Abstract
Rcl is a potential anti-angiogenic therapeutic target that hydrolyzes the N-glycosidic bond of 2′-deoxyribonucleoside 5′-monophosphate, yielding 2-deoxyribose 5-phosphate and the corresponding base. Its recently elucidated solution structure provided the first insight into the molecular basis for the substrate recognition. To facilitate the development of potent and specific inhibitors of Rcl, the active site was probed by site-directed mutagenesis and by the use of substrate analogs. The nucleobase shows weak interactions with the protein, and the deoxyribose binding pocket includes the catalytic triad Tyr-13, Asp-69, and Glu-93 and the phosphate binding site Ser-87 and Ser-117. The phosphomimetic mutation of Ser-17 to Glu prevents substrate binding and, thus, abolishes the activity of Rcl. The synthetic ligand-based analysis of the Rcl binding site shows that substitutions at positions 2 and 6 of the nucleobase as well as large heterocycles are well tolerated. The phosphate group at position 5 of the (deoxy)ribose moiety is the critical binding determinant. This study provides the roadmap for the design of small molecules inhibitors with pharmacological properties.
Keywords: Cancer Tumor Promoter, Enzyme Inhibitors, Hydrolases, Nucleoside Nucleotide Analogs, Protein Phosphorylation
Introduction
Rcl is a c-Myc target (1) that becomes tumorigenic when the metabolic environment of the cell is permissive (2). Its participation in tumorigenesis is supported by several pieces of evidence as follows. rcl is up-regulated in several cancers such as human prostate, breast cancers, and chronic lymphocytic leukemia (3–5). rcl is repressed in response to histone deacetylase inhibitors, anticancer agents that induce tumor cell death, differentiation, and/or cell cycle arrest (6). rcl is over-expressed in response to chronic administration of the synthetic glucocorticoid methylprednisolone or estrogen diethylstilbestrol. Altogether these data indicate that Rcl has a role in cell growth and/or cell proliferation (7, 8). Moreover, a causal role of Rcl in breast tumorigenesis was suggested as the up-regulation of Rcl correlates with the tumor grade (4). Thus, Rcl is a potential therapeutic target. However, its function remains to be determined.
We have shown that Rcl is a 2′-deoxynucleoside 5′-phosphate N-hydrolase that had not been previously described (9). Rcl hydrolyzes 2′-deoxyribonucleoside 5′-monophosphate (dNMP)4 to form a free nucleobase moiety (N) and 2-deoxyribose 5-phosphate. 2-Deoxyribose 5-phosphate may be converted to 2-deoxyribose, a downstream mediator of thymidine phosphorylase (also known as the angiogenic factor endothelial cell growth factor 1, ECGF1) that regulates tumor angiogenesis and progression (10). 2-Deoxyribose is also a substrate for glycolysis through its conversion to glyceraldehyde 3-phosphate, which may indirectly increase the development and malignancy of cancer cell overexpressing Rcl.
Rcl belongs to the nucleoside 2-deoxyribosyltransferase (NDT) family (EC 2.4.2.6; pfam 05014). NDT catalyzes the reversible transfer of the deoxyribosyl moiety from a deoxynucleoside donor to an acceptor nucleobase (11). On the contrary, Rcl is a hydrolase (or a transferase using only water molecule as an acceptor) (9). Both NDT and Rcl are specific for 2′-deoxyribose-containing substrates, whereas the corresponding riboside analogs are inhibitors. Their amino acid sequences display an overall low level of identity but share the catalytic triad composed of a tyrosine, an aspartate, and a glutamate, suggesting a similar mode of action (9, 12).
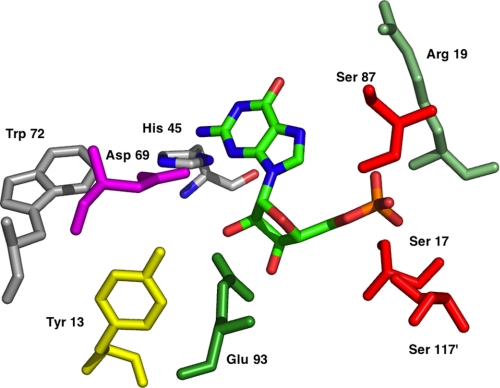
The structure of Rcl in solution was recently described (13, 14). Rcl is a symmetric homodimer; each monomer is composed of five buried β-strands alternating with five α-helices. The global architecture of the active site in Rcl resembles that of known NDTs. It is mainly composed of the N-terminal region from one monomer and a few residues from second monomer. The overall orientation of the nucleotide in Rcl appeared to be similar to that of the bound nucleoside in NDTs. It is composed of three main parts that recognize each moiety of the ligand; that is, the nucleobase, the deoxyribose, and the phosphate group (specific for Rcl).
The nucleobase shows weak interactions with the protein and is largely accessible to solvent. The deoxyribose binding pocket is central and includes the catalytic triad (Tyr-13, Asp-69, and Glu-93). The phosphate binding site is reminiscent of other phosphate binding proteins (15), and hydrogen bonds with Ser-17, Arg-19, Ser-87, and Ser-117 were suggested.
Here, we report the analysis of the contribution of the different amino acids participating at the active site. The binding determinants of the active site were probed with various synthetic nucleotide analogs. These studies defined chemical groups that are important for molecular recognition and will guide the rational design of inhibitors. Furthermore, a phosphomimetic mutation at serine 17 abolishes Rcl activity, suggesting that phosphorylation of Ser-17 plays an important role in Rcl regulation.
