Abstract
The CD300 family of myeloid immunoglobulin receptors includes activating (CD300b, CD300e) and inhibitory members (CD300a, CD300f), as well as molecules of uncertain function presenting a negative charge within their transmembrane domain (CD300c, CD300d). In this paper, we establish that CD300c is a functional immune receptor able to deliver activating signals upon ligation in RBL-2H3 mast cells. CD300c signaling is partially mediated by a direct association with the immune receptor tyrosine-based activation motif-bearing adaptor FcϵRγ. The existence of complementary transmembrane-charged residues in certain CD300 receptors suggested the formation of heterodimers within this family. Indeed, we proved the interaction between CD300b and CD300c in transfected COS-7 cells and demonstrated that it has important functional consequences. Unexpectedly, dimmer formation was dependent on the immunoglobulin domains rather than the charged transmembrane residues. Concordantly, all CD300 members were found to interact with each other, even with themselves, forming both homo- and heterodimers. We found that the combination of CD300 receptors in a complex differentially modulates the signaling outcome, strongly suggesting a new mechanism by which CD300 complexes could regulate the activation of myeloid cells upon interaction with their natural ligands.
Keywords: Adaptor Proteins, Cell Surface Receptor, Innate Immunity, Myeloid Cell, Signal Transduction
Introduction
The immune system is controlled by a precise balance between positive and negative signals mediated by receptors mostly present on the surface of leukocytes. Structure strongly determines the functional properties of immune receptors. In this regard, inhibitory receptors deliver intracellular signals directly due to the presence of immune receptor tyrosine-based inhibitory motifs within their long cytoplasmic tails. Upon receptor-ligand interaction, immune receptor tyrosine-based inhibitory motifs become tyrosine phosphorylated and thereafter recruit Src homology 2 bearing phosphatases such as Src homology 2 domain-containing phosphatase 1 and SHIP. This in turn starts a cascade of intracellular signaling events promoting cell inhibition (1, 2). On the contrary, activating receptors need to associate with specialized signal transduction transmembrane polypeptides because of the lack of functional signaling units within their sequences. This association takes place at the transmembrane region and is dependent on the formation of a non-covalent bond between oppositely charged amino acid residues in the receptor and the adaptor polypeptide. Ligand engagement by activating receptors results in the phosphorylation of tyrosine-based motifs present in the cytoplasmic tails of the associated adaptor molecules. This phosphorylation is required for further recruitment of protein-tyrosine kinases that will stimulate a series of intracellular events inducing cell differentiation, growth and survival, adhesion, migration, phagocytosis, cytokine production and/or cytotoxicity (3, 4). The DNAX-activating protein of 12 kDa (DAP-12), FcϵRγ, and CD3ζ adaptor molecules contain immune receptor tyrosine-based activation motifs (ITAMs)4 for the recruitment of Syk and ZAP-70 protein-tyrosine kinases, whereas DAP-10 displays a YXXM motif that serves as a docking site for PI3K (5, 6).
The immunoglobulin (Ig) fold is present in a wide range of molecules that includes not only cell surface antigen receptors but also co-receptors and co-stimulatory molecules, antigen presenting structures, cell adhesion molecules, and certain types of cytokine and growth factor receptors. Most of the cell surface antigen receptors are clustered within the genome. This is the case for the SIGLECs (sialic acid-binding Ig-like lectins), ILTs (Ig-like transcripts), KIRs (killer-cell Ig-like receptors), NCRs (natural cytotoxicty receptors), SIRPs (signal regulatory proteins), and TREMs (triggering receptors expressed by myeloid cells) (7–12). Ig-like domains from receptors in the same family present a high degree of homology among them, but sometimes the similarity is also maintained between receptors belonging to different families. This is likely a consequence of duplication processes driving the appearance of these families along evolution.
The CD300 leukocyte surface molecules are encoded by a cluster of genes on human chromosome 17q25.1 (13, 14). The CD300 family includes six molecules with the ability to deliver activating or inhibitory signals to the cells on which they are expressed that are predominantly from the myeloid origin. CD300a (IRp60) and CD300f (IREM-1) act as inhibitory receptors, whereas CD300b and CD300e (IREM-2) trigger activating signals (15–21). It is worth mentioning that CD300f presents a putative functional duality, as it has been recently shown to deliver activating signals though the recruitment of PI 3-kinase and FcϵRγ (22, 23). CD300c (CMRF-35) and CD300d function remains uncertain as a consequence of their particular structural properties (24). Both receptors combine a transmembrane region containing a negatively charged residue with a short cytoplasmic tail devoid of known signaling motifs.
In the present work, we establish CD300c as an activating immune receptor. Characterization of the signal transduction mechanisms of CD300c led us to identify the capability of CD300 receptors to bind together extracellularly forming homo- and hetero-signaling complexes. We show that the integration of CD300 molecules in complexes modifies the signaling properties of individual receptors allowing synergies at the same time as agonistic and/or antagonistic processes. This distinctiveness of CD300 receptors may represent a new mechanism of myeloid cells to precisely control immune responses in terms of intensity and duration.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
P815 cells were maintained in RPMI 1640/l-glutamine medium supplemented with 10% heat inactivated FBS, 25 mm HEPES, 2 mm glutamine, 100 IU/ml of penicillin, and 100 μg/ml of streptomycin. COS-7 and RBL-2H3 cells were grown in DMEM containing 10% heat inactivated FBS, 2 mm glutamine, 1 mm sodium pyruvate, 100 IU/ml of penicillin, and 100 μg/ml of streptomycin. Anti-hemagglutinin (HA).11 mAb ascites was from Covance. Anti-FLAG M2® mAb (purified and peroxidase-conjugated), rabbit anti-actin (clone 20–33), anti-2,4-dinitrophenol SPE7 mAb, mouse IgG purified immunoglobulin, and F(ab′)2 sheep anti-mouse IgG were from Sigma. Anti-rat high affinity IgE receptor mAb was obtained from BD Biosciences. Anti-HA12CA5 mAb and anti-Myc9E10 mAb were described before (18, 25). Rabbit anti-FcϵRγ subunit was from Millipore. Polyclonal antibody against a peptide mapping in the cytoplasmic tail of CD300c (SSRSRQNWPKGENQ) was raised in rabbit (Sigma). Polyclonal rabbit anti-mouse FITC was from DAKO. Streptavidin-HRP was purchased from Roche Applied Science. Sheep anti-mouse and donkey anti-rabbit IgG horseradish peroxidase-linked Abs were from GE Healthcare.
DNA Reagents
Constructions used in this study were generated by PCR amplification and standard cloning techniques, and further confirmed by DNA sequencing under Big DyeTM cycling conditions on an Applied Biosystems 3730xl DNA Analyzer (Macrogen Inc). Details have been summarized in supplemental Table S1.
Mammalian pDisplay-CD300b (19), -CD300f (18), and -CD300e (20) were described previously. Full-length CD300c (including 5′ and 3′ untranslated regions) was amplified from a human spleen cDNA library and cloned into pCDNA3.1-V5-His TOPO (Invitrogen). CD300c sequence lacking the signal peptide was cloned into pDisplay and pCDNA3-FLAG (19). pDisplay-CD300c E191V substitution mutant was generated with mutagenic oligonucleotides according to the instructions of the QuikChange Site-directed Mutagenesis kit (Stratagene). Two deletion mutants of CD300c affecting the cytoplasmic tail (CD300c Δcyto, del209–224aa) or the Ig domain (CD300c ΔIg, del1–132aa) were cloned into pDisplay. CD300b wild-type (WT), CD300b K158L, CD300b Δ1 (del 178–201 aa) (19), CD300e WT (20), and CD300f WT (18) were subcloned into pCDNA3-FLAG. A pBabePuro-2×Myc expression vector was generated by cloning the Igκ signal peptide followed by two Myc epitopes in tandem into the BamHI/EcoRI sites of pBabePuro. CD300c WT, CD300c E191V, CD300c Δcyto, and CD300f WT (18) were subcloned into pBabePuro-2×Myc. A punctual mutant of CD300b affecting Cys-50 (CD300b C50G) and a deletion mutant affecting the Ig domain (CD300b ΔIg, del1–122aa) were cloned into pCDNA3-FLAG. CD300b ΔIg was additionally cloned into pDisplay. Full-length rat CD300b was amplified from fresh peripheral blood mononuclear cell cDNA and cloned into pCDNA3.1-TOPO. The molecule without the signal peptide was further subcloned into pDisplay. Full-length CD300a was amplified from a phytohemagglutinin-activated peripheral blood mononuclear cell cDNA library (Clontech). CD300a devoid of the signal peptide was subcloned into pCDNA3-FLAG and pDisplay vectors. Full-length TREM-1 and CD28 were amplified from human isolated monocyte cDNA and fresh peripheral blood mononuclear cell cDNA, respectively. Both molecules were transferred to pCDNA3-FLAG upon removal of the signal peptide sequence. Soluble FLAG-tagged CD300b and TREM-1, and HA-tagged CD300c and CD300f were cloned into pEXP5-CT-TOPO. FcϵRγ transmembrane adaptor protein was transferred from pFLAG-CMV2 (20) to pCDNA3-FLAG. Two different FcϵRγ siRNA were cloned into pSilencer2.1-U6Hygro according to the manufacturer's instructions (Ambion).
