/CD4/CD8 labeling).
Immunofluorescence Laser Scanning Confocal Microscopy
For confocal immunofluorescence microscopy, cells were grown on glass coverslips (Menzel-Gläser), fixed for either 4 min in methanol at −20 °C or for 20 min in 3% fresh paraformaldehyde (Sigma) in PBS. Paraformaldehyde-fixed cells were washed in PBS and incubated with 50 mm ammonium chloride (Merck) for 15 min to quench free aldehydes. Permeabilization was performed by incubating cells with primary antibody for 45 min in the presence of 0.1% saponin (Sigma). Cells were then washed and incubated for 30 min with secondary antibody. Cells were then washed again and mounted over MOWIOL 4–88 (Calbiochem). Images were captured using a Zeiss LSM510 confocal laser-scanning microscope with an attached Zeiss inverted epifluorescence microscope under a Plan Apo BL ×63/1.4 numeric aperture oil-immersion objective. Fluorophores were excited using the 488-nm line of a Kr/Ar laser (Alexa Fluor 488), and the 543-nm line of a He/Ne laser (TRITC).
For time course studies, CHO-CCR5 cells were washed and cultured with medium without FCS for 120 min at 37 °C. For TGN38 and Lamp1 labeling, cells were preincubated in medium containing 0.2% BSA with 5 μg/ml 3A9 antibody for 45 min at 4 °C. Cells were washed and treated for different periods of time (0, 10, 20, 30, and 60 min) with 100 nm PSC-RANTES at 37 °C or 100 nm CCL5. For each time, cells were fixed in 3% fresh paraformaldehyde (Sigma) in PBS for 20 min and processed for immunofluorescence as above. For rhodamine-Tf labeling, cells were preincubated in medium containing 0.2% BSA with 50 μg/ml rhodamine-Tf for 30 min at 37 °C, then processed as with TGN38 and Lamp1 labeling except that anti-CCR5 antibody staining was carried out in the presence of 50 μg/ml rhodamine-Tf.
Image Analysis
Deconvolution is a computationally intensive image processing technique to improve signal-to-noise ratio and image resolution obtained by laser scanning confocal microscopy. Huygens Essential is an image processing software package tailored for deconvolution of microscopy images. Stacks were captured with a 0.37-μm z step and then restored with Huygens Essential software (Scientific Volume Imaging, Hilversum, The Netherlands), usually requiring 40 iterations of deconvolution and a maximum likelihood estimation algorithm. The deconvoluted images were compiled to produce maximum intensity projections using Imaris software (Bitplane AG, Zürich, Switzerland).
Western Blot Analysis
2 × 106 CHO-CCR5 cells were incubated for 120 min in the presence or absence of chemokines. Cells were then washed extensively with PBS and treated in lysis buffer (1% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl, pH 7.5) containing protease inhibitors (Roche). Samples were precleared by a 2-h incubation with a rabbit IgG anti-mouse IgG (DAKO) preadsorbed on protein G-Sepharose CL4B beads (Amersham Biosciences). The immunoadsorbates were removed by centrifugation (1200 × g, 5 min, using a table-top Eppendorf centrifuge) and the precleared supernatants incubated overnight with C20 antibody. Finally, the supernatants were incubated for 1 h with 50 μl of a 50% suspension of protein G-Sepharose beads. The resulting immunoadsorbates were collected by centrifugation and washed three times with lysis buffer. The washed beads were resuspended in reducing SDS-PAGE sample buffer, heated at 95 °C for 4 min, and subjected to SDS-PAGE in 18% Tris-glycine gels (Invitrogen). Gels were transferred to Immobilon-P membranes (Millipore), which were blocked with PBS supplemented with 5% (w/v) nonfat dried milk (Applichem) and 0.1% (w:v) Tween 20 (Sigma-Aldrich) for 90 min. Subsequently, the membranes were incubated for 90 min with biotinylated anti-human CCL5 antibody (BAF 278; R&D Systems), washed, and incubated with horseradish peroxidase-conjugated streptavidin (R&D Systems). Detection was performed using enhanced chemiluminescence (Pierce).
RESULTS
Validation of the Experimental System
In initial work we established that receptor down-modulation in the CHO-CCR5 cell line used in this study occurs to an extent comparable with that observed in previous studies by ourselves and others and with similar kinetics (9, 10, 21, 22). For both CCL5 and PSC-RANTES, plateau levels of CCR5 down-modulation are reached after 120-min treatment with 100 nm ligand (supplemental Fig. S1A). Additionally, in agreement with previously published work, we confirmed (i) that surface CCR5 down-modulation correlates with accumulation in a perinuclear region (10, 12–17) (supplemental Fig. S1B), (ii) that sequestered CCR5 does not accumulate in the degradative pathway (10) (supplemental Fig. S2), and (iii) that during desensitization CCR5 is mobile and accessible to the cell surface (13, 15) (supplemental Fig. S3).
