Abstract
The acid-sensing ion channels (ASICs) are proton-gated cation channels activated when extracellular pH declines. In rodents, the Accn2 gene encodes transcript variants ASIC1a and ASIC1b, which differ in the first third of the protein and display distinct channel properties. In humans, ACCN2 transcript variant 2 (hVariant 2) is homologous to mouse ASIC1a. In this article, we study two other human ACCN2 transcript variants. Human ACCN2 transcript variant 1 (hVariant 1) is not present in rodents and contains an additional 46 amino acids directly preceding the proposed channel gate. We report that hVariant 1 does not produce proton-gated currents under normal conditions when expressed in heterologous systems. We also describe a third human ACCN2 transcript variant (hVariant 3) that is similar to rodent ASIC1b. hVariant 3 is more abundantly expressed in dorsal root ganglion compared with brain and shows basic channel properties analogous to rodent ASIC1b. Yet, proton-gated currents from hVariant 3 are significantly more permeable to calcium than either hVariant 2 or rodent ASIC1b, which shows negligible calcium permeability. hVariant 3 also displays a small acid-dependent sustained current. Such a sustained current is particularly intriguing as ASIC1b is thought to play a role in sensory transduction in rodents. In human DRG neurons, hVariant 3 could induce sustained calcium influx in response to acidic pH and make a major contribution to acid-dependent sensations, such as pain.
Keywords: Acid Sensing Ion Channels (ASIC), Gene Structure, Ion Channels, Peptides, Toxins, Transcription, Calcium, Human, Proton-gated, Species Difference
Introduction
The acid-sensing ion channels (ASICs)4 are a family of proton-gated cation channels expressed in neurons throughout the central and peripheral nervous system (1). There are four ASIC genes (Accn1–4) that produce at least six individual ASIC subunits in rodents (ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3, and ASIC4). ASIC subunits have a characteristic topology with two transmembrane domains separated by a large cysteine-rich extracellular region (2, 3). Three individual ASIC subunits associate to form functional cation channels that are activated by decreases in extracellular pH (4, 5). ASIC currents are typically transient, inactivate even in the continued presence of acidic pH, and (except for ASIC1a) are not substantially permeable to calcium (6). The specific properties of an ASIC current, such as the pH necessary for activation and the kinetics of inactivation, are defined by the subunit composition of the channel (7). In the central nervous system, the ASIC1a subunit plays an important role (8). Genetic disruption or pharmacological inhibition of ASIC1a affects learning and memory, fear-related behaviors, pain, depression, and seizure duration in rodents (9–15). ASIC1a also contributes to neuronal damage after cerebral ischemia in mice and mediates neuronal death following prolonged acidosis (16–19). ASIC1a is thought to play a prominent role in neuronal death because it is uniquely permeable to calcium compared with other ASICs (16, 20, 21).
In rodents, the Accn2 gene encodes both ASIC1a and ASIC1b. ASIC1b is a transcript variant expressed predominately within the dorsal root ganglion (DRG) where it is thought to play a role in sensory transduction (22–24). ASIC1b and ASIC1a differ in the N-terminal third of their protein sequences, which are encoded by distinct exons (22, 23). This region of the protein encompasses the intracellular N terminus, first transmembrane domain, and a part of the extracellular domain. These regions are known to be important for kinase regulation, proton sensitivity, inactivation, and ion permeability (25–28). As expected, ASIC1b and ASIC1a display divergent channel properties and are distinct in their proton concentration-response curves, ion selectivity, and sensitivity to modulatory agents (22, 23, 29–31).
Although the distinct isoforms of rodent ASIC1 have been well studied, the transcript variants expressed from the human ACCN2 gene (which encodes ASIC1 subunits) have not been well defined (5, 23, 31, 32). It is clear that humans express a subunit homologous to mouse and rat ASIC1a (ACCN2 transcript variant 2), which has been referred to as both human ASIC1a and human ASIC1b (27, 32, 33). In addition, ACCN2 transcript variant 1 has been identified in humans (32). This variant is expected to produce a protein that differs from ASIC1a by the addition of 46 amino acids within the extracellular domain of the channel. The human genome sequence suggests the existence of a third ACCN2 transcript variant with a predicted amino acid sequence similar to rodent ASIC1b (23). In this article, we report the channel properties of the two unstudied human ACCN2 transcript variants 1 and 3. We find that many properties of ACCN2 transcript variant 3 (hVariant 3) are similar to rodent ASIC1b. However, hVariant 3 is calcium permeable and displays a small acid-dependent sustained current unlike rodent ASIC1b. Together, these results describe novel human acid-sensing ion channel isoforms and highlight the divergence of human and rodent ASICs.
EXPERIMENTAL PROCEDURES
ACCN2 Transcript Variants
The mouse ASIC1b (NCBI AB208022.1), human ACCN2 transcript variant 1 (NCBI NM_020039.2), and ACCN2 transcript variant 2 (NCBI NM_001095.2) were a generous gift from J. Wemmie, M. Price, and M. J. Welsh at the University of Iowa (13, 34–36). ACCN2 transcript variant 3 was identified using the TBLASTN program on the ENTREZ data base from NCBI to identify sequences within the human genome similar to the first 220 amino acids of mouse ASIC1b (23, 37). Primers were designed against the predicted start of this sequence and the shared stop region of hVariants 1 and 2 (forward 5′-aaaatgcccatccagatcttc-3′ and reverse 5′-tcagcaggtaaagtcctcgaac-3′) and were synthesized (Integrated DNA Technologies, Inc., Coralville, IA). Oligo(dT)-primed cDNA was made using the Invitrogen cDNA Synthesis Kit (Carlsbad, CA) from human DRG RNA purchased from Clontech (catalogue number 636150, Mountain View, CA). PCR was performed with the above primers as follows: 94 °C for 1 min, then 31 cycles of 94 °C for 30 s, 55 °C for 45 s, 72 °C for 4 min, and finally 72 °C for 2 min. This yielded a 1689-base pair fragment that was cloned into the pSTBlue-1 vector from Novagen (Merck KGaA, Darmstadt, Germany). The insert was then cloned into the pMT3 expression vector. Site-directed mutagenesis was used to generate the hVariant 3 (A93S) using the Stratagene QuikChange® mutagenesis kit (La Jolla, CA). All inserts were sequenced at the Plant-Microbe Genomics Facility at the Ohio State University prior to heterologous expression. Plasmid DNA was prepared from bacteria using Qiagen Midiprep kits (Valencia, CA).