Cell Transfections
COS-7 cells (6 × 105) were transiently transfected with LyoVecTM Reagent (InvivoGen) according to the manufacturer's instructions. For the generation of RBL-2H3 stable transfectants, 20 × 106 cells were electroporated in the presence of 20 μg of the appropriate linearized construct at 250 V, 960 μF, and 100 Ω in a Gene Pulser Electroporator (Bio-Rad). Transfectants were selected and maintained in culture with the appropriate selection antibiotic. G418 (BioWhittaker) and/or puromycin (Sigma) were used at 1 mg/ml and 1 μg/ml, respectively. Positive cells were further selected by immunostaining with the appropriate antibodies and sorting with magnetic Dynabeads® M-450 coated with sheep anti-mouse IgG (Dynal).
siRNA Silencing
RBL-2H3 transfectants were transiently electroporated with pSilencer-U6/Hygro containing FcϵRγ or scrambled siRNA (3 μg/106 cells). Forty-eight hours post-transfection silencing was assessed by Western blot analysis. The intensity of the bands in the blot was quantified using the GeneTools Program (SynGene).
Flow Cytometry
Cell surface expression of the desired molecules was tested by indirect immunofluorescence following standard techniques (19). Analysis was performed using a FACScalibur instrument and CellQuest software (BD Biosciences).
Expression of Recombinant Proteins
FLAG-tagged CD300b and TREM-1 and HA-tagged CD300c and CD300f soluble receptors were produced using the ExpresswayTM Cell-free Escherichia coli Expression System kit according to the manufacturer's instructions (Invitrogen).
Immunoprecipitation and Western Blot Analysis
Cells were lysed at 4 °C for 20 min using 1% Triton X-100 or 0.5% CHAPS-containing buffer as described previously (26). Cell lysates were clarified by centrifugation at 16,000 × g for 15 min at 4 °C. Crude lysates were precleared for 1 h at 4 °C using 20 μl of IgG-Sepharose 6 Fast Flow (GE Healthcare). Two additional preclearings were conducted for 30 min at 4 °C. For immunoprecipitations, precleared lysates were incubated with 30 μl of Protein G-Sepharose beads (GE Healthcare) and 1 μg of Ab for 3 h at 4 °C. Proteins in the crude lysates (2%) and immunoprecipitates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) filters (Millipore). Filters were blocked with 5% skim milk and then probed with the indicated Abs at appropriate dilutions. Bound Abs were detected using West Pico Supersignal kit (Pierce).
Luciferase Assays
RBL-2H3 transfectants were transiently electroporated with a luciferase reporter plasmid (pT81Luc) containing three tandem copies of the distal NFAT/AP-1 site of the murine IL-2 promoter (27) (0.5 μg/106 cells) and a TK Renilla construct (Promega) (0.1 μg/106 cells). Twenty-four hours post-transfection, 1.5 × 106 cells were stimulated for 7 h with the indicated antibodies using the murine mastocytoma P815 cell line as the presenting cell (1 × 106). Plastic-coated anti-2,4-dinitrophenol IgE (5 μg/ml) was used as positive control for RBL-2H3 cell stimulation. The P815 cell line cultured in supplemented RPMI 1640/l-glutamine medium alone was used as negative control. Postnuclear lysates were obtained as described (27) and luciferase activity was measured according to the Dual Luciferase Report kit manual (Promega) using a FB12 Luminometer (Berthold).
β-Hexosaminidase Release Assays
5 × 105 RBL-2H3 transfectant cells resuspended in 50 μl of Tyrode's buffer (28) were stimulated in 96-well plates for 1 h at 37 °C and 5% CO2 by plastic-coated Abs (5 μg/ml) previously cross-linked with sheep anti-mouse (5 μg/ml). 20 μl of supernatants were transferred to a new plate and incubated for an additional hour at 37 °C and 5% CO2 with 50 μl of 1 mm 4-nitrophenyl N-acetyl-β-d-glucosaminide (Sigma) in 0.05 m citrate buffer (pH 4.5). Reactions were stopped and color was developed by adding 100 μl of 0.2 m glycine buffer (pH 10.7). Optical density was measured at 405 nm. For determining the maximum degree of enzyme release, cells were lysed with Triton X-100 (1%) prior to incubation with the substrate. Results were expressed as a percentage of specific hexosaminidase release: (Abs test − Abs spont)/(Abs max − Abs spont) × 100, where Abs spont is the spontaneous release obtained with cells incubated in the absence of Abs and Abs max is the maximum release obtained with cells lysed using Triton X-100 (1%).
RESULTS
CD300c Molecule Is a Functional Triggering Receptor in RBL-2H3 Cells
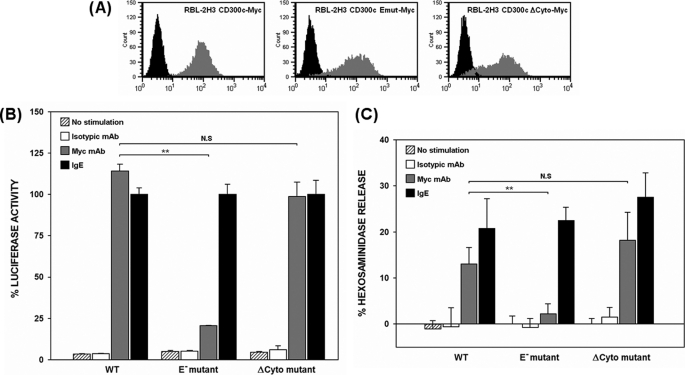
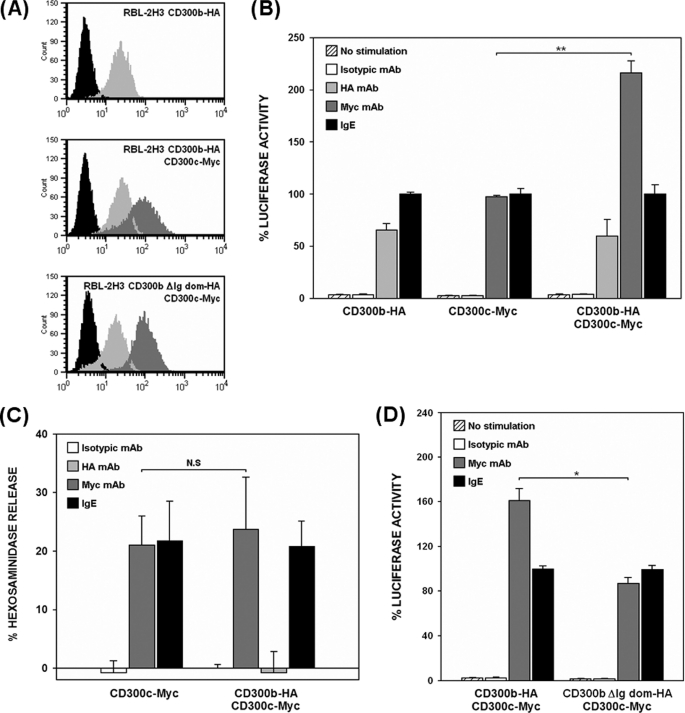
CD300c is a type I transmembrane protein that belongs to the CD300 locus. Despite being the first member of the family to be cloned, little information is known about this immune receptor. CD300c displays a short cytoplasmic tail in combination with a transmembrane region bearing a negatively charged residue. The unusual structure and the lack of experimental data regarding the function make CD300c classification uncertain (24). Additionally, the CD300c cell surface expression pattern has not been elucidated in detail because monoclonal antibodies against the molecule strongly cross-react with CD300a as a consequence of the high degree of extracellular homology between them (29, 30). This fact has also impaired the approach of CD300c behavior in primary cells. We wanted to address whether CD300c was a functional Ig receptor and characterize the nature of the signal triggered upon its engagement. We have extensively used the RBL-2H3 mast cell line as per its inducible exocytotic response of preformed cytoplasmic secretory granules and reporter-dependent transcriptional activity upon receptor cross-linking (19, 22). We stably expressed CD300c-2×Myc on the surface of RBL-2H3 cells (Fig. 1A). Cells were transiently transfected with a luciferase reporter gene under control of a NFAT/AP-1-dependent promoter and stimulated with anti-Myc mAb mimicking the CD300c natural ligand. Engagement of the receptor elicited an increase in promoter activity quantitatively comparable with that delivered by FcϵRI cross-linking (Fig. 1B). To determine whether the negative transmembrane charge in CD300c, the cytoplasmic region or both, were responsible for signal transduction of the receptor, we generated two RBL-2H3 stable transfectants expressing CD300c-2×Myc mutants affecting the transmembrane glutamic acid (E191V) or the cytoplasmic tail (ΔCyto) (Fig. 1A). The CD300c ΔCyto mutant mediated the same transcriptional activity as the wild type, suggesting that there are no signaling motifs within the cytoplasmic region of CD300c able to contribute to the activation of the reporter (Fig. 1B). Conversely, substitution of the glutamic acid by a hydrophobic residue led to a dramatic reduction in the reporter activity, thus establishing a role for the transmembrane region of the receptor (Fig. 1B). Equivalent results were obtained when assessing β-hexosaminidase granule release upon CD300c stimulation (Fig. 1C). No differences were observed among transfectants in the activating capacity and cell surface levels of FcϵRI (≥99% expression, data not shown).