CCR5 Sequestration: Excluding the Golgi Apparatus and ERC
To address the differences between previously published descriptions of the site of intracellular sequestration of CCR5 during desensitization (10, 13–17), we carried out a series of colocalization immunofluorescence experiments on cells treated under conditions (120-min incubation with 100 nm PSC-RANTES) that achieve plateau levels of CCR5 down-modulation (supplemental Fig. S1A).
In a first set of experiments, PSC-RANTES-treated CHO-CCR5 cells were fixed, permeabilized, and colabeled for CCR5 together with the Golgi markers CTR433 (medial/trans-Golgi) and GM130 (cis-Golgi). Colocalization with both markers was suggested in an initial experiment (Fig. 1, A and B). However, we were concerned about the potential for false positive results to arise due to a combination of (i) clustering of different subcellular compartments in the perinuclear region and (ii) the resolution limits of confocal microscopy, and we opted to carry out a follow-up experiment in which cells treated with PSC-RANTES were subsequently incubated with nocodazole (10 μm, 60 min at 37 °C) prior to fixing and staining. Nocodazole has the effect of dispersing the ensemble of perinuclear compartments (endosomes, TGN, and Golgi membranes) throughout the cytoplasm (23). Importantly, although the CCR5 and Golgi marker signals appear to codisperse after nocodazole treatment, they clearly do not overlap (Fig. 1, C and D). Similar results were obtained when cells were treated with the native ligand, CCL5 (data not shown). Hence, our observations are not consistent with sequestration of CCR5 in the Golgi apparatus during desensitization, as proposed previously (16, 17).
FIGURE 1.
CCR5 sequestered by PSC-RANTES is not associated with the Golgi apparatus. Single confocal sections (midsection of cells) are shown. Scale bars, 10 μm. Cells were incubated for 120 min with 100 nm PSC-RANTES at 37 °C and then fixed and costained with anti-CCR5 antibodies (C20; green) together with antibodies against the Golgi markers GM130 (A and B) and CTR433 (C and D). B and D, PSC-RANTES-treated cells were incubated with nocodazole (10 mm, 60 min, 37 °C) prior to fixing and staining. The images shown are representative of at least three independent experiments.
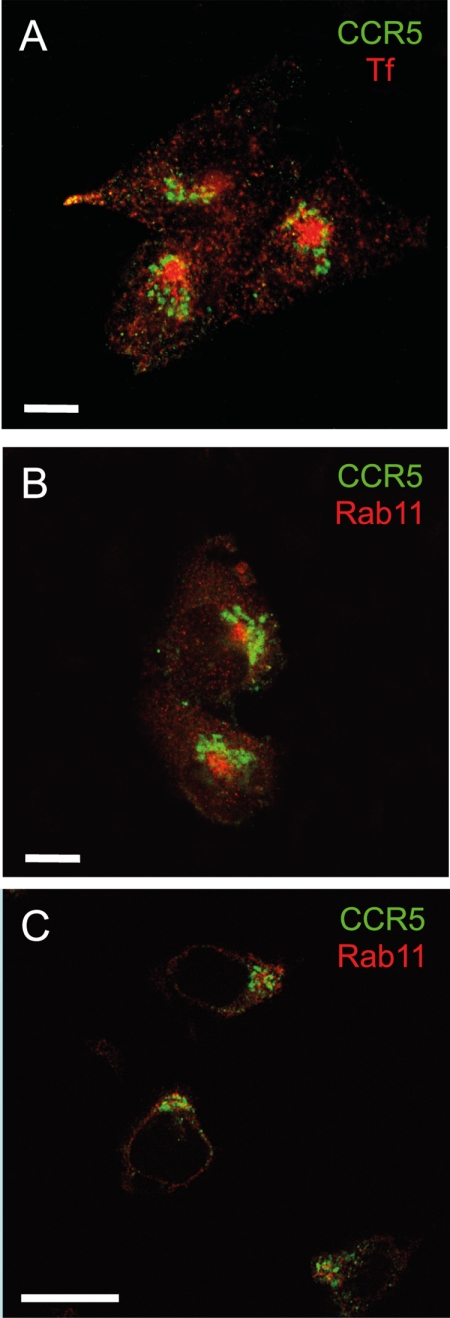
In a second series of experiments, PSC-RANTES-treated CHO-CCR5 cells were fixed, permeabilized, and labeled with CCR5 together with two markers for the ERC: anti-Rab11 and rhodamine-conjugated Tf. No significant colocalization between CCR5 and the ERC was detected under these conditions (Fig. 2, A and B). Similar results were obtained with human primary T cells treated with PSC-RANTES and costained for CCR5 and Rab11 (Fig. 2C), when CHO-CCR5 cells were treated with AOP-RANTES, CCL4, or CCL5 (supplemental Fig. S4, A–C) and when human primary T cells were treated with CCL5 and costained for CCR5 and Rab11 (supplemental Fig. S4D). Hence, our observations are not consistent with the ERC as the sequestration site for CCR5 during desensitiztaion as reported previously (10, 13–15).