Real Time Quantitative Polymerase Chain Reaction
Human DRG RNA was purchased from Clontech (catalog number 636150) and human total brain RNA was purchased from Agilent Technologies (Agilent Technologies, La Jolla, CA, catalog number 540005). Adult mouse total brain and DRG RNA were isolated using the Qiagen RNAeasy Mini kit. RNA was treated with the Ambion DNA-free Kit (Ambion, Austin, TX) to destroy possible contaminating genomic DNA. cDNA was then made from DNase-treated RNA using the Applied Biosystems SuperScript® VILOTM cDNA Synthesis Kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer's instructions. For each cDNA synthesis, a “no RT” reaction lacking reverse transcriptase was performed to produce template for the “−RT” control experiments. Real time PCR was performed using either Invitrogen SYBR GreenER qPCR SuperMix Universal (Invitrogen) or Applied Biosystems Power SYBR® Green PCR Master Mix in a 25-μl reaction volume. Primers used include: hVariant 2 forward 5′-tcagcttaccttccctgctg-3′ and reverse 5′-agctccccagcatgatacag-3′; hVariant 3 forward 5′-cccatccagatcttctgctc-3′ and reverse 5′-accccaaatatctcccaagg-3′; hVariant 1 forward 5′-gtggctccctatcatccaaaag-3′ and reverse 5′-atgtcaccaagacaacagggttt-3′; human GAPDH forward 5′-gatcatcagcaatgcctcct-3′ and reverse 5′-tgtggtcatgagtccttcca-3′; mouse ASIC1a forward 5′-ctgtaccatgctggggaact-3′ and reverse 5′-gctgcttttcatcagccatc-3′; mouse ASIC1b forward 5′-tgccagccatgtctttgtg-3′ and reverse 5′-cacaggaaggcacccagt-3′; mouse GAPDH forward 5′-acccagaagactgtggatgg-3′ and reverse 5′-ggatgcagggatgatgttct-3′. The PCR product amplified by each primer set was sequenced to ensure amplification of the target transcript. Real time PCR was performed using an Applied Biosystems StepOneplusTM Thermal Cycler and StepOneTM Plus software. Cycling conditions consisted of an initial activation at 95 °C for 10 min, followed by 40 cycles of two-step PCR; 95 °C for 15 s, 60 °C for 1 min, followed by a melt curve analysis. All reactions were performed in triplicate along with “no cDNA” controls (in which water was added instead of cDNA). A no RT control reaction was always done with each primer set on each batch of cDNA. Efficiency measurements were performed with all primer sets and primers displayed efficiencies calculated to be within 0.1 of each other using the equation e = 10(1/slope) − 1. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression to determine the ΔCt according to the manufacturer's instructions. Relative gene expression was determined using the ΔCt method.
Oocyte Expression and Two-electrode Voltage Clamp Electrophysiology
Xenopus laevis oocytes were harvested from female frogs purchased from Xenopus I (Dexter, MI), using standard procedures (38). Oocytes were stored in modified Barths solution with calcium (2.4 mm NaHCO3, 88 mm NaCl, 15 mm HEPES, 1 mm KCl, 0.8 mm MgSO4, 0.4 mm CaCl2, 0.3 mm Ca(NO3)2, and 125 units/liter of penicillin/streptomycin) for at least 2 h before injection. The animal pole of isolated oocytes was injected with plasmid DNA (∼5 ng from a 100 ng/μl of stock solution) using a PV820 pneumatic picopump (World Precision Instruments, Sarasota, FL). Injected oocytes were stored in modified Barths solution at 18 °C and recording was done 1–5 days after injection.
Whole cell macroscopic currents were recorded using two-electrode voltage clamp at a holding potential of −60 mV (38). Data were recorded using a Clamp OC-725 amplifier (Warner Instruments, Hamden, CT) and either a Powerlab 4SP digitizer with CHART software (ADInstruments, Colorado Springs, CO) or an Axon Digidata 1200 digitizer with pCLAMP-8 software (Molecular Devices, Sunnyvale, CA). Glass electrodes were pulled with a Sutter P-97 micropipette puller (Sutter Instrument Co., Novato, CA) and filled with 3 m KCl to yield 0.5–2 megaohm resistance. Acid did not induce currents in uninjected oocytes. Recordings determining the τinact, proton concentration-response, and steady-state desensitization were done in frog Ringers solution (116 mm NaCl, 2 mm KCl, 5 mm HEPES, 5 mm MES, 2 mm CaCl2, 1 mm MgCl2) adjusted to the indicated pH using 1 m NaOH. Recordings determining ion permeability in oocytes were performed in solutions containing 116 mm NaCl, LiCl, or KCl, 0.4 mm CaCl, 1 mm MgCl, 5 mm HEPES, 5 mm MES and adjusted to the indicated pH using 1 m NaOH, LiOH, or KOH as appropriate. During recordings, cells were incubated in basal pH 7.4 (unless otherwise indicated) for at least 30 s before activation. pH 5.0 solution produced maximal current for all ASICs recorded. Before experiments, multiple applications of pH 5.0 were done (generally 2–3) until the ASIC-current amplitude stabilized. For quantification, the current produced by test pH values was normalized to the average maximal current of flanking pH 5.0 applications to reduce the impact of potential tachyphylaxis (39). “Maximal” current was evoked by pH 5.0 from a holding potential of pH 7.4 for most experiments or 7.9 for experiments assessing steady-state desensitization. Big Dynorphin was synthesized from EZ Biolab (Carmel, IN). PcTx1 synthetic peptide was purchased from Peptide International Inc. (Louisville, KY).
Chinese Hamster Ovary Cell Transfection and Calcium Permeability Experiments
Calcium permeability studies in oocytes can be complicated by endogenous calcium-activated channels. Therefore, the whole cell patch clamp technique was used to measure calcium permeability of acid-evoked current in Chinese hamster ovary (CHO) cells transiently transfected with GFP and mASIC1b, hVariant 2, hVariant 3, hVariant 3 (A93S), or vector only (23). Briefly, trypsinized CHO cells (∼107 cells) were suspended in 0.4 ml of electroporation solution (120 mm KCl, 25 mm HEPES buffer, 10 mm K2HPO4, 10 mm KH2PO4, 2 mm MgCl2, 0.15 mm CaCl2, 5 mm EGTA, and 2 mm MgATP, pH 7.6) and mixed with 20 μg of plasmid DNA containing ASIC expression vector (or empty pcDNA3.1 vector as indicated) and pEGFP-C1 vector (Clontech) at a 3:2 ratio. Cells were electroporated with the Gene Pulser Xcell system (Bio-Rad) and plated at a density of 35 cells/mm2 onto 10-mm coverslips in a 35-mm culture dish. Cells were used for patch clamping 2–3 days after transfection. Transfected cells were identified by expression of green fluorescent protein (GFP) (35). Data were collected at 5 kHz using an Axopatch 200B amplifier, Digidata 1322A, and Clampex 9.0 (Molecular Devices). Extracellular solutions contained either 160 mm NaCl or 80 mm CaCl2 with 20 mm HEPES buffer and 10 mm glucose. The pH was adjusted with NaOH or Ca(OH)2 as appropriate. Intracellular pipette solution (pH 7.4) contained 160 mm NaCl, 10 mm HEPES, and 10 mm EGTA. Pipette solution was allowed to dialyze with cytosolic contents for at least 5 min after whole cell access was gained before ion permeability was measured. The membrane potential was held constant at −50 mV between voltage ramp protocols. ASIC current was evoked by application of acidic extracellular solutions (pH 5.3) and a voltage ramp (−150 to 50 mV) was initiated immediately before the current decayed into the plateau phase. To isolate background conductance, an identical voltage ramp was performed before and after ASIC activation at holding pH 7.6. Mean background conductance was subtracted from conductance following acid application and data were fitted to a linear equation to determine the reversal potential of ASIC current. PNa/PCa was calculated from the change in reversal potentials when Na+ was replaced with Ca2+ in the extracellular solution (see below) using the equations: ΔErev = Erev,Na − Erev,Ca = (RT/F) ln(PNa[Na+]o/4PCa′[Ca2+]o) and PCa′ = PCa/(1 + eEF/RT), where the gas constant (R), Faraday constant (F), and temperature constant (T) have their standard meanings.