FIGURE 1.
CD300c delivers activating signals in RBL-2H3 cells. A, RBL-2H3 cells were stably transfected with 2×Myc-tagged forms of CD300c. Cell surface expression of CD300c molecules was checked by flow cytometry using anti-Myc9E10 mAb (dark gray histogram) or an isotypic mAb as a negative control (black histogram). B, RBL-2H3 transfectants were transiently transfected with 3×NFAT/AP1-Luciferase and TK-Renilla plasmids. Luciferase activity was measured after stimulation for 7 h with the indicated antibodies. Data were normalized and expressed as a percentage of luciferase activity considering IgE stimulation as the top threshold of activation. Duplicates were performed for all the stimulations. The result is representative of three independent experiments. C, RBL-2H3 transfectants were stimulated with the indicated antibodies to induce cell degranulation. Percentage of β-hexosaminidase release was assessed by incubating the supernatant with 4-nitrophenyl N-acetyl-β-d-glucosaminide substrate. Each assay was set up in triplicate. The result is a mean of three independent experiments (**, p ≤ 0.01). N.S., non-significant. Error bars represent standard deviation.
CD300c Signaling Is Partially Mediated by Its Association to FcϵRγ
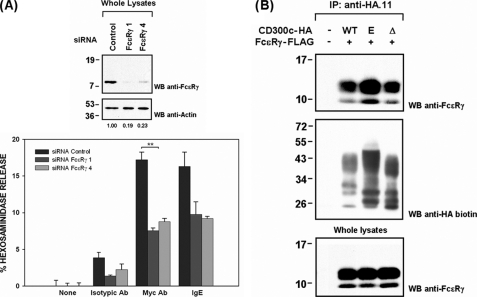
LMIR-4, also known as CLM-5 and MAIR-IV, is the murine structural ortholog of CD300c. This receptor has recently been shown to elicit triggering signals by means of FcϵRγ polypeptide to which it binds in transfected cell lines and purified blood neutrophils (31–33). However, the interaction between these two molecules has not been mapped. To study the role of FcϵRγ in CD300c signaling we reduced FcϵRγ expression in RBL-2H3 cells using siRNA technology. RBL-2H3 CD300c-2×Myc cells in which FcϵRγ was down-regulated, exhibited a marked reduction in hexosaminidase release upon receptor cross-linking (Fig. 2A). This result manifested the functional dependence of CD300c on FcϵRγ. FcϵR engagement was impaired likewise validating the specificity of the siRNAs employed. Next, we explored whether CD300c signaling by means of FcϵRγ occurred as a consequence of a direct interaction between both molecules. FcϵRγ immunoprecipitated together with the CD300c receptor in COS-7 cells (Fig. 2B). Surprisingly, the CD300c mutants affecting the transmembrane charge (E191V) and the cytoplasmic tail (ΔCyto) retained the ability to interact with the adaptor molecule (Fig. 2B). These results together with the functional blockade observed for the CD300c E191V mutant suggested the existence of a molecule/complex recruited by CD300c necessary for signaling through FcϵRγ in the RBL-2H3 system.
FIGURE 2.
CD300c signals through FcϵRγ adaptor molecule. A, RBL-2H3 CD300c-2×Myc cells were transiently transfected with control or FcϵRγ siRNAs. Forty-eight hours after transfection a subset of cells were lysed and proteins (2 μg) were run on 15% SDS-PAGE and transferred to PVDF filters. FcϵRγ interference was assessed by Western blot (WB). Values correspond to FcϵRγ expression, which was calculated by quantifying the intensity of FcϵRγ versus actin. Hexosaminidase release was assessed as described. Triplicates were performed for all the stimulations. The result is representative of two independent experiments. B, COS-7 cells were transiently transfected with HA-tagged CD300c WT or mutant forms (E191V and ΔCyto) in combination with FcϵRγ-FLAG. Cell lysates (0.5% CHAPS) were immunoprecipitated with anti-HA.11 mAb and analyzed by 15% SDS-PAGE under reducing conditions. Proteins were transferred to PVDF filter and probed with the indicated antibodies. Whole cell lysates (2%) were included as controls (**, p ≤ 0.01). Error bars represent standard deviation.
CD300c Associates with CD300b in Transfected COS-7 Cells
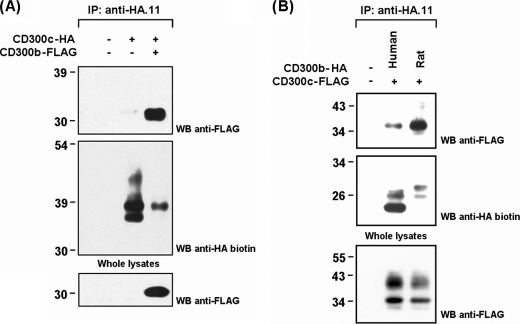
The CD300b receptor is a non-classical activating receptor able to deliver signals by associating with the transmembrane adaptor protein DAP-12 and the intracellular mediator Grb-2. The recruitment of these signaling molecules are two independent events. CD300b binds DAP-12 through a lysine residue within the transmembrane domain, whereas Grb-2 is recruited by means of a tyrosine-based motif present in the cytoplasmic tail of the receptor. Previous results in myeloid-derived cell lines suggested the existence of an unknown protein recruited by the CD300b transmembrane region necessary for signaling in the absence of DAP-12 (19, 34). The presence of oppositely charged amino acid residues embedded in the transmembrane regions of CD300b and CD300c, the equivalent functional dependence on unidentified transmembrane mediators, and the existence of multiple rat CD300 mRNA transcripts in the RBL-2H3 cell line suggested the binding between both receptors (19).5 To test this possibility, we transiently transfected COS-7 cells with CD300c-HA in the presence and absence of CD300b-FLAG. Immunoprecipitation experiments confirmed both molecules to be associated (Fig. 3A). Furthermore, Western blot analysis revealed that complex formation led to an uneven recovery of CD300c in the immunoprecipitates. Systematically, only the lower electrophoretic bands described for CD300c were retrieved in the immunoprecipitates when co-transfected with CD300b (Fig. 3A). It is noteworthy that this effect was not observed when assessing the interaction of CD300c with FcϵRγ (Fig. 2B).
FIGURE 3.