FIGURE 2.
Ligand-sequestered CCR5 is not associated with the ERC. Single confocal sections are shown; for CHO-CCR5 cells these correspond to a z plane above the nucleus. Scale bars, 10 μm. A, CHO-CCR5 cells were treated with 100 nm PSC-RANTES for 120 min at 37 °C, pulsed for 10 min with rhodamine-Tf at 4 °C, and then incubated for a further 15 min at 37 °C in the presence of rhodamine-Tf (red) prior to fixing and staining with anti-CCR5 antibody (3A9; green). B, CHO-CCR5 cells were treated with 100 nm PSC-RANTES for 120 min at 37 °C and then fixed and costained with anti-CCR5 antibody (3A9; green) together with anti-Rab11 antibody (red). C, human T lymphocytes were treated with 100 nm PSC-RANTES for 120 min at 37 °C and then fixed and co-stained with anti-CCR5 antibody (3A9; green) together with anti-Rab11 antibody (red). The images shown are representative of at least three independent experiments.
Sequestered CCR5 Accumulates in the TGN
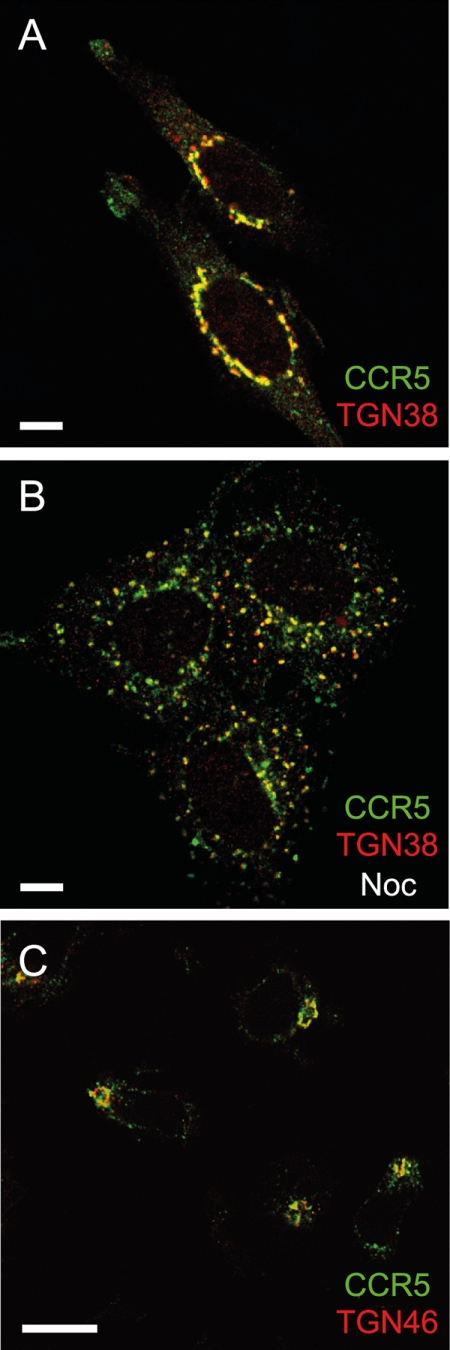
We next investigated the TGN as a potential site of perinuclear CCR5 sequestration during desensitization. PSC-RANTES-treated (100 nm, 120 min) CHO-CCR5 cells were fixed, permeabilized, and stained for CCR5 together with the TGN marker TGN38. Costaining revealed high levels of colocalization (Fig. 3A), which, importantly, were maintained after nocodazole treatment (Fig. 3B). Similar results were obtained when syntaxin-6 was used as TGN marker (data not shown). Importantly, similar results were obtained in costaining experiments using TGN46, the human ortholog of TGN38, to label the TGN of human primary T cells treated with PSC-RANTES (Fig. 3C). Colocalization of ligand-sequestered CCR5 with TGN markers was also apparent when CHO-CCR5 cells were treated with CCL4, CCL5, or AOP-RANTES (supplemental Fig. S5) and when human T cells were treated with CCL5 (supplemental Fig. S5).
FIGURE 3.