Data Analysis
Data were analyzed using either Axon Clampfit 9.0, or CHART and Excel (Microsoft, Redmond, WA). To measure the τ of inactivation (τinact), the decay phase of the indicated current was fit to the equation I = k0 + k1 × e−t/τinact. In proton concentration-response experiments the half-maximal pH (pH0.5) was calculated using the equation I/IpH,max = 1/{1 + (EC50/[H+])n} = 1/{1 + 10n(pH-pH0.5)}, where n is the Hill coefficient, EC50 is the proton concentration yielding half-maximal activation, and pH0.5 is the pH yielding half-maximal. Relative monovalent cation permeabilities of ASICs expressed in oocytes were determined using the equation: ΔErev = Erev(X) − Erev(Na+) = (RT/F)ln{Px[X]o/PNa+[Na+]o}. Erev(X) and Erev(Na+) are the experimentally determined reversal potentials of cation “X” and Na+, respectively, [X]o and [Na+]o are the extracellular concentrations of cation X and Na+, and the constants R, T, and F have their standard meanings. Statistical analysis was done with ANOVA (one-way) with post hoc Bonferroni's multiple comparison test or two-tailed Student's t test (paired or unpaired data) as indicated in the figure legends. A “p” value less than 0.05 was considered significant.
RESULTS
Human ACCN2 Transcript Variant Sequence and Expression
The human ACCN2 gene encodes two defined transcript variants: ACCN2 transcript variant 1 (GenBank NM_020039.2) and ACCN2 variant 2 (GenBank NM_001095.2) (Fig. 1). Human ACCN2 variant 2 (hVariant 2) is homologous to the ASIC1a subunit from rodents (5, 32). The properties of this human channel have been well studied in heterologous systems (13, 33, 40, 41). ACCN2 transcript variant 1 (hVariant 1) differs from hVariant 2 in apparent alternate 3′ splicing of coding exon 9, which results in a coding sequence containing an additional 138 in-frame nucleotides (Fig. 1, A and B) (32). hVariant 1 was originally isolated from human brain and is represented within the expressed sequence tag data base (AW015707) (32). hVariant 1 is predicted to produce a protein with an additional 46 amino acids located just prior to the proposed desensitization gate of the second transmembrane domain, a junction known to be critical for ASIC gating (Fig. 1C) (42–44). It is unknown how these additional amino acids are incorporated into the protein architecture and whether this subunit can form functional ASIC channels.
FIGURE 1.
Human ACCN2 transcript variants. A, schematic of the human ACCN2 gene located on chromosome 12 (at positions 9917749 to 9940609 of the genome assembly NW001838057.1). Coding exons are indicated by rectangles on a continuous line and numbered according to location. Common hVariant 1- and 2-specific exons are striped. The alternatively spliced portion of exon 9 (in 9b), which is unique in hVariant 1, is dotted. The hVariant 3-specific coding exon (3b) is dark gray. Note that the exon encoding the specific region of hVariant 3 is not yet annotated as an exon within the human genome reference sequence. B, schematic of the resulting transcript variants from the human ACCN2 gene. The numbers represent the coding exons illustrated above. hVariant 2 is homologous to rodent ASIC1a. hVariant 3 has sequence similarity to rodent ASIC1b. C, ClustalW alignment showing divergence in the predicted protein sequence of hVariant 1 and hVariant 2. Bold residues are conserved between hVariant 1 and hVariant 2. The amino acid sequence of the inserted region in hVariant 1 is illustrated at the top. The DEG residue, present just before the desensitization gate (at the “DIGG” sequence), is underlined in hVariant 2 (42). Notice the inclusion of an homologous glycine in the LG sequence (in bold) replicated within the inserted region just prior to the desensitization gate of hVariant 1. The predicted protein domains are indicated under the amino acid sequences. The gray bar represents the location of transmembrane domain 2, as predicted by the crystal structure (4, 42). The solid line represents the extracellular domain and the hatched box represents a β-sheet region (4). The intracellular region is indicated by the wavy line beneath the sequence. D, ClustalW alignment of the first third of the predicted protein sequences of hVariant 3, mouse ASIC1b (mASIC1b), and hVariant 2. Residues in bold are identical to amino acids in hVariant 3. Asterisk below represents identity between all three sequences. The colon or dot below represent conserved amino acids. Underline indicates the position of Ala93 in hVariant 3, which is a serine in polymorphism dbSNP: rs706792. The relative location of the intracellular region is indicated by the wavy line below the sequence. Transmembrane domain 1 is indicated by a gray box. Gray bars represent α helical regions and hatched bars represent β-sheet regions (4, 42).
To determine whether a homolog of mouse ASIC1b exists in humans, the first 220 amino acids of mouse ASIC1b (unique to 1b) were compared with the human genome using the TBLASTN program from NCBI. Similar to a previous report, a sequence that was 88% identical to the first 220 amino acids of mouse ASIC1b was identified on chromosome 12 (23). This sequence is located in the ACCN2 gene within a large region that separates the ASIC1a-specific coding exons 2 and 3 from coding exons 4–12 (Fig. 1A), suggesting it is an alternative coding exon for human ACCN2. However, this sequence is not currently present in the human expressed sequence tag data base, and therefore it is not known if the sequence is transcribed. Rodent ASIC1b is expressed predominantly in the DRG (22, 45). Therefore, we reasoned that this human transcript might also be expressed predominantly within the DRG. Using primers specific for the predicted start region of the alternate exon and the common stop region of ACCN2 transcript variants 1 and 2, we isolated a 1689-base pair PCR product from human DRG cDNA. This sequence displayed a continuous open reading frame of 563 amino acids (GenBank HM991481). We termed this transcript human ACCN2 transcript variant 3 (hVariant 3). The first 220 amino acids of the predicted hVariant 3 protein are 88% identical to the first 219 amino acids of mouse ASIC1b (Fig. 1D) and the remaining 343 amino acids are 100% identical to hVariant 2. Thus, humans have a novel transcript variant (hVariant 3) that is similar to rodent ASIC1b. End point PCR with primers specific to hVariant 3 suggests that it is expressed in a pattern similar to hVariant 2 in both neuronal and non-neuronal tissues (supplemental data and Fig. S1).
To determine the relative expression levels of each human ACCN2 variant, we employed quantitative real time PCR of cDNA made from human DRG and total brain (Fig. 2). hVariant 2 (the human homolog of mouse ASIC1a) is expressed within the DRG and, to a greater extent, in total brain (Fig. 2A). hVariant 3 is also expressed in human DRG and the relative expression level of hVariant 3 is 26.5 ± 2.6% of hVariant 2 (Fig. 2A). Thus, hVariant 3 represents a substantial proportion of the total ACCN2 transcripts in DRG. However, the relative expression level of hVariant 3 is significantly less in total brain (2.0 ± 0.1% of hVariant 2). hVariant 1 is expressed at a low level in both DRG and total brain (Fig. 2A). For comparison, the relative expression levels of mouse ASIC1a and ASIC1b transcripts were included (Fig. 2B). Like hVariant 2, mouse ASIC1a expression is more pronounced in total brain compared with DRG (Fig. 2B). Like hVariant 3, mouse ASIC1b is more abundantly expressed in the DRG compared with total brain (Fig. 2B). However, the relative level of mouse ASIC1b is 214.7 ± 23.8% of ASIC1a in the DRG. In fact, the level of ASIC1b exceeds ASIC1a in mouse DRG and, therefore, ASIC1b represents the dominant ASIC1 transcript in the DRG. This is in contrast to human Variant 3, which does not exceed hVariant 2 levels in DRG. Together, these data indicate that expression of hVariant 3 and mASIC1b are similar, as they are both substantially more abundant in the DRG compared with total brain. However, there are specific differences in the relative expression of ASIC1 transcripts between mice and humans.
FIGURE 2.