CD300c associates with both human and rat CD300b in transfected COS-7 cells. A, COS-7 cells were transiently transfected with HA-tagged CD300c alone or in combination with FLAG-tagged CD300b. Cell were lysed (1% Triton X-100) and immunoprecipitated (IP) with anti-HA.11 mAb. Blots were probed with the indicated antibodies. Whole cell lysates (2%) were included as controls. B, same experiment as in A to compare the recruitment of CD300b-FLAG from human and rat origin by CD300c-HA. WB, Western blot.
Human CD300b and CD300c molecules interact between them, but to explain the functional effects observed in the RBL-2H3 system the association should be conserved across species, allowing the transfected human receptors to recruit the endogenously expressed rat orthologs. We used the human CD300b cDNA sequence to perform a search in the Ensembl rat genome data base to identify its rat counterpart. The full-length transcript was amplified from peripheral blood mononuclear cell cDNA and after sequence validation was termed rat CD300b (GenBankTM accession GU054494) (supplemental Fig. S1A). Human and rat CD300b receptors share a high amino acid identity (55%), but present important structural differences. For instance, rat CD300b receptor is devoid of the cytoplasmic Grb2-interacting motif (supplemental Fig. 1B). Despite the divergence, both CD300b receptors were able to interact with CD300c from human origin, confirming the binding between ortholog molecules (Fig. 3B).
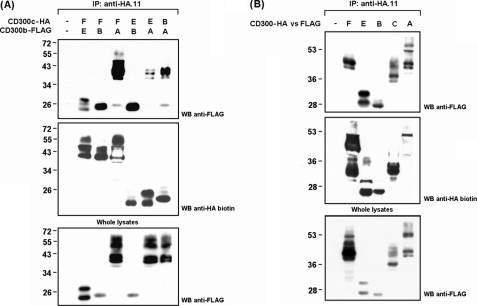
CD300c/CD300b Association Relies on Their Ig Domains
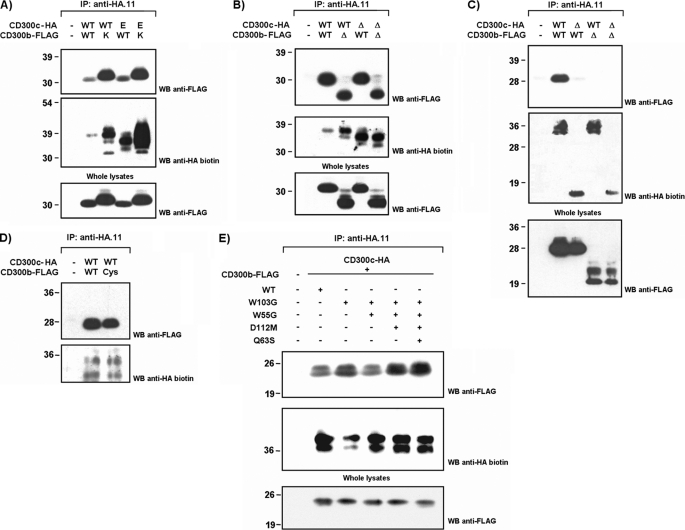
Considering the presence of oppositely charged residues in the transmembrane regions of CD300b and CD300c, we wanted to address whether the association between both receptors was dependent on the formation of a membrane-embedded salt bridge involving these amino acids. We generated substitution mutants affecting the acidic residue on CD300c (HA-tagged CD300c E191V) and the basic residue on CD300b (FLAG-tagged CD300b K158L). COS-7 cells were co-transfected following all possible combinations between WT and mutant forms. Surprisingly, CD300c-CD300b complex formation was not disrupted by substitution of transmembrane residues, neither individual nor in combination (Fig. 4A). To further map the interaction between both immune receptors a wider set of mutants were assessed in the co-transfection experiments. We conducted cytoplasmic tail deletions to create HA-CD300c Δcyto and FLAG-CD300b Δcyto mutant forms for their co-expression in COS-7 cells with WT forms. Intracellular association between both molecules was ruled out given that none of the cytoplasmic mutants could disrupt CD300c-CD300b complex formation (Fig. 4B). Next, we tested whether CD300c and CD300b were interacting through their extracellular portions, and more specifically through the Ig domain. COS-7 cells were transfected with epitope-tagged forms of the receptors in which the Ig domain was removed from the extracellular region. The membrane-proximal portion, also known as the stem region, was maintained. Ig domains were shown to be essential for the interaction between CD300b and CD300c as their removal impaired the binding of the receptors (Fig. 4C). CD300 family members, pIgRs (polymeric Ig receptors), TREMs, NKp44, CLM-1, and TLT-1 display in their Ig domains the pair of cysteine residues describing the Ig-V-type fold, but also a second pair of residues that has been proposed as a differential trade for a discrete evolutionary group of receptors stemming from the ancestral V-type Ig domain (35). We wanted to evaluate the role of this disulfide bridge in the formation of the CD300c-CD300b complex. The association between both receptors was not affected when a CD300b receptor carrying a substitution in Cys-50 was used instead of the WT form, indicating that the additional pair of cysteines in CD300 molecules is dispensable for the interaction observed (Fig. 4D). The crystal structure of CD300f revealed a prominent protrusion extending from the main immunoglobulin body in CD300 molecules. This protrusion creates in turn, a cavity with hydrophobic properties and a negative electrostatic potential, capable of accommodating ligands (36). To disturb the cavity, we designed FLAG-CD300b mutants affecting a combination of key structural residues lying off this area. Trp-55 (C-C′ strand) and Asp-112 (G strand) block the floor and the roof of the cavity, respectively, whereas Glu-63 (C′ strand) and Trp-103 (F strand) fill up the protrusion (supplemental Fig. S2). FLAG-CD300b mutants bound as efficiently as the WT to HA-CD300c upon co-transfection in COS-7 cells (Fig. 4E).
FIGURE 4.
CD300c and CD300b interact through their Ig-like domains. COS-7 cells were transiently transfected with HA-tagged CD300c in combination with FLAG-tagged CD300b. Wild type forms were tested against transmembrane substitution mutants (A), intracellular deletion mutants (B), extracellular deletion mutants (C), cysteine substitution mutants (D), or point mutants affecting the protruding body in the Ig fold (E) to map the interaction between both molecules. Cells were lysed (1% Triton X-100) and CD300c was immunoprecipitated (IP) with anti-HA.11 mAb in all cases. Western blots (WB) were conducted with the indicated Abs. Whole cell lysates (2%) were included as controls.
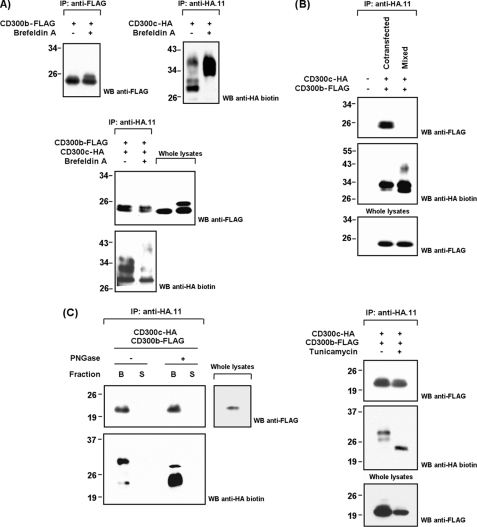
CD300c/b Are cis-Interacting Receptors Assembled Prior to Their Plasma Membrane Export: Association Is Not Mediated by Glycosylation
Co-immunoprecipitation techniques do not discriminate between cis- and trans-interactions in the case of integral membrane proteins binding extracellularly. Therefore, it was of interest to test whether CD300c and CD300b bound each other head-to-head as a receptor-ligand pair. We analyzed the binding of a soluble CD300b-mIgG2a fusion protein to non-hematopoietic cell lines stably transfected with CD300c. No staining was detected with this fusion protein on HeLa and CHO-K1 CD300c-expressing cells, concluding that the interaction observed occurs laterally between receptors co-expressed on the same cell (data not shown). It is remarkable that the CD300b-mIgG2a molecule used in these experiments is fully functional as it stains certain cell populations that are being screened for CD300b ligand identification.5 Next, we wanted to know whether the CD300b/c interaction occurred on the plasma membrane or if it could be established intracellularly. Brefeldin A is a fungal metabolite that causes disassembly of the Golgi apparatus and consequently protein accumulation in the endoplasmic reticulum (ER)/Golgi network. Retrograde transport from the plasma membrane to the lysosomes is not affected. Brefeldin A treatment caused a shift in the molecular weight of the receptors as a consequence of its retention in the cellular compartments where most post-translational modifications occur, thus allowing visualization of the fully mature proteins (Fig. 5A, upper panels). CD300c and CD300b co-transfected in brefeldin A-treated COS-7 cells co-precipitated as efficiently as in mock-treated cells, indicating that the interaction between both receptors occur before their export to the membrane (Fig. 5A, lower panel). Effectiveness of treatments was further assessed by tracking the disappearance of receptors from the cell surface upon transfection (data not shown). However, only the bands with low molecular weight were involved in complex formation indicating that the receptors assembly is established right after protein synthesis in ER. This entails that some epitopes susceptible to modification might become inaccessible as a consequence of protein interaction. Concordantly, CD300c and CD300b individually transfected and fully modified were not able to interact in solution (Fig. 5B). However, as the interaction was shown to occur through the Ig regions, it is also very likely that inclusion of the receptors in intracellular membranes is required to properly position the Ig fold for complex formation.