Ligand-sequestered CCR5 accumulates in the TGN in CHO-CCR5 cells and in primary T lymphocytes. Single confocal sections (midsection of cells) are shown. Scale bars, 10 μm. A and B, CHO-CCR5 cells were incubated for 120 min with 100 nm PSC-RANTES at 37 °C and then fixed and stained with anti-CCR5 antibody (3A9; green) together with anti-TGN38 (red). B, cells were treated with nocodazole (10 μm, 60 min, 37 °C) following incubation with PSC-RANTES and prior to fixing and staining. C, phytohemagglutinin/IL-2-activated human lymphocytes were incubated for 120 min with PSC-RANTES (100 nm, 37 °C) and then fixed and stained with antibodies recognizing CCR5 (3A9; green) and TGN46 (red). The images shown are representative of at least three independent experiments.
Together, these results indicate that the TGN is the major site of CCR5 sequestration during desensitization induced by both native ligands and potent anti-HIV chemokine analogs. Definitive identification of the TGN as the sequestration site would require ultrastructural studies such as coimmunogold labeling electron microscopy. Unfortunately, despite an exhaustive screen of all suitable and available antibodies, we were unable to establish experimental conditions necessary to obtain the required data.3
Kinetics and Route Taken into the TGN
In an attempt to elucidate the route taken by CCR5 to its sequestration site, we next performed time course experiments to follow CCR5 trafficking after a ligand pulse, colabeling the receptor with markers of either the TGN (TGN38), the ERC (rhodamine-Tf) or the degradative pathway (Lamp1). Having noted in preliminary experiments that the ERC and TGN compartments in CHO cells are located predominantly on different z axis planes (supplemental Fig. S6), we opted to make use of maximum intensity projections of cells obtained from deconvoluted stacks of confocal images in this work.
CHO-CCR5 cells were preloaded with 3A9 anti-CCR5 antibody at 4 °C, and as expected, CCR5 staining was localized to the plasma membrane at time zero (Fig. 4). The chase phase was initiated by raising the incubation temperature to 37 °C in the presence of PSC-RANTES or CCL5 (100 nm). Ten min later, CCR5 staining had moved inward and become apparent in punctate structures at the periphery of the cell. After 20 min, the majority of CCR5 had accumulated in the Tf-positive ERC. This accumulation was transient, however, with the bulk of CCR5 apparent in the TGN38-positive region from 30 min after the ligand pulse, where it remained up to the longest time point tested (60 min). Indistinguishable results (Fig. 4) were obtained when cells were pulsed with native CCL5, suggesting that both native chemokines and RANTES analogs induce trafficking of internalized CCR5 via the same route and with similar kinetics. We did not detect significant association of CCR5 with Lamp1 at any time point (Fig. 4), indicating that a route to the TGN involving passage through the degradative pathway is unlikely.
FIGURE 4.
Ligand-sequestered CCR5 passes through the ERC before accumulating in the TGN. Cell surface CCR5 on CHO-CCR5 cells was pulse-labeled by preincubation with 3A9 antibody (green) for 30 min at 4 °C. Cells were then treated with either CCL5 or PSC-RANTES (100 nm). At the indicated times, cells were immediately washed and fixed prior to revelation. Rhodamine-Tf, TGN38, and LAMP-1 (both red) were used to label the recycling endosome, the TGN, and lysosomes, respectively. Maximum intensity projections from stacks of deconvoluted confocal images are shown. Scale bars, 10 μm. The images shown are representative of at least three independent experiments.
CCR5 Sequestered by CCL5, but Not by PSC-RANTES, Can Be Resensitized by Pulsing Cells with TAK-779
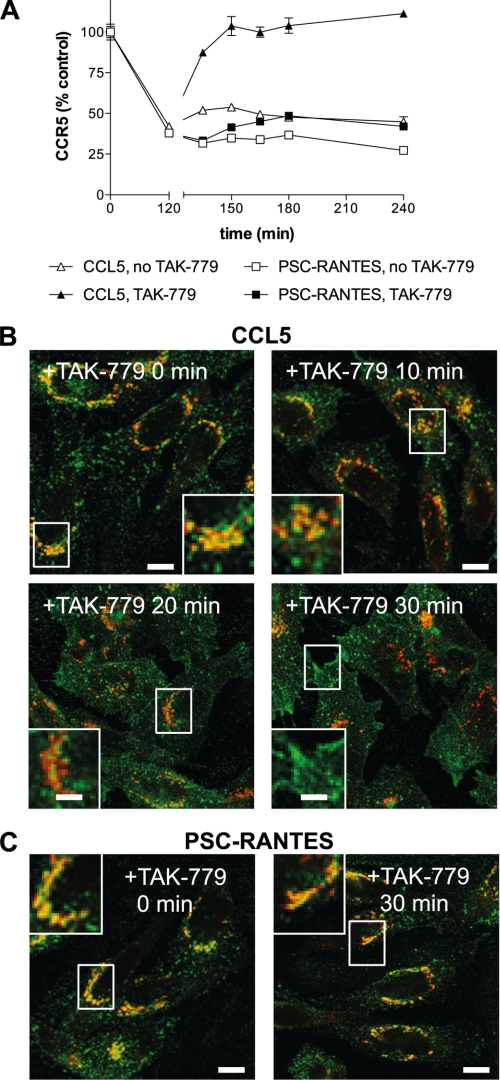