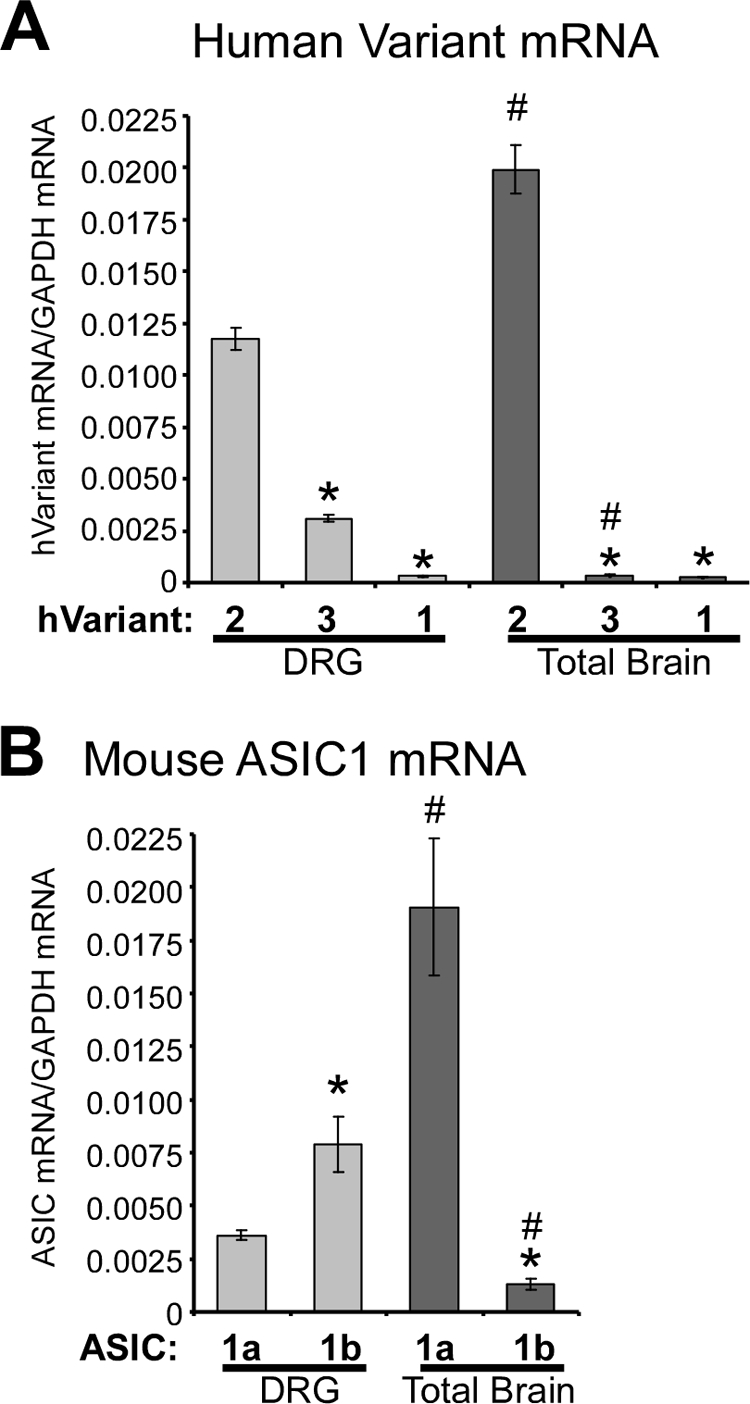
Expression analysis of human and mouse ASIC1 transcript variants. A, relative levels of human ACCN2 variant mRNA determined using quantitative real time PCR from human total brain and DRG. Data are presented as the relative level of the indicated transcript normalized to GAPDH (n = 3, see “Experimental Procedures” for details). B, relative levels of mouse ASIC1a and ASIC1b in mouse brain and DRG determined by quantitative real time PCR (n = 3). Error bars represent the mean ± S.E. The asterisk indicates p < 0.05 between hVariant 2 or mASIC1a in the same tissue; # indicates p < 0.05 compared with the same transcript in total brain using unpaired Student's t test.
Basic Properties of Human ACCN2 Transcript Variants
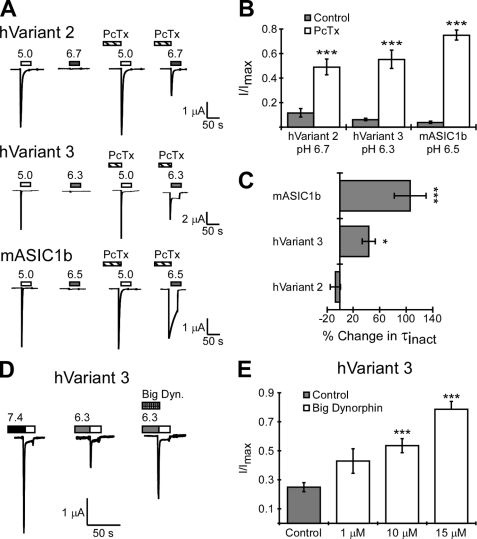
Expression plasmids encoding cDNA for each of the three human transcript variants from ACCN2 were injected into X. laevis oocytes and two-electrode voltage clamp was used to record acid-activated currents (38). Oocytes were also injected with mouse ASIC1b (mASIC1b) to compare with hVariant 3 because of their sequence similarity. hVariant 2, the human homolog of rodent ASIC1a, produced large inward sodium currents when the extracellular pH was reduced (Fig. 3, A and B). Oocytes injected with hVariant 1 never displayed acid-activated current (Fig. 3, A and B), even with extremely low pH values (pH 3.0 application, not shown). Because hVariant 1 failed to produce acid-activated current, it was not included in subsequent analysis. hVariant 3 produced acid-activated currents with a reduced amplitude on average compared with hVariant 2 (Fig. 3, A and B). Acid-gated currents from hVariant 3 also displayed distinctly different kinetics compared with acid-gated currents from hVariant 2. First, proton-gated currents from hVariant 3 inactivated faster than both hVariant 2 and mouse ASIC1b (τinact of hVariant 3 = 0.55 ± 0.04 s; τinact of hVariant 2 = 1.77 ± 0.10 s, and τinact of mASIC1b = 1.05 ± 0.09 s, p < 0.001 between hVariant 3 and 2 as well as between hVariant 3 and mASIC1b) (Fig. 3C). Currents from hVariant 3 also often showed incomplete inactivation distinguished by the presence of an acid-dependent sustained current following activation (Fig. 3D). However, this sustained current varied considerably between oocytes, similar to previous descriptions of other ASICs displaying incomplete inactivation (46), and did not achieve statistical significance. Monovalent ion permeability of hVariant 3 was also assessed in oocytes (Fig. 3, E–G). As with other ASICs, hVariant 3 showed a linear current/voltage relationship in the presence of potassium, lithium, or sodium. Quantification of the relative monovalent ion permeability revealed that hVariant 3 was more selective for sodium versus potassium (PNa/PK = 6.3 ± 0.6, n = 4) compared with hVariant 2 (PNa/PK = 4.2 ± 0.5, n = 4, p < 0.01 ANOVA). Like hVariant 3, the PNa/PK of mASIC1b was also more selective for sodium (PNa/PK mASIC1b = 7.6 ± 0.4), but was statistically different from hVariant 3 (n = 6, p < 0.05 compared with variant 3, ANOVA). The PNa/PLi was not different between the three subunits (PNa/PLi was 0.86 ± 0.09 for hVariant 3, 0.86 ± 0.03 for hVariant 2, and 1.07 ± 0.01 for mASIC1b).
FIGURE 3.