FIGURE 5.
CD300c and CD300b form cis-interacting complexes in the ER. A, COS-7 cells were transiently transfected with CD300 constructions. Forty-eight hours post-transfection cells were stimulated for 24 h with brefeldin A (1 μg/ml) or vehicle (70% EtOH). Cells were lysed (1% Triton X-100) and immunoprecipitated (IP) with the indicated antibodies. Western blots (WB) were conducted as described. Whole cell lysates (2%) were included as controls when assessing co-transfections. B, CD300c-HA and CD300b-FLAG were expressed both individually and simultaneously in COS-7. When expressed individually, clarificates where processed independently until the immunoprecipitation step with anti-HA.11 mAb. Western blots were conducted normally. C, immunoprecipitates from COS-7 co-transfected cells were subjected to peptide:N-glycosidase treatment in non-denaturing conditions. The eluted and immobilized fractions were analyzed separately (S = supernatant and B = beads) (left panel). COS-7 cells were transiently cotransfected with CD300 constructions. Eighteen hours post-transfection cells were stimulated for 48 h with tunicamycin (1 μg/ml) or vehicle (dimethyl sulfoxide). Cells were lysed and immunoprecipitated as described above (right panel).
Glycosylation is a common modification in cell surface molecules and has been shown to affect ligand binding properties. Accordingly, to establish whether the interaction observed between CD300c/b could be assisted by N-glycans we induced deglycosylation of N-linked sugars both in vitro and in vivo. Treatment of co-immunoprecipitates with peptide:N-glycosidase F under non-denaturing conditions could not disrupt the CD300b-CD300c interaction (Fig. 5C, left panel). Tunicamycin treatment of co-transfected COS-7 cells, an inhibitor of bacterial and eukaryote N-acetylglucosamine transferases, led to equivalent results (Fig. 5C, right panel). O-Glycosylation does not have specific acceptor motifs, but it is well known that this modification occurs mainly in serine and threonine residues localized in proline-rich areas. The CD300 membrane-proximal region fit this consensus. Unfortunately, removal or inhibition of O-sugars is a difficult task because the diversity of glycotransferases is as big as the number of O-sugars with different structures (37). Therefore, to evaluate the role of O-glycans in CD300 association, we produced recombinant soluble receptors using an E. coli expression system for in vitro transcription and translation. This method allows the use of naked polypeptidic backbones. O-Linked glycans were not required for CD300c-CD300b complex formation (supplemental Fig. S3). Therefore, the interaction seems to depend strictly on the peptide-peptide contacts.
CD300b Enhances CD300c-mediated Transcriptional Activity
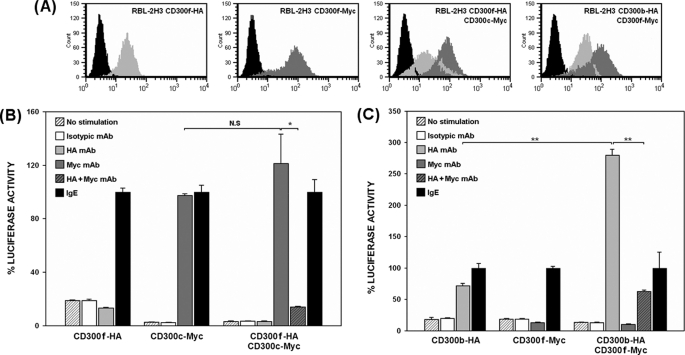
To explore whether the association between CD300c and CD300b modified their functional properties, we performed transcriptional activity assays comparing the activation induced by single and double RBL-2H3 stable transfectants upon individual cross-linking of the receptors (Fig. 6A). CD300b-HA and CD300c-2×Myc transfectants lead to a similar activation of the NFAT/AP-1 reporter gene, whereas cells transfected with both CD300b-HA and CD300c-2×Myc exhibited a 2-fold increase in transcriptional activity when stimulated through the CD300c receptor (Fig. 6B). The increase was not observed when the double transfectant was stimulated through the CD300b receptor. The activating capacity and cell surface levels of FcϵRI did not change as per the double transfection (≥99% expression, data not shown). Hexosaminidase release in cells simultaneously expressing CD300b and CD300c did not exhibit the same increase when stimulated under the equivalent experimental conditions (Fig. 6C). CD300b receptor was shown previously to be unable to elicit exocytotic responses in the absence of DAP-12, an adaptor molecule that is not endogenously expressed by RBL-2H3 cells (19). Therefore, amplification of the positive signal generated by the complex was likely to be propagated through CD300b which, as described before, displays a functional cytoplasmic tyrosine-based motif (19). CD300c-dependent transcriptional activity returned to the levels of the single CD300c transfectant when the receptor was stimulated in the presence of a CD300b molecule lacking the Ig domain (Fig. 6D). Ig domain deletion did not modify access of these molecules to the plasma membrane (Fig. 6A). This result confirmed functionally the biochemical experiments showing that the association between both receptors occurs at the extracellular level.
FIGURE 6.
CD300b acts as a modifier of CD300c signaling. A, RBL-2H3 cells stably expressing CD300c-Myc on the cell surface were transfected with CD300b-HA or CD300b ΔIg-HA. Cell surface expression of the different transfectants was checked by flow cytometry using anti-Myc9E10 for CD300c (light gray histogram) and anti-HA12CA5 mAb for CD300b (dark gray histogram). Isotypic mAb was used as negative control (black histogram). B and D, RBL-2H3 transfectants were transiently transfected with 3×NFAT/AP1-Luciferase and TK-Renilla plasmids. Luciferase activity was measured after stimulation for 7 h with the indicated antibodies. Data were normalized as described before. Duplicates were performed for all stimulations. Results are representative of five independent experiments. C, RBL-2H3 transfectants were stimulated with the indicated Abs for assessing hexosaminidase granule release. Data are expressed as a percentage of specific release. Each stimulation point was set up in triplicate. The result is a mean of three independent experiments (*, p ≤ 0.05; **, p ≤ 0.01). N.S., non-significant. Error bars represent standard deviation.
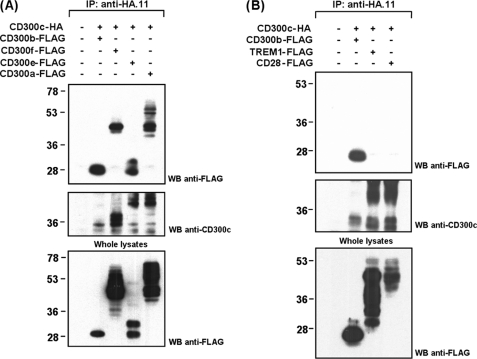
CD300c Is Able to Interact in COS-7 Cells with All CD300 Family Members but Not with Other Ig-like Immune Receptors
Given the high level of homology among CD300 Ig folds, we wanted to address whether CD300c was capable of complexing with CD300 members beyond CD300b. COS-7 cells were transiently co-transfected with CD300c-HA and FLAG-tagged forms of CD300a, CD300e, or CD300f. All CD300 members were recruited by the CD300c receptor (Fig. 7A). To determine specificity of the detected interactions, we evaluated whether CD300c could bind to other Ig superfamily receptors such as TREM-1, which is closely related to CD300 receptors regarding homology among the V-type Ig fold and cell surface distribution; and CD28, that despite evolving differentially in terms of expression pattern shares very similar structural features with CD300 family members (12, 38). Neither TREM-1 nor CD28 were able to bind to CD300c (Fig. 7B). Therefore, the interactions observed occur specifically within immune receptors belonging to the CD300 family. The high variability affecting the CD300c electrophoretic pattern is remarkable. Co-transfection of CD300c with receptors to which it does not associate resulted in an electrophoretic pattern comparable with that obtained when the molecule was transfected alone or in combination with the FcϵRγ (see Figs. 2B, 3A, and 7B). On the contrary, when CD300c was co-expressed with CD300 receptors, to which it binds, the electrophoretic pattern of the molecule changed, being variable and dependent on the co-transfected partner (Fig. 7A). These observations are in agreement with the formation of the complexes at the ER-Golgi level, which determines the acquisition of post-translational modifications.