EXPERIMENTAL PROCEDURES
Chemicals
2′-Deoxyguanosine 5′-monophosphate, adenosine 3′,5′-cyclic-monophosphate, adenosine 5′-monosulfate, 8-bromoadenosine 5′-monophosphate, and xanthosine 5′-monophosphate were purchased from Sigma. 2-Fluoroadenine-9-β-d-arabinofuranoside 5′-monophosphate (Fludara®) was from Schering. N2-Ethyl-2′-deoxyguanosine 5′-monophosphate, N7-methyl-2′-deoxyguanosine 5′-monophosphate, glyoxal-2′-deoxyguanosine 5′-monophosphate, and O6-methyl-2′-deoxyguanosine 5′-monophosphate were from Jena Bioscience GmbH. 5-Aminoimidazole-4-carboxamide-1-β-d-ribofuranoside 5′-monophosphate and acyclovir monophosphate were from Carbosynth. Adenosine 5′-monophosphorothioate was from BioLog.
General Synthetic Methods
Reagents and anhydrous solvents were obtained from commercial suppliers and used without further purification. Flash chromatography was performed using silica gel 60 (Merck). Preparative HPLC were carried out on a PerkinElmer Life Sciences system (200 Pump) with a C18 reverse phase column (Kromasil, 5 μm 100 Å, 150 × 4.5 mm) using a flow rate of 5.5 ml/min and a linear gradient of acetonitrile (A) in 10 mm triethylammonium acetate buffer (B) at pH 7 over 20 min. 1H, 13C, and 31P NMR spectra were recorded on a Bruker Avance 400 spectrometer operating at 400.13, 100.62, and 161.62 MHz, respectively. High resolution mass spectra were recorded on a Waters Q-TOF micro MS instrument using a mobile phase of acetonitrile/water with 0.1% formic acid. Structure of synthesized compounds was confirmed by NMR and mass spectrometry.
2′,3′-O-Isopropylidene adenosine 5′-monophosphate (1) was synthesized via phosphorylation of commercial 2′,3′-O-isopropylidene adenosine according to reported procedures (16). The product (50 mg, 0.16 mmol) was coevaporated three times with dry pyridine, then a 1 m solution of 2-cyanoethylphosphate in pyridine (320 μl, 0.32 mmol) was added, and the mixture was coevaporated twice with dry pyridine. The residue was dissolved in dry pyridine (1.6 ml) and N,N′-dicyclohexylcarbodiimide (198 mg, 0.96 mmol) was added at room temperature. The mixture was kept stirring for 24 h at room temperature. Water was added to the mixture, insolubles were filtered, and the solvent was removed under vacuum. The cyanoethyl group was removed by treatment with a 1% solution of NaMeO in methanol at room temperature for 2 h. The mixture was neutralized by the addition of a resin Dowex H+, filtered, and concentrated. The crude product was purified by reversed phase HPLC (10–20% A in B) to give, after repeated freeze-drying, compound 1 in a 65% yield.
Adenosine-5′-carboxylic acid (2) was synthesized via oxidation of the 5′-hydroxyl group of 2′,3′-O-isopropylidene adenosine as reported (17). Isopropylidene was then removed by treatment with a solution of HCOOH/H2O (70:30) at 35 °C for 12 h. Purification by reversed phase HPLC (5–30% A in B) afforded compound 2 in a 85% yield.
N7-Deaza-adenosine 5′-monophosphate (N7-deaza-AMP) was prepared from commercial 7-deaza-adenosine (tubercidin). First, the 2′ and 3′ hydroxyl groups were protected by an isopropylidene acetal group with 2,2-dimethoxypropane and catalytic amounts of p-toluene sulfonic acid as previously described (18). Then, 7-deaza-2′,3′-O-isopropylidene-adenosine was 5′-phosphorylated as for compound 1, and the cyanoethyl and isopropylidene groups were removed as described above. Purification by reversed phase HPLC (5–15% A in B) afforded N7-deaza-AMP. N9-Deaza-dGMP was a gift of Professor Ioan Lascu (Université de Bordeaux-2, Institut de Biochimie et Génétique Cellulaires.
Construction of the Rcl Mutants
RclE93A, Rcl D69A, and Rcl D69N were previously described in Ghiorghi et al. (9) and Yang et al. (13) respectively. Oligonucleotides + (Y13A+, Y13F+, S17A+, S17E+, R19A+, H45A+, S87A+, S87D+, E93Q+, S117A+, S117N+) and T7term, oligonucleotides − (Y13A−, Y13F−, S17A−, S17E−, R19A−, H45A−, S87A−, S87D−, E93Q−, S117A−, S117N−) and T7prom (Table 1) were used in two separate PCR reactions using plasmid pET28aRcl as DNA template. The parameters used 1 cycle of 5 min at 95 °C, 25 cycles of 30 s at 95 °C, 30 s at 53 °C, and 30 s at 72 °C, and 1 cycle of 10 min at 72 °C. The annealing temperature was dependent on the pairs of oligonucleotides used. Oligonucleotides T7prom and T7term were used in a second PCR using aliquots of the first one using the same parameters as above with the exception of an annealing temperature of 61 °C.
TABLE 1.