Basic properties of ACCN2 transcript variants. A, representative traces of proton-gated current using the two-electrode voltage clamp of X. laevis oocytes injected with hVariant 2, hVariant 1, hVariant 3, and mouse ASIC1b (mASIC1b). Channels were activated with pH 5.0 solutions (white bars) from a holding pH of 7.4. B, quantification of peak current amplitude of pH 5.0-activated currents in oocytes injected with human ACCN2 transcript variants and mouse ASIC1b. Note that hVariant 1 failed to produce appreciable proton-gated currents (n = 14- 20 oocytes). C, inactivation kinetics of ACCN2 transcript variants. The τ of inactivation was calculated by fitting the decay phase of pH 5.0-activated current to an exponential equation (n = 24- 45 oocytes). D, quantification of the acid-dependent sustained current in oocytes expressing ACCN2 transcript variants. Sustained current was measured during the plateau phase after pH 5.0-induced activation and normalized to the peak current amplitude (n = 13–19 oocytes). E–G, I/V plots of hVariant 2 (E), hVariant 3 (F), and mASIC1b (G) currents activated in the presence of different extracellular ions. pH 5.0-activated currents were measured at the indicated holding potential using solutions with either 116 mm Na+, 116 mm Li+, or 116 mm K+. I/I[Na+]max is the peak current amplitude evoked by pH 5.0 at the given holding potential normalized to the current amplitude evoked by pH 5.0 in the Na+ solution at a holding potential of −60 mV (n = 4–6). Error bars are mean ± S.E. The asterisk indicates p < 0.05; double asterisk indicates a p < 0.01; and a triple asterisk indicates a p < 0.001 using an ANOVA (one-way).
Proton Sensitivity of Human ASIC1 Variants
Most ASICs are activated by protons alone and display proton concentration-response curves that are specific to individual channel subtypes (5, 36, 47–49). Therefore, we analyzed the pH dependence of hVariant 2, hVariant 3, and mASIC1b gating. Similar to previous reports, hVariant 2 showed an appreciable current with pH 6.5 application (Fig. 4A). hVariant 3, however, produced little activation with pH 6.5. This response was similar to mASIC1b, which also required more acidic pH values for activation (Fig. 4A). A concentration-response curve showed that both hVariant 3 and mASIC1b were less sensitive to protons compared with hVariant 2 (Fig. 4B). The calculated pH to produce 50% activation (pH0.5act) of hVariant 3 was significantly more acidic compared with hVariant 2 (pH0.5act = 5.92 ± 0.03, n = 12 for hVariant 3; pH0.5act of 6.43 ± 0.03, n = 6 for hVariant 2; p < 0.001, ANOVA). Although mASIC1b is also less proton sensitive than hVariant 2, the pH0.5act of hVariant 3 was also significantly lower than that of mASIC1b (pH0.5act = 6.13 ± 0.03 for mASIC1b, n = 9, p < 0.001 compared with hVariant 3, ANOVA).
FIGURE 4.
pH-dependent activation and desensitization of ACCN2 transcript variants. A, representative trace of the pH dependence of activation in oocytes expressing hVariant 2, hVariant 3, and mASIC1b. The activating pH is indicated by bars above the trace. B, quantification of the pH dependence of current activation. Current was produced by perfusion of bath solution from pH 7.4 to the indicated pH. I/Imax is the current produced by application of the test pH normalized to current evoked by pH 5.0 solutions (n = 5–14). C, representative traces showing steady-state desensitization of hVariant 2, hVariant 3, and mASIC1b. Basal pH was 7.9. The shaded bars above the trace indicate the conditioning pH (applied for 2 min), and the white bars indicate activating pH 5.0. D, concentration-response curve of steady-state desensitization. I/Imax is the current evoked from experimental conditioning pH normalized to pH 5.0-evoked current evoked from a conditioning pH of 7.9 (n = 5–21). Error bars are mean ± S.E.
In addition to activation, ASIC subunits also display a difference in proton concentration dependence for steady-state desensitization (31, 38, 49). Steady-state desensitization is induced when channels are exposed to mildly acidic pH values (typically insufficient for channel activation). After exposure to such conditioning pH, the channels can enter a desensitized state and fail to activate when a more acidic stimulus is encountered. For hVariant 2, steady-state desensitization was induced with pH 6.9 conditioning, and complete desensitization was observed with pH 6.7 conditioning (Fig. 4C). hVariant 3 was not desensitized with pH 6.7, and complete steady-state desensitization was not induced even with pH 6.3 (Fig. 4C). mASIC1b also requires more acidic pH values compared with hVariant 2, but pH 6.3 completely desensitized the channel (Fig. 4C). A concentration-response curve indicated that more acidic pH values were required to induce steady-state desensitization of hVariant 3 compared with hVariant 2 (Fig. 4D). In fact, the pH0.5 for induction of steady-state desensitization (pH0.5des) was distinctly different between hVariant 2 and hVariant 3 (pH0.5des = 6.88 ± 0.02, n = 7 for hVariant 2; pH0.5des = 6.36 ± 0.03, n = 17 for hVariant 3, p < 0.001 ANOVA). mASIC1b also required more acidic pH values for steady-state desensitization compared with hVariant 2 (pH0.5des = 6.52 ± 0.01 for mASIC1b, n = 8, p < 0.001 ANOVA). Yet, the pH0.5des of hVariant 3 was also significantly lower than mASIC1b (p < 0.001 ANOVA). Thus, hVariant 3, like mASIC1b, requires a greater concentration of protons for activation and desensitization compared with the human homolog of ASIC1a (hVariant 2). However, there are distinct differences between hVariant 3 and mouse ASIC1b.
PcTx1 and Big Dynorphin Action on Human ACCN2 Transcript Variant 3
hVariant 3 displays characteristics similar to mouse ASIC1b. In fact, the amino acid sequence, inactivation kinetics, and apparent proton sensitivity appear more similar to mASIC1b than to hVariant 2. To further explore these similarities, we tested PcTx1 modulation. The venom peptide PcTx1 inhibits hVariant 2 (the human homolog of ASIC1a) by shifting the proton sensitivity of steady-state desensitization such that the channel becomes desensitized at neutral pH (30, 50, 51). However, PcTx1 can also enhance the apparent proton sensitivity of rodent ASIC1a, hVariant 2, and rodent ASIC1b activation such that the channels activate at less acidic pH values (30, 51) (Fig. 5A). Our data indicate that PcTx1 promotes activation of hVariant 3 with a less acidic pH, allowing the channels to activate at pH 6.3 (Fig. 5, A and B). This was observed even with low concentrations of PcTx1 (20 nm, data not shown). As has been reported previously, PcTx1 also slows inactivation of rodent ASIC1b (30), but did not affect inactivation of hVariant 2 (Fig. 5, A and C). Inactivation of hVariant 3 was also slowed by PcTx1 (Fig. 5, A and C).
FIGURE 5.