FIGURE 7.
CD300c interacts with all CD300 family members but not with other Ig-like receptors. CD300c-HA was transiently transfected in COS-7 cells in combination with FLAG-tagged CD300 receptors (CD300a, CD300b, CD300e, and CD300f) (A) or CD300-related Ig-like receptors (TREM-1 and CD28) (B). Cell lysates (1% Triton X-100) were immunoprecipitated (IP) with anti-HA.11 mAb. Filters were probed with the indicated antibodies. Whole cell lysates (2%) were included as controls. WB, Western blot.
CD300 Receptors Form All Possible Combinations of Heterodimers but Also Are Able to Homodimerize
According to our data, it was reasonable that all members of the CD300 family exhibited the same interacting capacity as CD300c. To validate our hypothesis, COS-7 cells were co-transfected with CD300 receptors following all possible combinations. Co-precipitations occurred independently of the combination of receptors used, indicating that all CD300 receptors are able to interact between them (Fig. 8A). This experiment further confirmed the electrophoretic variability of CD300 receptors when associated with different partners. Because the establishment of disulfide bonds was dispensable for CD300 complex formation, homodimerization of CD300 receptors was also detected when transfecting each receptor with itself but differentially tagged (Fig. 8B).
FIGURE 8.
CD300 hetero- and homodimerization. CD300a, CD300b, CD300c, CD300e, and CD300f (HA- or FLAG-tagged) were transiently cotransfected in COS-7 cells following all possible combinations for the detection of hetero- (A) and homodimers (B). Immunoprecipitations (IP) were performed against HA-tagged receptors with anti-HA.11 mAb after cell lysis (1% Triton X-100). Filters were proved with the indicated antibodies. Whole cell lysates (2%) were included as controls. WB, Western blot.
Signaling Properties of CD300 Receptors Are Dependent on Complex Composition
As complex formation could be extended to all CD300 family members we wanted to examine the functional behavior of individual receptors when forming parts of different complexes. We generated supplementary RBL-2H3 CD300 double transfectants, and their matched single transfectants, to assay the NFAT/AP1-dependent transcriptional activity. More specifically, we generated CD300c/CD300f and CD300b/CD300f transfectants to cover all the combinations between the three archetypical structures present within the CD300 family of receptors: positively and negatively charged activating receptors versus inhibitory receptors (Fig. 9A). Stimulation and co-stimulation were conducted in parallel to allow the observation of either agonistic or antagonistic effects. CD300c-mediated transcriptional activity was not modified by the presence of CD300f (Fig. 9B). In contrast, CD300b signaling was highly enhanced when co-expressed with the same inhibitory receptor (Fig. 9C). The overall activating capacity and cell surface levels of FcϵRI were conserved between transfectants (≥99% expression, data not shown). In both cases, the stimulating signal led by CD300c or CD300b engagement could be reduced by CD300f when receptors were cross-linked simultaneously.
FIGURE 9.
CD300 functional behavior is modified based on complex composition. A, CD300 receptors (Myc or HA-tagged) stably transfected on RBL-2H3 cell surface were assessed by flow cytometry using anti-Myc9E10 (light gray histograms) and anti-HA12CA5 mAb (dark gray histograms). Isotypic mAb was used as negative control (black histograms). B and C, RBL-2H3 transfectants were transiently transfected with 3×NFAT/AP1-Luciferase and TK-Renilla plasmids. Luciferase activity was measured after stimulation for 7 h with the indicated antibodies. Data are normalized as described before. Duplicates were performed for all stimulations. Results are representative of three and two independent experiments respectively (*, p ≤ 0.05; **, p ≤ 0.01). N.S., non-significant. Error bars represent standard deviation.
DISCUSSION
In this paper we have shown that the CD300c molecule was able to induce triggering signals in RBL-2H3 cells. Its stimulation through epitopes promoted NFAT/AP-1-dependent transcriptional activity and release of β-hexosaminidase granules. The CD300c cytoplasmic tail did not contribute to signaling despite the fact that it displays a very high proportion of polar amino acids that could mediate electrostatic interactions with intracellular signaling mediators. FcϵRγ was involved in CD300c signaling by means of a direct interaction. However, CD300c transmembrane glutamic acid, which was shown to be essential for the functional properties of the receptor, was not responsible for FcϵRγ recruitment. This suggested the existence of an unknown CD300c-interacting protein displaying a positive charge in its transmembrane domain. To date there are no experimental evidences of transmembrane ITAM/YXXM bearing-like adaptor polypeptides fitting these structural requirements that could develop functions similar to those exhibited by DAP-12, FcϵRγ, CD3ζ, and DAP-10 (39). Bioinformatic tools have not been productive either, as there are no reports in the literature in which EST databases have been screened to identify genes that could match this new type of transmembrane adaptors. The equivalent and complementary functional scenario of CD300b and CD300c receptors, together with the fact that multiple rat CD300 transcripts were detected in the RBL-2H3 cell line, led us to hypothesize about its physical interaction. CD300c was found to associate with CD300b in COS-7-transfected cells but surprisingly, the binding between both molecules occurred extracellularly and was dependent on the lateral interaction of the Ig domains and not due to transmembrane charge complementation. This unexpected result left unexplained the role of the CD300c transmembrane negative charge in the receptor function. The human high affinity receptor for IgE (FcϵRI) presents two alternative forms. The trimeric form is composed by the IgE-binding α subunit and a disulfide-linked homodimer of γ chains, whereas the tetrameric form contains an additional tetraspanning β chain (40). Interestingly, the α chain binding the homodimeric γ chains presents a negative charge within its transmembrane domain. It is likely that both complexes, FcϵRI and CD300c-FcϵRγ, could be assembled similarly and deliver activating signals using the same mechanisms. It is noteworthy that down-regulation of FcϵRγ in RBL-2H3 likewise impaired hexosaminidase release upon CD300c and FcϵRI engagement. However, we cannot discard that this residue could help stabilize on the cell surface the complexes with other CD300 family members.
In unraveling CD300c signal transduction mechanisms, we have found that CD300 receptors are able to bind each other through their Ig domains. There are several examples in literature describing cis-interactions between Ig-containing molecules, but none pertaining to families of activating/inhibitory immune receptors. Some of these interactions have been mapped and are known to occur by means of disulfide bridges in the proximal transmembrane regions. This is the case for homodimeric structures such as CD28 and CTLA-4 and heterodimeric complexes between the α/β chains of the CD8 co-receptor or the Igα/Igβ molecules from the B-cell receptor (41–43). However, in other cases such as CD4 homodimerization, integrin assembly, or MHC class I and II complex formation, the interactions have not been elucidated in detail (44–46). We demonstrated previously that individual CD300 receptors do not dimerize or oligomerize through disulfide bonds, indicating that other residues different from cysteines should be involved in the lateral binding between CD300 Ig domains (18–20). However, crystallization of the CD300f receptor revealed that the distinctive second disulfide bridge of pIgR, NKp44, TREM-2, and CD300 receptors is within a loop defining the bottom of a solvent-exposed cavity suitable for protein-protein interactions (35, 36, 47). Notably, these residues are not present in TREM-1 and CD28, which failed in recruiting CD300c. The tertiary structure adopted by the Ig-fold, which consists of two sheets of antiparallel β-strands, does not allow the creation of deletion mutants (48). Thus, our first attempt to disturb the cavity was the substitution of the second pair of cysteine residues. Unfortunately, its mutation did not prevent CD300 complex formation. Similar results were observed when assessing mutants affecting key structural residues lying off the same area. In the future, we will also explore the possibility that the cavity could accommodate metal coordinates. Metal ion binding sites have been shown to modulate in the case of TIM proteins (T-cell Ig and mucin) to the cell surface conformation, trafficking, and more importantly the homophilic interactions (49). However, we cannot discard that this groove constitutes a ligand recognition structure. The mapping of the interaction will also help determine the stoichiometry of complexes. So far, we have detected homo- and heterodimers, but if CD300 Ig domains bear more than one surface for lateral contacts, it is feasible that complexes integrating more than two receptors might get formed.