Oligonucleotides used to generate the Rcl variants
| Y13A+ GGCTCCATGCTCCGTGGCCTTCTGCGGGAGC |
| Y13A− GCTCCCGCAGAAGGCCACGGAGCATGGAGCC |
| Y13F+ CCATGCTCCGTGTTCTTCTGCGGGAGCATC |
| Y13F− GATGCTCCCGCAGAAGAACACGGAGCATGG |
| S17A+ CCGTGTACTTCTGCGGGGCCATCCGCGGCGGGCGCG |
| S17A− CGCGCCCGCCGCGGATGGCCCCGCAGAAGTACACGG |
| S17E+ GTGTACTTCTGCGGGGAAATCCGCGGCGGGCGCG |
| S17E− CGCGCCCGCCGCGGATTTCCCCGCAGAAGTACAC |
| R19A+ TTCTGCGGGAGCATCGCCGGCGGGCGCGAGGAC |
| R19A− GTCCTCGCGCCCGCCGGCGATGCTCCCGCAGAAG |
| H45A+ GGTGCTCACTGAGGCCGTGGCTGATGCTGAG |
| H45A− CTCAGCATCAGCCACGGCCTCAGTGAGCACC |
| S87A+ GGAAGTGACACAGCCAGCCTTGGGTGTTGGC |
| S87A− GCCAACACCCAAGGCTGGCTGTGTCACTTCC |
| S87D+ GGAAGTGACACAGCCAGACTTGGGTGTTGGC |
| S87D− GCCAACACCCAAGTCTGGCTGTGTCACTTCC |
| E93Q+ GGGTGTTGGCTATCAACTGGGCCGGG |
| E93Q− CCCGGCCCAGTTGATAGCCAACACCC |
| S117A+ GTCTGGCCGAGTGCTTGCCGCCATGATCCGCGG |
| S117A− CCGCGGATCATGGCGGCAAGCACTCGGCCAGAC |
| S117N+ GTCTGGCCGAGTGCTTAACGCCATGATCCGCGG |
| S117N− CCGCGGATCATGGCGTTAAGCACTCGGCCAGAC |
| S158A GCGGATCCTCAGGCACTTGGGTGACTGGCGGAAGCCGTCTTCTG |
| S158E CCCAAGCTTTCAGGCACTTGGGTGACTCTCGGAAGCCGTCTTCTGAGG |
The amplified DNA fragments were purified by using the QIAquick PCR purification kit (Qiagen) and then digested with NdeI and BamHI enzymes over 2 h at 37 °C and repurified. Each PCR product was ligated with plasmid pET28a that had been digested with the same restriction enzymes. The ligation mixtures were used to transform strain DH5α. Plasmids with the correct sequence were used to transform strain Bli5.
The construction of the RclS158A and RclS158E mutants was done by using T7prom and oligonucleotides S158A or S158E, respectively. Each pair of oligonucleotides was used in a PCR reaction, and the amplified products were digested with NdeI and BamHI and with NdeI and HindIII, respectively, purified, and cloned into plasmid pET28a that had been digested with the same enzymes.
Overexpression and Purification of the N-terminal His-tagged Rcls
The different pET28aRcl plasmids were used to transform strain Bli5. Culture conditions and induction were performed as described by Ghiorghi et al. (9). Frozen cells resuspended in 40 ml of extraction/wash buffer (50 mm Na2HPO4, NaH2PO4, 300 mm NaCl pH 7.0) were broken by using a French press at 14,000 p.s.i. The lysate was centrifuged at 25,000 × g for 30 min at 4 °C. The supernatant was loaded on a column containing 6 ml of TALON (BD Bioscience) resin that had been previously equilibrated with the same buffer. After washing, Rcl was eluted with 150 mm imidazole. Fractions containing Rcl were pooled and dialyzed against sodium phosphate buffer (50 mm Na2HPO4, NaH2PO4, pH 6.0). The purity was checked by SDS-PAGE electrophoresis and by measuring the specific activity. Purified His-tagged Rcls in 50 mm sodium phosphate buffer, pH 6.0, were stored at −20 °C.
Kinetic Measurements
The enzyme activity was determined spectrophotometrically by incubating the enzyme with dGMP and by following the production of 2-deoxyribose 5-phosphate (one of the reaction products) as described previously (9). The initial velocity of the reaction was measured either at a variable concentration of dGMP both in the absence and presence of inhibitors or at a fixed concentration of dGMP and variable concentrations of inhibitors, allowing the investigation of the nature of the inhibition.
Isothermal Titration Calorimetry (ITC)
ITC was performed in a MicroCal VP-ITC calorimeter at 25 °C. Protein samples were prepared as indicated above. After thermal equilibration, titrant additions were made at 600-s intervals to the 1.41-ml protein samples by adding 5-μl aliquots of 10 mm GMP to protein samples ([RCLwt] = 257 μm, [Y13F] = 259 μm, [D69N] = 440 μm, [E93Q] = 250 μm, [S11A] = 400 μm, and [S17E] = 507 μm) in 25 mm Na2HPO4, NaH2PO4, pH 6.0, 25 mm NaCl, 2 mm β-mercaptoethanol.
The heat of dilution obtained from injecting the ligand into buffer was subtracted before fitting. Heat effects were integrated with ITC Origine 7.0 software, and a best fit was found using a single binding-site model.