Modulation of human ACCN2 transcript variant 3 by PcTx1 and Big Dynorphin. A, representative traces showing PcTx1 enhancement of ASIC1 activation. Proton-gated currents were activated by the indicated pH (white bar indicates pH 5.0, gray bars indicate pH 6.7, 6.3, or 6.5 as indicated) in the presence or absence of PcTx1 (60 nm). The pH used for activation was different between channels. This is because PcTx1 enhances the current by shifting the apparent proton sensitivity of activation. Because the proton sensitivity of each channel is different, we chose a pH that produces <15% maximal activation on average (pH 6.7 induces 11.5% current from hVariant 2, pH 6.3 induced 6.1% current from hVariant 3, pH 6.5 induces 3.8% activation from mASIC1b). B, quantification of PcTx1 (60 nm) effect on activation by submaximal pH applications (n = 5–7). Triple asterisks indicates a p < 0.001 between PcTx1 and control conditions using Student's t test. C, quantification of the change in the τ of inactivation of pH 5.0-evoked current in the presence of PcTx1. Fractional change represents the difference in τinact (in seconds) with and without PcTx1 modulation normalized to the τinact without PcTx1. A value of zero would indicate no change with PcTx1 application. D, representative trace of Big Dynorphin (15 μm) modulation of hVariant 3 steady-state desensitization induced by conditioning in pH 6.3 and activating with pH 5.0 (white bar). E, quantification of Big Dynorphin modulation of steady-state desensitization of hVariant 3. I/Imax is current from control or Big Dynorphin normalized to control currents (evoked from pH 6.3 in the absence of any intervention). Triple asterisks indicates a p < 0.001 between Big Dynorphin (at the given concentration) and control conditions using Student's t test (n = 4–19). Error bars represent the mean ± S.E.
Another peptide modulator of ASIC1 activity that affects steady-state desensitization is Big Dynorphin (52). Big Dynorphin limits steady-state desensitization of mASIC1a and mASIC1b, allowing the channels to remain closed even during incremental decreases in pH. Thus, when sufficiently acidic pH is applied to activate the channel following conditioning, greater current is observed in the presence of Big Dynorphin (52). We found that micromolar concentrations of Big Dynorphin limited steady-state desensitization of hVariant 3 (Fig. 5, D and E). Thus, hVariant 3 and mASIC1b are similarly enhanced by peptide modulators.
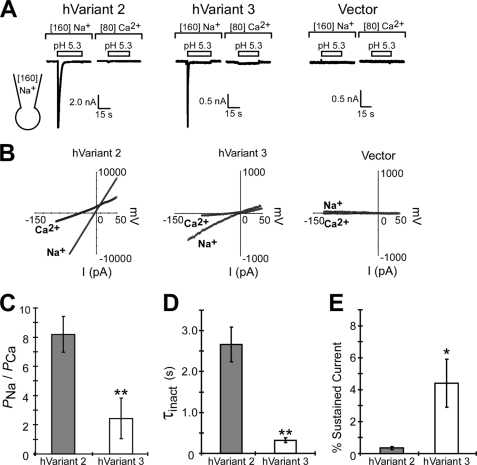
Calcium Permeability of ACCN2 Transcript Variant 3
Other ASICs have been reported to be relatively impermeant to calcium, making ASIC1a unique (5, 7, 23). To measure calcium permeability, we expressed hVariant 2 and hVariant 3 in CHO cells and used whole cell patch clamping to the measure current in extracellular solutions containing sodium or calcium as the permeant ion. In the calcium solutions, both hVariant 2 and hVariant 3 displayed a small transient current (Fig. 6A). hVariant 3 also displayed a small sustained current (Fig. 6A). No acid-evoked currents were observed in cells transfected with vector alone. Given that the extracellular solution contained calcium and the intracellular solution contained 160 mm sodium, the small inward currents suggest that both hVariant 2 and hVariant 3 are permeable to calcium. To test this hypothesis, the current/voltage relationship of proton-activated currents was measured in both the sodium and calcium-containing extracellular solutions and relative ion permeability was calculated (Fig. 6, B and C). hVariant 2 (the human homolog of rodent ASIC1a) had a PNa/PCa of 8.2 ± 1.2 (n = 7). Surprisingly, hVariant 3 had a PNa/PCa of 2.4 ± 1.4 (n = 9) indicating that this variant is substantially more calcium permeable than hVariant 2 (p = 0.009, Student's t test) (Fig. 6C). We observed similar calcium permeability with hVariant 3 (A93S), a polymorphism of hVariant 3 common in individuals of European decent (supplemental data and Fig. S2). Calcium permeability of mASIC1b under these conditions was expectedly low (PNa/PCa = 17.9 ± 4.7, n = 3, p = 0.002 compared with hVariant 3 using Student's t test, data not shown). The fact that hVariant 3 displays such low PNa/PCa is in striking contrast to rodent ASIC1b, which has been reported to be substantially less permeable to calcium (23). hVariant 3 currents in CHO cells also inactivated rapidly and incompletely indicating that transcript variants expressed in CHO cells produced currents with properties consistent with our previous oocyte studies (Fig. 6, D and E). In fact, the sustained acid-dependent current was more obvious when hVariant 3 was expressed in CHO cells and was statistically different from hVariant 2 (Fig. 6E). In addition, the fact that this calcium-permeable current is sustained (Fig. 6A), suggests that hVariant 3 could induce a continuous calcium influx in response to acidic extracellular pH in vivo.
FIGURE 6.
Calcium permeability of ACCN2 transcript variant 3 expressed in CHO cells. A, representative trace of proton-gated currents in CHO cells transfected with hVariant 2, hVariant 3, and vector (pMT3). Recordings were done using whole cell patch clamp with extracellular solutions containing 160 mm NaCl or 80 mm CaCl. Pipette solution contained 160 mm NaCl. B, representative I/V plot of voltage ramp (−100 to +50 mV) applied during the transient (inactivating) phase of pH 5.3-activated current. C, quantification of the average calculated sodium/calcium permeability ratio (PNa/PCa) of acid-activated currents from CHO cells expressing hVariant 2 and hVariant 3 (n = 7–9). D, quantification of the τ of inactivation (τinact) calculated from the decay phase of pH 5.3-activated proton-gated currents from CHO cells transfected with human hVariant2 or -3 in sodium-containing solutions. E, quantification of the sustained phase of pH 5.3-activated currents in CHO cells transfected with hVariants 2 or 3 in sodium-containing solutions. “% Sustained” current was calculated by measuring the residual current 10 s after activation and normalizing it to peak current amplitude. Asterisk indicates a p < 0.05 and double asterisk indicated a p < 0.02 using the Student's t test. Error bars are mean ± S.E.
DISCUSSION
Three human ACCN2 transcript variants are readily identifiable within the ENTREZ data base. hVariant 2 has been well studied and is homologous to ASIC1a in other species (5, 23, 31, 32). We report that hVariant 1, which contains a 46-amino acid insertion in the gating region near transmembrane domain 2, does not produce proton-gated currents when expressed alone in Xenopus oocytes. The area in which the additional 46 amino acids are inserted is known to be a site that profoundly affects gating of DEG/ENaCs, the superfamily of channels that includes ASICs (53, 54). In particular, the “DEG” residue is located adjacent to this region (Fig. 1C). When mutated, the DEG residue causes certain channels to remain constitutively open, resulting in neurodegeneration (43, 44, 55). Thus, the presence of the additional 46 amino acids in this location is particularly surprising. Whether this additional sequence would completely disrupt channel assembly or alter channel function is unknown. However, it is difficult to imagine how the channel could function with such a large addition within a region so critical for channel gating. Yet, transcripts similar to hVariant 1 are predicted to be present in other primate species (Pan troglodytes GenBank XM_509053.2; Macaca mulatta GenBank XM_002798568.1), but not in rodents. Interestingly, rabbits have an ASIC1 transcript variant with a similar 45-amino acid insertion at this same location (GenBank XM_002711143.1). For all species, the additional 45–46-amino acid sequence has no obvious similarity to other known mammalian proteins and the function for this additional sequence is not known. Our finding that hVariant 1 does not produce proton-gated currents under normal conditions suggests that either: 1) this subunit modulates other ASIC currents when present in heteromeric channels; 2) hVariant 1 is activated by another ligand yet to be discovered; or 3) hVariant 1 has an entirely different function unrelated to ion conduction. In addition, our data indicate that hVariant 1 is expressed within the CNS at very low levels. Thus, the role of hVariant 1 remains unknown.