In previous work it has been shown that the pool of CCR5 sequestered following treatment with CCL5 can be rapidly relocalized to the plasma membrane by exposing cells to the small molecule CCR5 inhibitor TAK-779 (15). We carried out similar experiments, pretreating cells with either CCL5 or PSC-RANTES (37 °C, 100 nm, 120 min) to induce receptor sequestration, then washing with medium prior to exposure to TAK-779 (400 nm). Cells treated in this way were analyzed at time points both by flow cytometry to quantify cell surface levels of CCR5 (Fig. 5A) and by immunofluorescence microscopy to localize internalized receptor (Fig. 5B). In agreement with the results of Signoret et al. (15), we found that within 30 min of treatment with TAK-779, CCR5 sequestered by CCL5 was completely relocalized to the plasma membrane (Fig. 5A). During this time period, CCR5 can be seen leaving the TGN38-positive perinuclear zone, apparently passing through punctate cytoplasmic structures en route to the plasma membrane (Fig. 5B). Interestingly, TAK-779 treatment, either at 400 nm (Fig. 5A) or at higher concentrations up to 40 μm (data not shown), did not cause relocalization to the plasma membrane of CCR5 sequestered by PSC-RANTES; the receptor remained sequestered in the TGN38-positive perinuclear zone (Fig. 5C). Only receptors sequestered by the native ligand but not those sequestered by PSC-RANTES can be induced to relocalize to the cell surface by extracellular addition of the small molecule inhibitor.
FIGURE 5.
TAK-779 induces rapid plasma membrane relocalization of CCR5 internalized by CCL5 but not by PSC-RANTES. CHO-CCR5 cells were treated with 100 nm PSC-RANTES or 100 nm CCL5 for 120 min at 37 °C to internalize CCR5. Cells were then washed and incubated in the presence or absence of TAK-779 (400 nm) at 37 °C. A, at the indicated times, the cells were washed at 4 °C, stained with phycoerythrin-conjugated 3A9, and analyzed by flow cytometry. Data shown correspond to mean cell surface CCR5 (n = 3; error bars, S.E.) expressed as a percentage of control levels and are representative of three independent experiments. B and C, at the indicated times, cells were washed, fixed, and labeled with anti-CCR5 (3A9; green) and anti-TGN38 (red). Maximum intensity projections from stacks of deconvoluted confocal images are shown. Scale bars, 10 μm. The images shown are representative of two independent experiments.
CCL5 Is Removed from the Cell during Desensitization, but PSC-RANTES Remains Colocalized with CCR5
In an attempt to investigate why CCR5 sequestered by native CCL5 is sensitive to resensitization by TAK-779 while CCR5 sequestered by PSC-RANTES is not, we used Cy5-labeled chemokines to study the fate of the ligand during receptor sequestration. As expected, (i) following incubation for 30 min at 4 °C, both CCL5-Cy5 and PSC-RANTES-Cy5 (200 nm) efficiently labeled the surface of CHO-CCR5 cells (Fig. 6, A and B) and (ii) when cells were then subjected to a 120-min incubation at 37 °C to allow receptor internalization, both native CCL5-Cy5 and PSC-RANTES-Cy5 induced perinuclear sequestration of CCR5 (Fig. 6, C–F). However, although the fluorescence signal for the PSC-RANTES-Cy5 persisted in the same perinuclear location as the sequestered CCR5 120 min after ligand addition (Fig. 6, D and F), the fluorescence signal for the CCL5-Cy5 was almost completely absent (Fig. 6, C and E). To eliminate the possibility that loss of the fluorescence signal was due to cleavage of the C-terminal fluorescent label, we carried out a subsequent experiment in which, following treatment of CHO-CCR5 cells with native CCL5 or PSC-RANTES, cells were lysed and immunoprecipitated with an anti-CCR5 antibody, and the precipitates were subjected to Western blot with a polyclonal anti-CCL5 antibody capable of recognizing both native CCL5 and PSC-RANTES (Fig. 6G). Although the CCR5 immunoprecipitate from cells treated with PSC-RANTES gave a strong signal corresponding to a protein of the expected size, no signal was obtained from the CCR5 immunoprecipitate of cells treated with native CCL5. Hence the loss in CCL5-Cy5 fluorescence signal seen in the immunofluorescence experiment relates to degradation and/or dissociation of the ligand from CCR5 rather than truncation of the C-terminal fluorochrome.