RESULTS
Catalytic Activities of Rcl Mutants
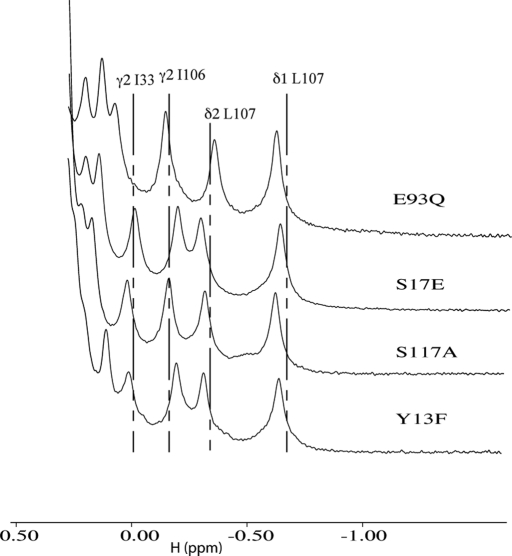
Based on the NMR structure of Rcl complex with GMP, the amino acids that participate at the active site (Fig. 1) and are potentially involved in the catalytic mechanism of the hydrolysis of 2′-deoxynucleoside 5′-monophosphate were chosen for mutagenesis. The residues Tyr-13, Ser-17, Arg-19, His-45, Asp-69 (previously described in Yang et al. (13)), Trp-72, Ser-87, Glu-93, and Ser-117 were replaced either by an alanine or by the most conservative amino acid. One-dimensional NMR spectra of Rcl and of the different mutants were recorded to control their structure (Fig. 2). The shifted methyl resonances of the Rcl mutants are very similar to those observed in Rcl wild type, indicating that the core domains of the proteins make similar residue-residue interactions. Furthermore, small angle x-ray scattering experiments indicate that Rcl mutants display the same overall shape and dimeric form as the wild type (supplemental data and supplemental figure). As dGMP was the best substrate of the native enzyme, it was chosen to measure the activity of the mutants. The results are summarized in Table 2. Mutants Y13A, D69A, S87A, and E93A did not abolish the catalysis but caused a large decrease in enzyme activity (≥98%). The side chains of Ser-17, Arg-19, and His-45 did not significantly influence the catalytic activity. Mutant W72F has a reduced catalytic activity due to an increase of Km for dGMP, indicating that it may be involved in the base stacking but is not essential for catalytic activity. Mutants E93Q and S117A showed no detectable activity. kcat values showed less than a 2-fold variation if Asp-69 and Ser-87 were excepted (4- and 5-fold). In contrast, the Km values varied from 50 μm to 10 mm.
FIGURE 1.
Schematic representation of the active site of Rcl.
FIGURE 2.
600 MHz one-dimensional NMR spectra of E93Q, S17E, S117A, and Y13F Rcl mutants. The shifted methyl resonances chemical shifts of Rcl wild type are indicated by the dotted lines.
TABLE 2.
Kinetic parameters of wild-type and Rcl mutants with dGMP as substrate
Vmax and Km were obtained from double reciprocal plots of initial velocity measurements. At least five different concentrations of dGMP were used. ND, not detectable.
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 | μm | m−1·s−1 | |
| WT | 0.0298 | 48 | 621 |
| Y13A | 0.0429 | 10,000 | 4.5 |
| Y13F | 0.0267 | 4,000 | 6.3 |
| S17A | 0.0273 | 74 | 545 |
| R19A | 0.0168 | 50 | 569 |
| H45A | 0.0412 | 109 | 885 |
| D69A | 0.0033 | 260 | 10.7 |
| D69N | 0.0066 | 890 | 7.4 |
| W72F | 0.0309 | 171 | 180 |
| S87A | 0.0042 | 435 | 9.1 |
| E93A | 0.0495 | 8,500 | 4.3 |
| E93Q | ND | ND | ND |
| S117A | ND | ND | ND |
Binding of GMP
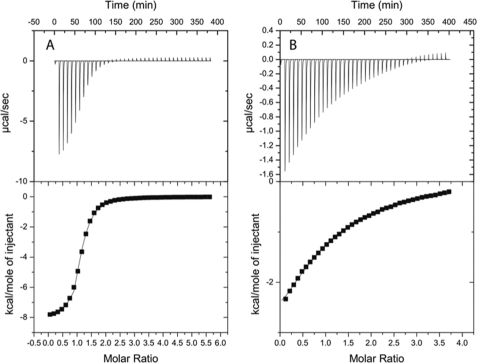
To further characterize the mutants and in particular those without any detectable activity (E93Q and S117A), the dissociation constant of Rcl and mutants with GMP was determined by ITC (Table 3). The Kd value of Rcl with GMP was 10.6 ± 0.4 μm (Fig. 3), similar to that described by Doddapaneni et al. (14). The Y13F mutant shows a Kd value comparable with Rcl wild type, whereas their Km values for dGMP differ by a factor of 80. Asp-69 contributes marginally to the binding of GMP as the Kd value was only reduced by a factor of three as compared with the wild-type enzyme. For both mutants E93Q and S117A the Kd value was enhanced by a factor of 14 and 30, respectively. These low Kd values indicate that these two amino acids are the major contributors to the GMP binding.
TABLE 3.
Dissociation constants Kd of Rcl and of different mutants
All energies reported as kcal/mol. ND, not detectable.
| Kd | ΔH | ΔS | ΔG | |
|---|---|---|---|---|
| μm | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | |
| RCL WT | 10.6 ± 0.4 | −8.11 ± 0.05 | −4.44 | −6.79 |
| Y13F | 11.0 ± 0.5 | −7.96 ± 0.05 | −4.00 | −6.78 |
| D69N | 33.3 ± 1.3 | −6.96 ± 0.06 | −2.86 | −6.11 |
| E93Q | 143.7 ± 8.6 | −4.84 ± 0.18 | +1.33 | −5.24 |
| S117A | 363.8 ± 14.6 | −4.61 ± 0.18 | +0.29 | −4.70 |
| S17E | ND | ND | ND | ND |
FIGURE 3.
ITC for Rcl binding to GMP. A, Rcl wild type is shown. The protein concentration in the calorimeter cell was 257 μm, and the titrant was 10 mm. B, Rcl S117A is shown. The protein concentration was 400 μm, and the GMP was 10 mm. Both experiments were carried out in 25 mm phosphate sodium, pH 6.0, 25 mm NaCl, 2 mm β-mercaptoethanol at 25 °C.