We show that a third ACCN2 transcript variant, hVariant 3, is expressed in human DRG and encodes an ASIC subunit that produces functional homomeric proton-gated channels when expressed in heterologous systems. We also assessed the activity of a common human polymorphism in hVariant 3 that alters amino acid 93 (A93S). The allele containing this A93S polymorphism occurs in the majority of individuals of European descent but is not abundantly present in African or Asian populations (GenBank, dbSNP: rs706792 and rs706793). We find that the A93S substitution does not affect the basic biophysical characteristics of hVariant 3 when expressed in heterologous systems (supplemental data and Fig. S2). However, additional analyses are required to test for other effects of this common polymorphism.
Overall, our data support the conclusion that hVariant 3 is the human homolog of rodent ASIC1b. hVariant 3 shares sequence identity with rodent ASIC1b. hVariant 3 displays multiple biophysical properties that are more similar to rodent ASIC1b than hVariant 2 (ASIC1a). Furthermore, like rodent ASIC1b, hVariant 3 is more abundant in human DRG compared with the brain. However, there are some key differences between hVariant 3 and rodent ASIC1b, which suggest that hVariant 3 may play a distinct role in human physiology.
Among the ASIC subunits, only ASIC1a (hVariant 2) is reported to show appreciable calcium permeability. Here, we report that hVariant 3 is even more permeable to calcium than hVariant 2. This suggests that hVariant 3 is different from rodent ASIC1b, which displays very low permeability to calcium (23). The pre-transmembrane domain 1 region is responsible for the difference in calcium permeability between rodent ASIC1a and ASIC1b (23). Interestingly, there are multiple amino acid differences between hVariant 3 and mouse ASIC1b within this region (Fig. 1D) that may account for the difference in ion permeability. However, the specific amino acids that mediate calcium permeability are not known. Calcium-mediated current of hVariant 3 is also sustained and maintained as long as acidic conditions are present. Interestingly, such sustained currents are also observed in shark ASIC1b and mammalian ASIC3, another ASIC expressed more abundantly in DRG compared with brain (56–58). For ASIC3, the presence of sustained currents is mediated by the N-terminal intracellular domain and transmembrane domain 1 (59). Shark ASIC1b and ASIC3 also display “window currents,” sustained acid-dependent currents that occur at more neutral pH and are due to steady-state activation (56, 59). We saw no evidence of such currents with hVariant 3. However, hVariant 3 would be expected to produce a small, albeit continuous calcium influx under more acidic conditions. Such a current could have dramatic consequences for acid-induced signaling in DRG neurons.
In the DRG, acid-dependent sustained currents have been implicated in nociception (1, 57, 58). Local acidosis often accompanies many conditions that result in pain (inflammation, infection, ischemia, arthritis, bone disorders, and tumors) (1). Cutaneous acid-dependent pain in humans is inhibited by amiloride, a nonspecific blocker of ASIC channels (60). ASIC3, which is more abundantly expressed in the DRG than central nervous system, plays a role in acid-induced pain in rodents (57, 58, 62–65). ASIC1b in rodents has also been proposed to play a nociceptive role (22, 61). However, it should be noted that rodent ASIC1b is expressed in both nociceptive and non-nociceptive DRG neurons (24). Therefore, ASIC1b likely contributes to multiple types of sensory transduction in rodents. Our study suggests that hVariant 3 may not only affect DRG excitability through acid-dependent depolarization, but might also affect DRG neurons through changes in intracellular calcium. Furthermore, the relative levels of hVariant 3 in human DRG is different from ASIC1b in mouse DRG suggesting differential cellular expression between mice and humans. This might be expected if hVariant 3 displays increased calcium permeability and mediates different signaling pathways compared with mouse ASIC1b. In the central nervous system, ASIC1a-mediated calcium influx has been linked to neuronal death and the role of ASIC1a in neuronal signaling under physiological conditions (5, 8, 21, 66, 67). Calcium influx mediated by ASIC1a activation in central neurons affects dendritic spine density via calmodulin KII-dependent signaling mechanisms (68). In the DRG, calcium-permeable ion channels such as the TRPs can cause a multitude of effects and are involved in nociceptive neuron sensitization (69, 70). Calcium influx through hVariant 3 in DRG neurons could have similar dramatic consequences for sensory transduction. To date, there are few publications measuring acid-activated current in human DRG neurons (71, 72). Human neurons display transient acid-evoked responses that are not inhibited by TRPV1 antagonists (71, 72). In many neurons, this transient response is followed by a small acid-dependent sustained response (71). More interestingly, in some human DRG neurons, a second ionic conductance is observed after the cell is returned to neutral pH. This has been hypothesized to represent possible activation of a calcium-activated chloride channel, suggesting that activation of the acid-gated current altered intracellular calcium levels (71). However, the involvement of ASICs, and specifically hVariant 3, in these transient proton-gated currents in human DRG has not been directly assessed.
hVariant 3 may also mediate acid-induced calcium entry in non-neuronal cells. Interestingly, ASIC1b has been implicated in vascular smooth muscle cell physiology (73). Prolonged acidification affects vascular smooth muscles and contributes to baseline blood pressure and vascular disease (74). In rodents, ASIC1b is expressed in these cells and may mediate a depolarizing response to acidosis (73). We did observe some hVariant 3 expression in non-neuronal tissues (supplementary data and Fig. S1). However, the specific expression of hVariant 3 within human vascular smooth muscle cells has not been assessed. Yet, it is exciting to speculate that if hVariant 3 is expressed in smooth muscle cells, it could make a dramatic contribution to vascular tone through acid-evoked calcium entry (75).
Our results indicate that hVariant 3 is similar to rodent ASIC1b, but displays some unique characteristics. This suggests that the channel could have a novel role in human physiology compared with its rodent counterpart. Similar results have been observed for other ion channels expressed in dorsal root ganglion neurons. In particular, GABA receptors and voltage-gated sodium channels in human DRG neurons display unique channel properties distinct from currents measured in rodent neurons (76, 77). Thus, there are likely distinct differences between the ion channels that mediate sensory transduction in humans and rodents. Our data suggest that acid-induced signaling in human sensory neurons may be more complicated than in rodent neurons. Because of this, studies of human neurons and human tissues (or generation of knock-in mice) are essential to fully understand the role of the hVariant 3 and provide a foundation for further studies addressing the roles of ASIC1 in human health and disease.
Supplementary Material
Acknowledgments
We thank M. Welsh, J. Wemmie, and M. Price of the University of Iowa for kindly providing human ACCN2 transcript Variant 1, ACCN2 transcript Variant 2, and the mouse ASIC1b clones. Thanks to J. Enyeart for assistance with expression analysis of human variants. We also thank K. Mykytyn for editorial comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 NS062967.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. S1 and S2.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HM991481.
- ASIC
- acid-sensing ion channels
- ACCN
- amiloride-sensitive cation channel, neuronal
- hVariant
- human ACCN2 transcript variant
- DRG
- dorsal root ganglion
- ANOVA
- analysis of variance.