FIGURE 6.
PSC-RANTES, but not native CCL5, remains intact and associated with CCR5 during desensitization. A–F, CHO-CCR5 cells were incubated for 30 min at 4 °C with either 200 nm Cy5-CCL5 (blue; A, C, and E) or 200 nm Cy5-PSC-RANTES (blue; B, D, and F). Cells were either fixed immediately and stained with anti-TGN38 antibody (red; A and B) or washed extensively and incubated for 120 min in the absence of chemokines, then fixed and stained with either anti-TGN38 antibody (red; C and D) or anti-CCR5 antibody 3A9 (red; E and F). G, CHO-CCR5 cells were incubated for 120 min at 37 °C with medium alone (lane 1), 100 nm PSC-RANTES (lane 2), or 100 nm CCL5 (lane 3) and then lysed. Total cell lysates were immunoprecipitated using anti-CCR5 antibody, and immunoprecipitates were subjected to Western blotting using an anti-CCL5 antibody. The images shown are representative of two independent experiments.
DISCUSSION
Studies of CCR5 trafficking have led to the establishment of a mechanistic model (11, 13, 24) in which agonist-activated CCR5 is internalized through clathrin-coated pits and delivered to a perinuclear compartment from where it is subsequently recycled to the plasma membrane in a resensitized form. Although there is general agreement about the steps involved in receptor internalization, details of the sequestration and resensitization processes and how they relate to the inhibitory mechanisms of potent anti-HIV chemokine analogs have yet to be elucidated.
Reconciling Differences in Current Models for CCR5 Trafficking
Past work has led to conflicting proposals for the site of sequestration during desensitization (10, 12–14, 16, 17). In this study we conducted a thorough investigation of CCR5 desensitization using both physiological chemokines and anti-HIV chemokine analogs as ligands. Like the previous studies of CCR5 trafficking, our work centered on immunofluorescence microscopy with CCR5 colabeled with markers for different subcellular compartments. However, important differences included our use, where required, (i) of nocodazole treatment to eliminate apparent colocalization due to close clustering of compartments in the perinuclear region; (ii) of maximum intensity projections compiled from stacks of deconvoluted confocal images enabling us to take account of colocalization the whole of the cell rather than in individual z axis planes; (iii) of pulse-chase studies enabling us to follow the intracellular localization of CCR5 as trafficking progresses rather than at a single time point.
Adopting this “bigger picture” approach allowed us to make observations that could explain and reconcile the different results obtained in previous studies of the site of CCR5 sequestration during desensitization. Our experiments show that CCR5 passes through the ERC during internalization at a time point ∼20 min following ligand engagement (Fig. 4). This observation could explain the reported colocalization between sequestered CCR5 and ERC markers (10, 13, 14). However, CCR5 subsequently moves out of the ERC en route to the TGN (Fig. 4). The close proximity of the TGN to the Golgi in the perinuclear region of the cell could account for the reported colocalization of internalized CCR5 with markers of the Golgi region (16, 17). We were able to exclude the Golgi region by performing colocalization studies with Golgi markers following nocodazole treatment (Fig. 1). Interestingly, the opposed, closely associated CCR5 and GM130 labeling that we observed is reminiscent of the labeling pattern seen by Neubrand et al. (25) in Golgi ministacks in cells treated with nocodazole prior to labeling with GM130 and p230, a marker of the TGN.
Retrograde Sequestration, from the Cell Surface to the TGN via the ERC
Our study is the first to implicate the TGN as the sequestration site for CCR5 during desensitization. Such a location for an internalized GPCR would be unusual but not be without precedent because internalized somatostatin type 2 receptors have recently been shown to traffic to the TGN prior to recycling back to the plasma membrane in neurons (26, 27). There is accumulating evidence to suggest the existence of connecting routes between the endocytic and the biosynthetic/secretory pathways, and several proteins have been shown to be trafficked from the cell surface via early endosomes to the TGN (28). Two different retrograde pathways have been implicated, the first involving transport via the ERC, exemplified by TGN38/46 (29) and Shiga B toxin (30), and the second involving transport to late endosomes, exemplified by the mannose 6-phosphate receptor (31). Our results (Fig. 4) indicate that the former pathway (i.e. via the ERC rather than late endosomes) is the route taken by CCR5.
GPCRs are currently divided into two classes according to their fate following internalization (2). Receptors of the first group are targeted to lysosomes, with or without ligand activation, leading to proteolytic degradation. Receptors of the second group dissociate rapidly from their ligands following endocytosis and are recycled back to the cell surface in a resensitized form via the ERC. Our results, together with those describing trafficking of the somatostatin type 2 receptor (26, 27), provide evidence for the existence of a third pathway, in which GPCRs destined for recycling take a retrograde trafficking pathway leading to sequestration within the TGN.