The GMP binding is enthalpy-driven with ΔH values from 7 to 8 kcal·mol−1 for the Rcl wild-type, Y13F, and D69N mutants. There is a loss in ΔH of ∼3–4 kcal·mol−1 for both S117A and E93Q mutants, consistent with their weaker binding interactions. In the latter, the positive ΔS value indicates an increased degree of freedom in the complex.
Effect of Replacement of the Phosphorylated Serine Residues of Rcl by Alanine or Glutamate (Phosphomimetic)
Phosphorylated peptides corresponding to human Rcl were identified by mass spectrometry in several proteomic studies (19–23). Three phosphorylated peptides on three different serine residues (indicated in bold in Fig. 4) were identified. Two of these, serine Ser-17 and Ser-158, are conserved in the rat Rcl. Replacement of the two serines by alanine did not affect either the affinity for dGMP or the catalytic efficiency. A similar observation was made with the S158E change. On the contrary, the S17E change has a profound effect on the enzyme activity, which was no longer detectable (Table 4). In addition, no GMP binds to the S17E mutant as determined by microcalorimetry (Table 3). This suggests that phosphorylation of Ser-17 negatively regulates the activity of Rcl.
FIGURE 4.
Conservation of Rcl amino-acids sequences in eucaryotes. The catalytic triad YDE is indicated in bold characters. The phosphorylated serine residues in the human Rcl are shadowed and underlined.
TABLE 4.
Kinetic parameters of wild-type and Rcl mutated at putative phosphorylated site
ND, not detectable.
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 | μm | m−1·s−1 | |
| WT | 0.0297 | 48 | 621 |
| S17A | 0.0273 | 74 | 545 |
| S17E | ND | ND | ND |
| S158A | 0.0184 | 48 | 385 |
| S158E | 0.0171 | 59 | 313 |
Modified Deoxyribonucleotides as Substrates
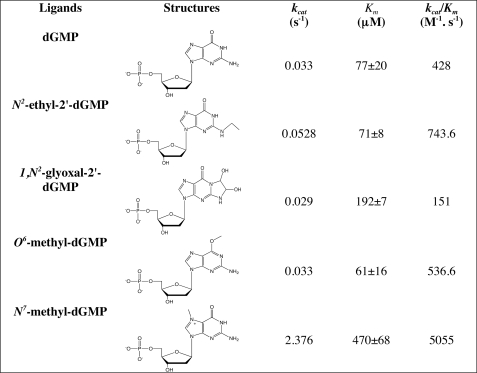
As guanine showed little interaction with Rcl, different purines modified by endogenous oxidants, alkylating agents, or endogenous aldehydes (24), found in cancerous cells as DNA adducts, were tested. We previously showed that 8-oxo-dGMP was neither a substrate nor an inhibitor of Rcl (9). As an example of alkylated dNMPs, N2-ethyl-dGMP, O6-methyl-dGMP, and N7-methyl-dGMP were chosen in addition to 1,N2-glyoxal adduct (Table 5). kcat values varied only by a factor of 2 at the exception of N7-methyl-dGMP (factor 70). Km values were also very close except for glyoxal-dGMP (factor 3) and N7-methyl-dGMP (factor 7). It is concluded that Rcl may accommodate several substitutions at positions 2, 6, and 7 and also large heterocycles. The enhanced kcat value with N7-methyl-dGMP is explained by the protonation of the nucleobase, creating a permanent positive charge and increasing the C1′-N9 bond cleavage reactivity.
TABLE 5.
Modified purine deoxyribonucleotides as Rcl substrates
Rcl Inhibition by Nucleotides with Various Nucleobases
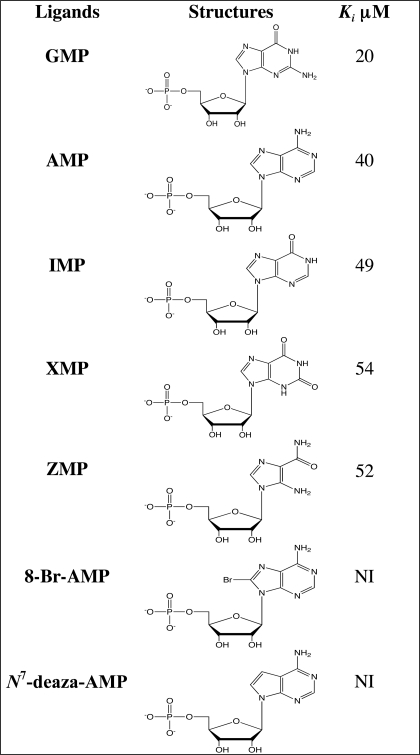
GMP and 6-methylthio-GMP were previously reported as inhibitors of the hydrolysis of dGMP by Rcl with Ki values of 20 and 10 μm, respectively. Four other naturally ribonucleotides of the de novo purine nucleotides pathway, AMP, IMP, xanthosine 5′-monophosphate, and 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranosyl 5′-monophosphate, display similar inhibitory constants around 50 μm (Table 6).
TABLE 6.
Ribonucleosides 5′-monophosphate as potential ligands; modifications on the nucleobase
NI, no inhibition; XMP, xanthosine 5′-monophosphate; ZMP, 5-aminoimidazole-4-carboxamide 1-β-D-ribofuranosyl 5′-monophosphate.
As N9-deaza-dGMP is not a substrate for Rcl, it was tested as a potential inhibitor in addition to N7-deaza-AMP. The absence of inhibition by N7-deaza-AMP (Table 6) indicates that N7 may participate in the binding. The absence of inhibition by N9-deaza-dGMP (not shown) may be explained either by the presence of a proton at position 7, which again indicates that N7 may participate in the binding, or by the shortening (of about 10%) of the C1′-C9 bond in N9-deaza-dGMP versus the C1′-N9 bond. In both cases the determining factor may be ionization and/or electron densities.