Integral membrane proteins undergo a secretory pathway that starts in the rough ER and proceeds through multiple compartments of the Golgi apparatus and fusing vesicles toward the plasma membrane. Accumulation of post-translational modifications such as glycosylation, occurs throughout this process. Some activating immune receptors are known to be pre-assembled with their transmembrane adaptor modules in the ER (50, 51). By using brefeldin A we have shown that CD300 complexes take place after protein synthesis in ER. The continuous changes in the electrophoretic mobility of CD300 molecules when co-transfected also supported this fact. These changes are most probably produced by the presence or absence of post-translational modifications, including N- and O-glycosylation. Nevertheless, we have demonstrated that these modifications do not constitute the interacting motifs between receptors. The variability in the electrophoretic pattern also sustains the idea of complexes of more than two CD300 molecules. For instance, the three differential electrophoretic patterns of CD300c depending on the co-transfected partner strongly suggest the existence of more than one lateral binding site in the CD300c Ig domain. The fact that interaction between CD300 receptors can occur before their export to the plasma membrane provides an important level of regulation. Presynthesized granules containing CD300 complexes could be stored in the cell cytoplasm and exported to the cell membrane when required. In fact, it has recently been observed that CD300a is rapidly up-regulated in LPS or GM-CSF-stimulated neutrophils by an increase in the translocation rate of presynthesized receptor (52).
According to our data CD300 Ig domains would be able to establish two different kinds of interactions: first, a trans-interaction with their ligands, and second, a cis-interaction with other CD300 molecules. The nature of CD300 ligands remains to be determined. It would be interesting to study whether the ligands for individual receptors are able to act on complexes or if receptor clustering promotes the formation of new recognition surfaces and as consequence, binding of alternative ligands. If the first scenario occurs, it is likely that ligand affinities vary depending on the receptor composition of the complex. This could have important functional implications, as partial or incomplete receptor cross-linking has been shown to invert the signaling nature of certain receptors. This is the mechanism by which some ITAM-associated activating immune receptors are thought to promote inhibitory signals (5, 53).
We have shown that in COS-7-transfected cells all members within the family are competent to form heterocomplexes and that all combinations are possible, yet this does not mean that all heterocomplexes might exist, might be present on the cell surface, or occur in a constitutive manner. Future experiments will concentrate in evaluating the interactions described in cells expressing the endogenous versions of these receptors rather than in transfected cell lines. It is of note that some associations will never take place due to the differential myeloid expression pattern affecting CD300 family members. For instance, CD300e-CD300f heterocomplexes could only be observed on peripheral blood monocytes or myeloid dendritic cells. The interactions of CD300f in granulocytes, where it is also expressed, should be with CD300 members different from CD300e, as this receptor is not found in those cell types. Moreover, the expression of CD300 family members can be modulated upon stimulation and/or differentiation processes. Thus, interactions taking place in a particular scenario could be disrupted as a consequence of a change in the surrounding environment and the other way around, nonexistent interactions could be created upon alteration of the system. In this line, CD300f-CD300e heterocomplexes in monocytes would be easily disrupted upon stimulation with IL-4, as CD300e is drastically down-regulated from the cell surface under these conditions (20). On the contrary, CD300c markedly increases following FcϵRI cross-linking in mast cells (54). It will be necessary to address the composition of the CD300 complexes within the different myeloid cell types and their differentiation/activation intermediates. At the present time this task is limited by the availability of monoclonal antibodies recognizing CD300 members specifically. Moreover, as CD300 clustering affects the acquisition of post-translational modifications, the recognition of all these antibodies should be re-evaluated.
CD300 receptors could function as individual receptors recruiting transmembrane and/or cytoplasmic mediators but also, as part of complexes with other CD300 receptors, and more importantly with their respective signaling molecules. From all the CD300 interactions detected biochemically we have focused at a functional level on those involving CD300b, CD300c, and CD300f receptors. We observed an increased signaling capacity of CD300c when complexed with CD300b. This increase could be explained by the usage of CD300c signaling mediators by CD300b: cross-linking of CD300c in the CD300b/c transfectant would lead to FcϵRγ recruitment and subsequent ITAM phosphorylation by Lyn tyrosine kinase, which is abundantly expressed in RBL-2H3 cells. This in turn would induce Syk to bind the FcϵRγ-phosphorylated ITAMs and the propagation of the triggering signal to the cytoplasm. In this environment, in which tyrosine kinases accumulate at the inner cell membrane, the CD300b Grb2-recruiting motif would be more easily phosphorylated by increasing the positive signaling. Something similar could be happening in the CD300b/CD300f transfectant. CD300f receptor is able to promote both activating and inhibitory events, but the way the duality is regulated is not understood yet (22). CD300b signaling was highly increased in the presence of CD300f. We have suggested that CD300f would be preferentially associated to activating mediators in basal conditions.5 Thus, CD300b signaling on CD300b/f complexes could be higher as per the cross-talk with CD300f triggering mediators: PI3K and FcϵRγ (22, 23).
Altogether, our results indicate that the CD300 family of immune receptors may constitute a new mechanism by which myeloid cells could precisely control immune responses. It is widely known that the formation of multisubunit receptor complexes represents an effective way to modify the nature, intensity, and duration of cell processes. In this context, CD300 inhibitory clusters would facilitate tonic inhibition processes, CD300 activating receptor complexes would be useful for creating long lasting stimulating signals and/or increase their potency, whereas the formation of mixed activating and inhibitory complexes would make possible their termination or attenuation.
Supplementary Material
Acknowledgments
We thank Dr. Aroa Ejarque and Dr. Jordi Minguillón for helpful discussion and critically reading the manuscript and Dr. Roser Corominas for assistance with statistical analysis.
This work was supported in part by Plan Nacional I+D Grants SAF2004-00972 and SAF2009-07548, Fondo de Investigaciones Sanitarias Grant PI080366, and Agencia de Gestio d'Ajuts Universitaris i de Recerca de Catalunya (AGAUR) (2009 SGR 493).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
Á. Martínez-Barriocanal, E. Comas-Casellas, S. Schwartz, Jr., M. Martín, and J. Sayós, unpublished results.
- ITAM
- immune receptor tyrosine-based activation motif
- DAP-12
- DNAX-activating protein of 12 kDa
- ER
- endoplasmic reticulum
- TREM
- triggering receptor expressed by myeloid cells
- Ab
- antibody.