CCR5 Sequestration: Cycling Receptors Passing through a Molecular Bottleneck
Previously studies have indicated that CCR5 sequestered by both native ligands and RANTES analogs is mobile and accessible to the cell surface (13, 15), and our work confirms that this is also the case for CCR5 sequestered by PSC-RANTES (supplemental Fig. S3). Building on the existing model for CCR5 trafficking during desensitization (13, 15) and incorporating our new results (Figs. 4 and 5), we propose that CCR5 follows a trafficking circuit that takes the receptor from the cell surface to the ERC via clathrin-mediated endocytosis (24), from the ERC to the TGN (Fig. 4), and then from the TGN back to the cell surface (Fig. 5). When cells are at rest, the slowest section of the circuit is receptor endocytosis, and so receptors accumulate at the cell surface. When agonists engage CCR5, the rate of clathrin-dependent endocytosis is increased, and the slowest section in the circuit becomes transport from the TGN to the cell surface, resulting in receptor accumulation in the TGN (Fig. 3). Addition of TAK-779, an allosteric CCR5 inhibitor, causes redistribution of CCR5 to the cell surface, presumably capturing receptors returning to the cell surface and inducing them to adopt an inactive conformation so that receptor endocytosis reverts to being the slowest section in the circuit.
Implications for the Inhibitory Mechanism of Anti-HIV Chemokine Analogs
Potent anti-HIV chemokine analogs such as AOP-RANTES (8) and PSC-RANTES (9) owe their potent anti-HIV activity to their capacity to induce more profound and more prolonged intracellular CCR5 sequestration than the native ligands from which they are derived (9, 10, 18). The results presented in this study provide new insights into this unusual inhibitory mechanism. First, our time course experiment in which cells were pulsed with either PSC-RANTES or CCL5 (Fig. 4) provides confirmation that, in agreement with previously published work (13), the enhanced CCR5 sequestration achieved by analogs does not occur because they induce more rapid receptor internalization than the native ligands. Second, the CCR5 colocalization experiments that we performed on cells treated with either chemokine analogs or physiological chemokines indicate that enhanced sequestration is not a consequence of CCR5 sequestered by analogs being directed to a subcellular compartment different from that used for CCR5 sequestered by native ligands (Figs. 1 and 2 and supplemental Figs. S4 and S5). Instead, our results suggest that enhanced sequestration occurs because analogs have the capacity to achieve longer duration occupancy of CCR5 during the desensitization process (Figs. 5 and 6).
The simplest explanation for the prolonged association between PSC-RANTES and CCR5 is that the analog binds more tightly to CCR5 than the native chemokine, although no significant differences in CCR5 binding affinity between native CCL5 and PSC-RANTES have been seen in previous studies (9, 22). Among other possible explanations are (i) that PSC-RANTES may be more resistant than native CCL5 to enzymatic processes postulated to be required for ligand-receptor dissociation (15) or (ii) that as a superagonist (22), PSC-RANTES is capable of “locking” CCR5 into a conformation that prevents ligand-receptor dissociation thereby blocking resensitization. Further investigation of this phenomenon may help explain the molecular and cellular details of the inhibitory mechanism of RANTES analogs and aid in the elaboration of new strategies to discover further optimized anti-HIV molecules that act via blockade of CCR5.
Supplementary Material
Acknowledgments
We thank P. Cosson, J. Gruenberg, V. Mercanti, and M. Marsh for critical comments on the manuscript; M. Bornens, B. Eipper, B. Hoflack, J. Gruenberg, and S. Méresse for provision of antibodies; G. Porcheron-Berthet for technical assistance, and O. Brun for expert advice on confocal microscopy.
This work was supported by grants from the Swiss National Science Foundation (3100A0-110042 to O. H.; 310000-120280/1 to M. F.), NIH/NIAID (U19 AI076981), and the Swedish International Development Agency (7500037313 to O. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. S1–S6, and additional references.