Substituted derivatives at position 8 of the nucleobase are not allowed, as illustrated by 8-Br-AMP and previously by 8-oxo-dGMP (9). According to the structure, any substituent at position 8 would point toward the phosphate group, leading to steric clashes.
Rcl Inhibition by Nucleotides with Modified Sugar
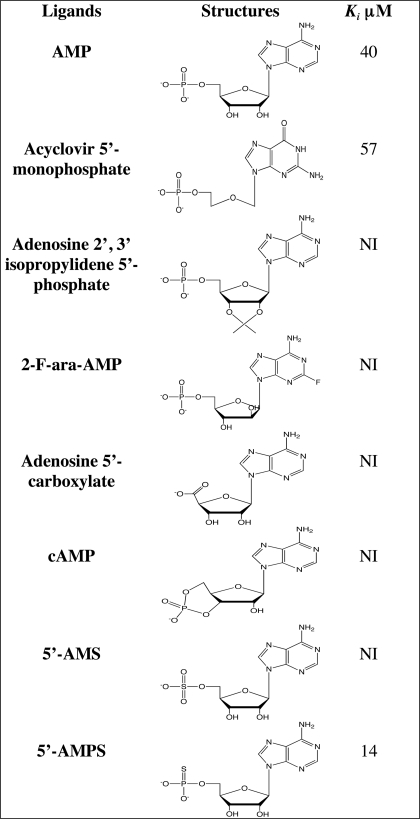
As previously shown, substitution of ribose by an acyclic chain diminishes the Ki by a factor of three, indicating that the 2′ and 3′ OH groups of the ribose contribute to the binding but are not essential. The addition of an isopropylidene group crossing at the 2′ and 3′ positions of the ribose abolishes the inhibition.
The orientation of the OH group at position 2′ of the deoxyribose is important because the chemotherapeutic agent 2-F ara-AMP, used for the treatment of chronic lymphocytic leukemia, is neither a substrate nor an inhibitor of the reaction. This may not be due to the presence of the fluor atom at position 2 of the nucleobase as Rcl accepts substitution at position 2 as demonstrated with N2-ethyl-dGMP (Table 7). The presence of the phosphate group is also important because cAMP is neither a substrate nor an inhibitor of Rcl activity and because phosphate cannot undergo substitution with a carboxylate.
TABLE 7.
Ribonucleosides 5′-monophosphate as potential ligands; modifications on the sugar part
NI, no inhibition; 2-F-ara-AMP, fludarabine; 5′-AMS, adenosine 5′-monosulfate; 5′-AMPS, adenosine 5′-monophosphorothioate.
Sulfate is considered to be a good phosphate mimic as it is isosteric and has a similar charge distribution and a net negative charge at physiological pH. However, adenosine 5′-monosulfate has no inhibitory effect. This result may be explained either by the differences of pKa between the sulfate and the phosphate group or by the difference of size between sulfur and phosphorus. Substitution of the α oxygen of AMP by sulfur (5′-AMPS) results in a diminution of the Ki by a factor of 3, which might be due to their difference of pKa: 6.8 (AMP) versus 5.3 (5′-AMPS).
DISCUSSION
The anti-apoptotic protein Rcl was described as an enzyme with unprecedented activity. Rcl is a hydrolase that catalyzes the cleavage of the N-glycosidic bond of dNMP yielding 2-deoxyribose 5-phosphate and the corresponding nucleobase. Rcl may be considered as another catabolic enzyme that cleaves dNMP, contributing to the maintenance of a balanced pool of nucleotides in addition to 5′-nucleotidases (25) but with a different activity. We hypothesize that understanding Rcl substrate specificity and catalytic mechanism should help in elucidating its cellular role. Its solution structure was recently solved and has provided insights into the molecular basis for substrate recognition (13, 14). Tyr-13, Glu-93, and His-45 are located in the vicinity of the 3′-OH of the ribose. The side chains of Asp-69 and Trp-72 are in close contact with the 2′ position. A positively charged pocket formed by Ser-87, Ser-117′, Ser-17, and Arg-19 may constitute the phosphate binding site. Mutagenesis of these amino acids to alanine or to conservative amino acids clearly shows that Glu-93 and Ser-117 are the main contributors to substrate binding and to catalysis. The differences of Kd values between variants Rcl E93Q and Rcl S117A for GMP suggest that phosphate binding is primordial.
The low catalytic activity of Rcl with different 2′-deoxyribonucleoside 5′-monophosphates as substrates led us to investigate other ligands. DNA damage plays a major role in mutagenesis, carcinogenesis, and aging. DNA is subject to modifications by various endogenous and exogenous chemicals, and modified guanine, resulting from oxidation or alkylation processes, is frequently found in cancerous cells (26, 27). These DNA lesions are subject to a network of DNA repair mechanisms that generally excise the modified base (28). However, in some cases the modified base can be recycled as illustrated by 8-oxo-2′-deoxyguanosine and 8-oxo-guanine (29). We hypothesized that modified dNMPs could be present during this cycle and that Rcl could be involved in their catabolism. The presence of an ethyl or a glyoxal group at position 2 or of a methyl at position 6 of the guanine ring has no significant influence on the affinity and on the catalytic efficiency. The addition of a methyl group at position 7 results in the formation of a reactive cation that enhances depurination of the 2′-deoxyribonucleotide. Thus, none of the modified deoxyribonucleoside 5′-monophosphates tested is a specific substrate of Rcl. The question of the role of Rcl in the maintenance of a balanced pool of nucleotides or in the catabolism of anomalous deoxynucleotides remains unanswered.