REFERENCES
- 1.Veillette A., Latour S., Davidson D. (2002) Annu. Rev. Immunol. 20, 669–707 [DOI] [PubMed] [Google Scholar]
- 2.Long E. O. (2008) Immunol. Rev. 224, 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomasello E., Bléry M., Vély F., Vivier E. (2000) Semin. Immunol. 12, 139–147 [DOI] [PubMed] [Google Scholar]
- 4.Humphrey M. B., Lanier L. L., Nakamura M. C. (2005) Immunol. Rev. 208, 50–65 [DOI] [PubMed] [Google Scholar]
- 5.Underhill D. M., Goodridge H. S. (2007) Trends Immunol. 28, 66–73 [DOI] [PubMed] [Google Scholar]
- 6.Chang C., Dietrich J., Harpur A. G., Lindquist J. A., Haude A., Loke Y. W., King A., Colonna M., Trowsdale J., Wilson M. J. (1999) J. Immunol. 163, 4651–4654 [PubMed] [Google Scholar]
- 7.Crocker P. R. (2005) Curr. Opin. Pharmacol. 5, 431–437 [DOI] [PubMed] [Google Scholar]
- 8.Dietrich J., Nakajima H., Colonna M. (2000) Microbes Infect. 2, 323–329 [DOI] [PubMed] [Google Scholar]
- 9.Vilches C., Parham P. (2002) Annu. Rev. Immunol. 20, 217–251 [DOI] [PubMed] [Google Scholar]
- 10.Moretta L., Moretta A. (2004) EMBO J. 23, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barclay A. N., Brown M. H. (2006) Nat. Rev. Immunol. 6, 457–464 [DOI] [PubMed] [Google Scholar]
- 12.Colonna M. (2003) Nat. Rev. Immunol. 3, 445–453 [DOI] [PubMed] [Google Scholar]
- 13.Speckman R. A., Wright Daw J. A., Helms C., Duan S., Cao L., Taillon-Miller P., Kwok P. Y., Menter A., Bowcock A. M. (2003) Hum. Genet. 112, 34–41 [DOI] [PubMed] [Google Scholar]
- 14.Clark G. J., Ju X., Azlan M., Tate C., Ding Y., Hart D. N. (2009) Immunobiology 214, 730–736 [DOI] [PubMed] [Google Scholar]
- 15.Cantoni C., Bottino C., Augugliaro R., Morelli L., Marcenaro E., Castriconi R., Vitale M., Pende D., Sivori S., Millo R., Biassoni R., Moretta L., Moretta A. (1999) Eur. J. Immunol. 29, 3148–3159 [DOI] [PubMed] [Google Scholar]
- 16.Bachelet I., Munitz A., Moretta A., Moretta L., Levi-Schaffer F. (2005) J. Immunol. 175, 7989–7995 [DOI] [PubMed] [Google Scholar]
- 17.Munitz A., Bachelet I., Eliashar R., Moretta A., Moretta L., Levi-Schaffer F. (2006) Blood 107, 1996–2003 [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Errico D., Aguilar H., Kitzig F., Brckalo T., Sayós J., López-Botet M. (2004) Eur. J. Immunol. 34, 3690–3701 [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Barriocanal A., Sayós J. (2006) J. Immunol. 177, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 20.Aguilar H., Alvarez-Errico D., García-Montero A. C., Orfao A., Sayós J., López-Botet M. (2004) J. Immunol. 173, 6703–6711 [DOI] [PubMed] [Google Scholar]
- 21.Brckalo T., Calzetti F., Pérez-Cabezas B., Borràs F. E., Cassatella M. A., López-Botet M. (2010) Eur. J. Immunol. 40, 722–732 [DOI] [PubMed] [Google Scholar]
- 22.Alvarez-Errico D., Sayós J., López-Botet M. (2007) J. Immunol. 178, 808–816 [DOI] [PubMed] [Google Scholar]
- 23.Izawa K., Kitaura J., Yamanishi Y., Matsuoka T., Kaitani A., Sugiuchi M., Takahashi M., Maehara A., Enomoto Y., Oki T., Takai T., Kitamura T. (2009) J. Immunol. 183, 925–936 [DOI] [PubMed] [Google Scholar]
- 24.Jackson D. G., Hart D. N., Starling G., Bell J. I. (1992) Eur. J. Immunol. 22, 1157–1163 [DOI] [PubMed] [Google Scholar]
- 25.Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. (1985) Mol. Cell. Biol. 5, 3610–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayós J., Martín M., Chen A., Simarro M., Howie D., Morra M., Engel P., Terhorst C. (2001) Blood 97, 3867–3874 [DOI] [PubMed] [Google Scholar]
- 27.Hedin K. E., Bell M. P., Kalli K. R., Huntoon C. J., Sharp B. M., McKean D. J. (1997) J. Immunol. 159, 5431–5440 [PubMed] [Google Scholar]
- 28.Geng L., Pfister S., Kraeft S. K., Rudd C. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11527–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark G. J., Cooper B., Fitzpatrick S., Green B. J., Hart D. N. (2001) Tissue Antigens 57, 415–423 [DOI] [PubMed] [Google Scholar]
- 30.Clark G. J., Green B. J., Hart D. N. (2000) Tissue Antigens 55, 101–109 [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto M., Takatsu H., Ohno H. (2006) Int. Immunol. 18, 1499–1508 [DOI] [PubMed] [Google Scholar]
- 32.Izawa K., Kitaura J., Yamanishi Y., Matsuoka T., Oki T., Shibata F., Kumagai H., Nakajima H., Maeda-Yamamoto M., Hauchins J. P., Tybulewicz V. L., Takai T., Kitamura T. (2007) J. Biol. Chem. 282, 17997–18008 [DOI] [PubMed] [Google Scholar]
- 33.Nakano T., Tahara-Hanaoka S., Nakahashi C., Can I., Totsuka N., Honda S., Shibuya K., Shibuya A. (2008) Mol. Immunol. 45, 289–294 [DOI] [PubMed] [Google Scholar]
- 34.Yamanishi Y., Kitaura J., Izawa K., Matsuoka T., Oki T., Lu Y., Shibata F., Yamazaki S., Kumagai H., Nakajima H., Maeda-Yamamoto M., Tybulewicz V. L., Takai T., Kitamura T. (2008) Blood 111, 688–698 [DOI] [PubMed] [Google Scholar]
- 35.Cantoni C., Ponassi M., Biassoni R., Conte R., Spallarossa A., Moretta A., Moretta L., Bolognesi M., Bordo D. (2003) Structure 11, 725–734 [DOI] [PubMed] [Google Scholar]
- 36.Márquez J. A., Galfré E., Dupeux F., Flot D., Moran O., Dimasi N. (2007) J. Mol. Biol. 367, 310–318 [DOI] [PubMed] [Google Scholar]
- 37.Marth J. D., Grewal P. K. (2008) Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sansom D. M., Walker L. S. (2006) Immunol. Rev. 212, 131–148 [DOI] [PubMed] [Google Scholar]
- 39.Chiesa S., Tomasello E., Vivier E., Vély F. (2005) Mol. Immunol. 42, 477–484 [DOI] [PubMed] [Google Scholar]
- 40.Rivera J., Fierro N. A., Olivera A., Suzuki R. (2008) Adv. Immunol. 98, 85–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudd C. E., Schneider H. (2003) Nat. Rev. Immunol. 3, 544–556 [DOI] [PubMed] [Google Scholar]
- 42.Zamoyska R. (1998) Curr. Opin. Immunol. 10, 82–87 [DOI] [PubMed] [Google Scholar]
- 43.Wienands J. (2000) Curr. Top. Microbiol. Immunol. 245, 53–76 [DOI] [PubMed] [Google Scholar]
- 44.Wu H., Kwong P. D., Hendrickson W. A. (1997) Nature 387, 527–530 [DOI] [PubMed] [Google Scholar]
- 45.Springer T. A., Wang J. H. (2004) Adv. Protein Chem. 68, 29–63 [DOI] [PubMed] [Google Scholar]
- 46.Shields M. J., Assefi N., Hodgson W., Kim E. J., Ribaudo R. K. (1998) J. Immunol. 160, 2297–2307 [PubMed] [Google Scholar]
- 47.Dimasi N., Flot D., Dupeux F., Márquez J. A. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bork P., Holm L., Sander C. (1994) J. Mol. Biol. 242, 309–320 [DOI] [PubMed] [Google Scholar]
- 49.Santiago C., Ballesteros A., Tami C., Martínez-Muñoz L., Kaplan G. G., Casasnovas J. M. (2007) Immunity 26, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanier L. L., Corliss B., Wu J., Phillips J. H. (1998) Immunity 8, 693–701 [DOI] [PubMed] [Google Scholar]
- 51.Smith K. M., Wu J., Bakker A. B., Phillips J. H., Lanier L. L. (1998) J. Immunol. 161, 7–10 [PubMed] [Google Scholar]
- 52.Alvarez Y., Tang X., Coligan J. E., Borrego F. (2008) Mol. Immunol. 45, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrow A. D., Trowsdale J. (2006) Eur. J. Immunol. 36, 1646–1653 [DOI] [PubMed] [Google Scholar]
- 54.Nakajima T., Inagaki N., Tanaka H., Tanaka A., Yoshikawa M., Tamari M., Hasegawa K., Matsumoto K., Tachimoto H., Ebisawa M., Tsujimoto G., Matsuda H., Nagai H., Saito H. (2002) Blood 100, 3861–3868 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.