M. Foti, unpublished results.
- GPCR
- G protein-coupled receptor
- CCR5
- CC chemokine receptor 5
- CCL
- CC chemokine ligand
- CHO
- Chinese hamster ovary
- ERC
- endosome recycling compartment
- Tf
- transferrin
- TGN
- trans-Golgi network
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 2.Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lederman M. M. (1995) Ann. Intern. Med. 122, 218–222 [DOI] [PubMed] [Google Scholar]
- 4.Kuhmann S. E., Hartley O. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 425–461 [DOI] [PubMed] [Google Scholar]
- 5.Cocchi F., DeVico A. L., Garzino-Demo A., Arya S. K., Gallo R. C., Lusso P. (1995) Science 270, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 6.Trkola A., Paxton W. A., Monard S. P., Hoxie J. A., Siani M. A., Thompson D. A., Wu L., Mackay C. R., Horuk R., Moore J. P. (1998) J. Virol. 72, 396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhatib G., Locati M., Kennedy P. E., Murphy P. M., Berger E. A. (1997) Virology 234, 340–348 [DOI] [PubMed] [Google Scholar]
- 8.Simmons G., Clapham P. R., Picard L., Offord R. E., Rosenkilde M. M., Schwartz T. W., Buser R., Wells T. N., Proudfoot A. E. (1997) Science 276, 276–279 [DOI] [PubMed] [Google Scholar]
- 9.Hartley O., Gaertner H., Wilken J., Thompson D., Fish R., Ramos A., Pastore C., Dufour B., Cerini F., Melotti A., Heveker N., Picard L., Alizon M., Mosier D., Kent S., Offord R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16460–16465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack M., Luckow B., Nelson P. J., Cihak J., Simmons G., Clapham P. R., Signoret N., Marsh M., Stangassinger M., Borlat F., Wells T. N., Schlöndorff D., Proudfoot A. E. (1998) J. Exp. Med. 187, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oppermann M. (2004) Cell. Signal. 16, 1201–1210 [DOI] [PubMed] [Google Scholar]
- 12.Amara A., Gall S. L., Schwartz O., Salamero J., Montes M., Loetscher P., Baggiolini M., Virelizier J. L., Arenzana-Seisdedos F. (1997) J. Exp. Med. 186, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signoret N., Pelchen-Matthews A., Mack M., Proudfoot A. E., Marsh M. (2000) J. Cell Biol. 151, 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollok-Kopp B., Schwarze K., Baradari V. K., Oppermann M. (2003) J. Biol. Chem. 278, 2190–2198 [DOI] [PubMed] [Google Scholar]
- 15.Signoret N., Christophe T., Oppermann M., Marsh M. (2004) Traffic 5, 529–543 [DOI] [PubMed] [Google Scholar]
- 16.Longden J., Cooke E. L., Hill S. J. (2008) Br. J. Pharmacol. 153, 1513–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiss D. L., Longden J., Fechner G. A., Avery V. M. (2009) Cell. Mol. Biol. Lett. 14, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastore C., Picchio G. R., Galimi F., Fish R., Hartley O., Offord R. E., Mosier D. E. (2003) Antimicrob. Agents Chemother. 47, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Offord R. E., Gaertner H. F., Wells T. N., Proudfoot A. E. (1997) Methods Enzymol. 287, 348–369 [DOI] [PubMed] [Google Scholar]
- 20.Gaertner H. F., Offord R. E., Cotton R., Timms D., Camble R., Rose K. (1994) J. Biol. Chem. 269, 7224–7230 [PubMed] [Google Scholar]
- 21.Gaertner H., Cerini F., Escola J. M., Kuenzi G., Melotti A., Offord R., Rossitto-Borlat I., Nedellec R., Salkowitz J., Gorochov G., Mosier D., Hartley O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17706–17711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaertner H., Lebeau O., Borlat I., Cerini F., Dufour B., Kuenzi G., Melotti A., Fish R. J., Offord R., Springael J. Y., Parmentier M., Hartley O. (2008) Protein Eng. Des. Sel. 21, 65–72 [DOI] [PubMed] [Google Scholar]
- 23.Sakai T., Yamashina S., Ohnishi S. (1991) J. Biochem. 109, 528–533 [DOI] [PubMed] [Google Scholar]
- 24.Signoret N., Hewlett L., Wavre S., Pelchen-Matthews A., Oppermann M., Marsh M. (2005) Mol. Biol. Cell 16, 902–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neubrand V. E., Will R. D., Möbius W., Poustka A., Wiemann S., Schu P., Dotti C. G., Pepperkok R., Simpson J. C. (2005) EMBO J. 24, 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csaba Z., Lelouvier B., Viollet C., El Ghouzzi V., Toyama K., Videau C., Bernard V., Dournaud P. (2007) Traffic 8, 820–834 [DOI] [PubMed] [Google Scholar]
- 27.Lelouvier B., Tamagno G., Kaindl A. M., Roland A., Lelievre V., Le Verche V., Loudes C., Gressens P., Faivre-Baumann A., Lenkei Z., Dournaud P. (2008) J. Neurosci. 28, 4336–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannes L., Popoff V. (2008) Cell 135, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 29.Ghosh R. N., Mallet W. G., Soe T. T., McGraw T. E., Maxfield F. R. (1998) J. Cell Biol. 142, 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., Johannes L. (2002) J. Cell Biol. 156, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganley I. G., Espinosa E., Pfeffer S. R. (2008) J. Cell Biol. 180, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.