Nevertheless, the fact that they are all substrates, even if they have different affinities, confirms that the active site is open, making relatively smooth interactions with the nucleobase. The Ki values for Rcl inhibition by nucleotides with various nucleobases lead to similar conclusions. As observed by Holguin and Cardinaud (30) with purine as substrates for trans-N-deoxyribosylase, substitutions at positions 2 and 6 are of minor importance, whereas the imidazole moiety is less tolerant. The position 8 does not accept substituents for steric reasons.
The recognition of the sugar part displays some similitude with NDT but also important differences. Although not absolutely required, the OH groups at positions 2′ and 3′ of the ribose are important. If they can be suppressed, as in the case of acyclovir 5′-monophosphate, no additional groups can be introduced at these two positions. The configuration of the sugar is also crucial as fludarabine does not compete with dGMP. This differs from the situation of NDT where 2′-fluoro-2′-deoxyarabinonucleosides inhibit the transferase activity. 2′-Fluoro-2′-deoxyarabinonucleosides have allowed trapping of the NDT-DFDAP (2,6-diamino-9-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-9H-purine) covalent intermediate and identification of Glu-98 as the nucleophile (31), whereas no Rcl-deoxyribose 5-phosphate intermediate was found.
A positively charged pocket composed of Ser-87, Ser-117′, Ser-17, and Arg-19 was proposed to form hydrogen bonds with the negative charges of the phosphate group. The selectivity of the recognition of the phosphate group is high as it cannot be replaced by a carboxylate or a sulfate group, and the strength of the binding is influenced by the net charge.
Several proteomic studies have shown that Rcl is phosphorylated on three different serine residues Ser-12, Ser-28, and Ser-169 (19–23). According to the rat Rcl structure, Ser-12 and Ser-169 are located in flexible regions that do not interact with the core structure of the protein. In rat these two flexible regions can be deleted without affecting the enzymatic activity (13). According to this, the phosphomimetic S158E has the same affinity for dGMP as the wild type and a comparable catalytic efficiency. Whether these flexible regions and the corresponding phosphorylated serine are involved in protein-protein interactions (22), protein stability, or cellular localization remains to be determined. Ser-28 is conserved in all Rcl identified so far. Mutation of the corresponding rat serine (Ser-17) to an alanine has no consequence, whereas its change to glutamic acid, to mimic phosphorylation, completely abolishes Rcl activity. Ser-17 is located in the loop between the β1 strand and the α1 helix and is, thus, accessible to protein kinases. CKII was predicted to phosphorylate rat Ser4–158, cdc2, or ATM (ataxia telangiectasia mutated) human S169 (32). As no protein kinase was proposed for the phosphorylation of the two other serines, further studies are, thus, required to identify them. It will also be important to determine whether the expression of Rcl is cell cycle-regulated and if its phosphorylation state varies during the cycle. The identification of a phosphorylated peptide in only M phase-arrested cells supports this hypothesis (19).
The cell deoxyribonucleotide triphosphate pools are regulated by a network of enzymes involved in their synthesis (de novo and salvage pathways) and in their degradation (nucleotidases). Deregulation of this control leads to imbalance pools, which has consequences on DNA replication fidelity, maintenance of the nuclear DNA, and cell death (33, 34). dNTPs levels are generally elevated in actively dividing cells compared with normal cells (35), and their accumulation could be one of the events involved in the mutator phenotype in cancer (36).
An increase in the nucleotide pool during phase S is essential for cell proliferation (37). c-Myc directly activates genes involved in purine and pyrimidine biosynthesis, and a deregulation of c-Myc leads to an increase in nucleotide pools (38) (39). Paradoxically, c-Myc also activates rcl expression (1), whose product hydrolyzes dNMP.
However, it has to be mentioned that dNTP pools are also regulated by enzymes of the salvage pathway. Deoxycytidine (dCK) and deoxyguanosine kinases are constitutively expressed (40), and dCK activity is positively regulated by phosphorylation (41). Both enzyme activities are significantly elevated in cell lines as they start to proliferate (42), and deoxyguanosine kinase was shown to be relocated from the mitochondrial matrix to the cytosol at the early step of apoptosis (43). Thus, the anti-apoptotic role of Rcl could be attributed to a counter activity of the deoxycytidine and deoxyguanosine kinases to maintain cellular homeostasis.
Supplementary Material
Acknowledgments
We thank Valérie Huteau, Amandine Cohen, Wen Luo for their technical contribution, Gilles Labesse for heplful discussions and small angle x-ray scattering experiments, Pr. Ioan Lascu for N9-deaza-dGMP, and Jason Hargreaves and Yves Janin for proofreading. Small angle x-ray scattering experiments were recorded on the beamline SWING in SOLEIL (Saint-Aubin, France) with the kind help of Javier Perez.
This work was supported by the Institut Pasteur (Direction des applications de la recherche et des relations industrielles) and CNRS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and a figure.
- dNMP
- 2′-deoxyribonucleoside 5′-monophosphate
- NDT
- nucleoside 2-deoxyribosyltransferase
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1.Lewis B. C., Shim H., Li Q., Wu C. S., Lee L. A., Maity A., Dang C. V. (1997) Mol. Cell. Biol. 17, 4967–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis B. C., Prescott J. E., Campbell S. E., Shim H., Orlowski R. Z., Dang C. V. (2000) Cancer Res. 60, 6178–6183 [PubMed] [Google Scholar]
- 3.Rhodes D. R., Barrette T. R., Rubin M. A., Ghosh D., Chinnaiyan A. M. (2002) Cancer Res. 62, 4427–4433 [PubMed] [Google Scholar]
- 4.Shin S., Bosc D. G., Ingle J. N., Spelsberg T. C., Janknecht R. (2008) J. Cell. Biochem. 105, 866–874 [DOI] [PubMed] [Google Scholar]
- 5.Gorgun G., Ramsay A. G., Holderried T. A., Zahrieh D., Le Dieu R., Liu F., Quackenbush J., Croce C. M., Gribben J. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6250–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peart M. J., Smyth G. K., van Laar R. K., Bowtell D. D., Richon V. M., Marks P. A., Holloway A. J., Johnstone R. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3697–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almon R. R., DuBois D. C., Jusko W. J. (2007) Endocrinology 148, 2209–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miki Y., Suzuki T., Tazawa C., Ishizuka M., Semba S., Gorai I., Sasano H. (2005) Cancer Lett. 220, 197–210 [DOI] [PubMed] [Google Scholar]
- 9.Ghiorghi Y. K., Zeller K. I., Dang C. V., Kaminski P. A. (2007) J. Biol. Chem. 282, 8150–8156 [DOI] [PubMed] [Google Scholar]
- 10.Brown N. S., Bicknell R. (1998) Biochem. J. 334, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminski P. A. (2002) J. Biol. Chem. 277, 14400–14407 [DOI] [PubMed] [Google Scholar]
- 12.Armstrong S. R., Cook W. J., Short S. A., Ealick S. E. (1996) Structure 4, 97–107 [DOI] [PubMed] [Google Scholar]
- 13.Yang Y., Padilla A., Zhang C., Labesse G., Kaminski P. A. (2009) J. Mol. Biol. 394, 435–447 [DOI] [PubMed] [Google Scholar]
- 14.Doddapaneni K., Mahler B., Pavlovicz R., Haushalter A., Yuan C., Wu Z. (2009) J. Mol. Biol. 394, 423–434 [DOI] [PubMed] [Google Scholar]
- 15.Hirsch A. K., Fischer F. R., Diederich F. (2007) Angew. Chem. Int. Ed. Engl. 46, 338–352 [DOI] [PubMed] [Google Scholar]
- 16.Tener G. M. (1961) J. Am. Chem. Soc. 83, 159–168 [Google Scholar]
- 17.Epp J. B., Widlanski T. S. (1999) J. Org. Chem. 64, 293–295 [DOI] [PubMed] [Google Scholar]
- 18.Mizuno Y., Ikehara M., Watanabe K. A., Suzaki S., Itoh T. (1963) J. Org. Chem. 28, 3329–3331 [Google Scholar]
- 19.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 22.Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 23.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 24.Burcham P. C. (1999) Mutat. Res. 443, 11–36 [DOI] [PubMed] [Google Scholar]
- 25.Hunsucker S. A., Mitchell B. S., Spychala J. (2005) Pharmacol. Ther. 107, 1–30 [DOI] [PubMed] [Google Scholar]
- 26.De Bont R., van Larebeke N. (2004) Mutagenesis 19, 169–185 [DOI] [PubMed] [Google Scholar]
- 27.Neeley W. L., Essigmann J. M. (2006) Chem. Res. Toxicol. 19, 491–505 [DOI] [PubMed] [Google Scholar]
- 28.Barnes D. E., Lindahl T. (2004) Annu. Rev. Genet. 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 29.Hah S. S., Mundt J. M., Kim H. M., Sumbad R. A., Turteltaub K. W., Henderson P. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11203–11208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holguin J., Cardinaud R. (1975) Eur. J. Biochem. 54, 515–520 [DOI] [PubMed] [Google Scholar]
- 31.Porter D. J., Merrill B. M., Short S. A. (1995) J. Biol. Chem. 270, 15551–15556 [DOI] [PubMed] [Google Scholar]
- 32.Huang H. D., Lee T. Y., Tzeng S. W., Horng J. T. (2005) Nucleic Acids Res. 33, W226–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunz B. A., Kohalmi S. E., Kunkel T. A., Mathews C. K., McIntosh E. M., Reidy J. A. (1994) Mutat. Res. 318, 1–64 [DOI] [PubMed] [Google Scholar]
- 34.Bebenek K., Roberts J. D., Kunkel T. A. (1992) J. Biol. Chem. 267, 3589–3596 [PubMed] [Google Scholar]
- 35.Traut T. W. (1994) Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 36.Loeb L. A. (2001) Cancer Res. 61, 3230–3239 [PubMed] [Google Scholar]
- 37.Mathews C. K. (2006) FASEB J. 20, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 38.Liu Y. C., Li F., Handler J., Huang C. R., Xiang Y., Neretti N., Sedivy J. M., Zeller K. I., Dang C. V. (2008) PLoS One 3, e2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mannava S., Grachtchouk V., Wheeler L. J., Im M., Zhuang D., Slavina E. G., Mathews C. K., Shewach D. S., Nikiforov M. A. (2008) Cell Cycle 7, 2392–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson S., Munch-Petersen B., Johansson K., Eklund H. (2002) Cell. Mol. Life Sci. 59, 1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smal C., Vertommen D., Bertrand L., Ntamashimikiro S., Rider M. H., Van Den Neste E., Bontemps F. (2006) J. Biol. Chem. 281, 4887–4893 [DOI] [PubMed] [Google Scholar]
- 42.Fyrberg A., Albertioni F., Lotfi K. (2007) Biochem. Biophys. Res. Commun. 357, 847–853 [DOI] [PubMed] [Google Scholar]
- 43.Jüllig M., Eriksson S. (2001) J. Biol. Chem. 276, 24000–24004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.