REFERENCES
- 1.Dubé G. R., Elagoz A., Mangat H. (2009) Curr. Pharm. Des. 15, 1750–1766 [DOI] [PubMed] [Google Scholar]
- 2.Benos D. J., Stanton B. A. (1999) J. Physiol. 520, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 4.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 5.Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 6.Waldmann R. (2001) Adv. Exp. Med. Biol. 502, 293–304 [DOI] [PubMed] [Google Scholar]
- 7.Waldmann R., Lazdunski M. (1998) Curr. Opin. Neurobiol. 8, 418–424 [DOI] [PubMed] [Google Scholar]
- 8.Wemmie J. A., Price M. P., Welsh M. J. (2006) Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 9.Wemmie J. A., Chen J., Askwith C. C., Hruska-Hageman A. M., Price M. P., Nolan B. C., Yoder P. G., Lamani E., Hoshi T., Freeman J. H., Jr., Welsh M. J. (2002) Neuron 34, 463–477 [DOI] [PubMed] [Google Scholar]
- 10.Wemmie J. A., Askwith C. C., Lamani E., Cassell M. D., Freeman J. H., Jr., Welsh M. J. (2003) J. Neurosci. 23, 5496–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziemann A. E., Allen J. E., Dahdaleh N. S., Drebot I. I., Coryell M. W., Wunsch A. M., Lynch C. M., Faraci F. M., Howard M. A., 3rd, Welsh M. J., Wemmie J. A. (2009) Cell 139, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coryell M. W., Wunsch A. M., Haenfler J. M., Allen J. E., Schnizler M., Ziemann A. E., Cook M. N., Dunning J. P., Price M. P., Rainier J. D., Liu Z., Light A. R., Langbehn D. R., Wemmie J. A. (2009) J. Neurosci. 29, 5381–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wemmie J. A., Coryell M. W., Askwith C. C., Lamani E., Leonard A. S., Sigmund C. D., Welsh M. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coryell M. W., Ziemann A. E., Westmoreland P. J., Haenfler J. M., Kurjakovic Z., Zha X. M., Price M., Schnizler M. K., Wemmie J. A. (2007) Biol. Psychiatry 62, 1140–1148 [DOI] [PubMed] [Google Scholar]
- 15.Ziemann A. E., Schnizler M. K., Albert G. W., Severson M. A., Howard M. A., 3rd, Welsh M. J., Wemmie J. A. (2008) Nat. Neurosci. 11, 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Z. G., Zhu X. M., Chu X. P., Minami M., Hey J., Wei W. L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., Simon R. P. (2004) Cell 118, 687–698 [DOI] [PubMed] [Google Scholar]
- 17.Pignataro G., Simon R. P., Xiong Z. G. (2007) Brain 130, 151–158 [DOI] [PubMed] [Google Scholar]
- 18.Friese M. A., Craner M. J., Etzensperger R., Vergo S., Wemmie J. A., Welsh M. J., Vincent A., Fugger L. (2007) Nat. Med. 13, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 19.Gao J., Duan B., Wang D. G., Deng X. H., Zhang G. Y., Xu L., Xu T. L. (2005) Neuron 48, 635–646 [DOI] [PubMed] [Google Scholar]
- 20.Hey J. G., Chu X. P., Seeds J., Simon R. P., Xiong Z. G. (2007) Stroke 38, Suppl. 2, 670–673 [DOI] [PubMed] [Google Scholar]
- 21.Xiong Z. G., Chu X. P., Simon R. P. (2006) J. Membr. Biol. 209, 59–68 [DOI] [PubMed] [Google Scholar]
- 22.Chen C. C., England S., Akopian A. N., Wood J. N. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10240–10245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bässler E. L., Ngo-Anh T. J., Geisler H. S., Ruppersberg J. P., Gründer S. (2001) J. Biol. Chem. 276, 33782–33787 [DOI] [PubMed] [Google Scholar]
- 24.Ugawa S., Ueda T., Yamamura H., Shimada S. (2005) Brain Res. Mol. Brain Res. 136, 125–133 [DOI] [PubMed] [Google Scholar]
- 25.Coric T., Zhang P., Todorovic N., Canessa C. M. (2003) J. Biol. Chem. 278, 45240–45247 [DOI] [PubMed] [Google Scholar]
- 26.Li T., Yang Y., Canessa C. M. (2009) J. Biol. Chem. 284, 4689–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashari E., Qadri Y. J., Zhou Z. H., Kapoor N., Anderson S. J., Meltzer R. H., Fuller C. M., Benos D. J. (2009) Am. J. Physiol. Cell Physiol. 296, C372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coscoy S., de Weille J. R., Lingueglia E., Lazdunski M. (1999) J. Biol. Chem. 274, 10129–10132 [DOI] [PubMed] [Google Scholar]
- 29.Vukicevic M., Weder G., Boillat A., Boesch A., Kellenberger S. (2006) J. Biol. Chem. 281, 714–722 [DOI] [PubMed] [Google Scholar]
- 30.Chen X., Kalbacher H., Gründer S. (2006) J. Gen. Physiol. 127, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babini E., Paukert M., Geisler H. S., Grunder S. (2002) J. Biol. Chem. 277, 41597–41603 [DOI] [PubMed] [Google Scholar]
- 32.García-Añoveros J., Derfler B., Neville-Golden J., Hyman B. T., Corey D. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunthorpe M. J., Smith G. D., Davis J. B., Randall A. D. (2001) Pflugers Arch. 442, 668–674 [DOI] [PubMed] [Google Scholar]
- 34.Askwith C. C., Cheng C., Ikuma M., Benson C., Price M. P., Welsh M. J. (2000) Neuron 26, 133–141 [DOI] [PubMed] [Google Scholar]
- 35.Askwith C. C., Wemmie J. A., Price M. P., Rokhlina T., Welsh M. J. (2004) J. Biol. Chem. 279, 18296–18305 [DOI] [PubMed] [Google Scholar]
- 36.Benson C. J., Xie J., Wemmie J. A., Price M. P., Henss J. M., Welsh M. J., Snyder P. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2338–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gründer S., Geissler H. S., Bässler E. L., Ruppersberg J. P. (2000) Neuroreport 11, 1607–1611 [DOI] [PubMed] [Google Scholar]
- 38.Sherwood T. W., Askwith C. C. (2008) J. Biol. Chem. 283, 1818–1830 [DOI] [PubMed] [Google Scholar]
- 39.Chen X., Gründer S. (2007) J. Physiol. 579, 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho J. H., Askwith C. (2007) Am. J. Physiol. Cell Physiol. 292, C2161–C2174 [DOI] [PubMed] [Google Scholar]
- 41.Sherwood T., Franke R., Conneely S., Joyner J., Arumugan P., Askwith C. (2009) J. Biol. Chem. 284, 27899–27907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzales E. B., Kawate T., Gouaux E. (2009) Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Champigny G., Voilley N., Waldmann R., Lazdunski M. (1998) J. Biol. Chem. 273, 15418–15422 [DOI] [PubMed] [Google Scholar]
- 44.Waldmann R., Champigny G., Voilley N., Lauritzen I., Lazdunski M. (1996) J. Biol. Chem. 271, 10433–10436 [DOI] [PubMed] [Google Scholar]
- 45.Ugawa S., Ueda T., Takahashi E., Hirabayashi Y., Yoneda T., Komai S., Shimada S. (2001) Neuroreport 12, 2865–2869 [DOI] [PubMed] [Google Scholar]
- 46.Lingueglia E., de Weille J. R., Bassilana F., Heurteaux C., Sakai H., Waldmann R., Lazdunski M. (1997) J. Biol. Chem. 272, 29778–29783 [DOI] [PubMed] [Google Scholar]
- 47.Bassilana F., Champigny G., Waldmann R., de Weille J. R., Heurteaux C., Lazdunski M. (1997) J. Biol. Chem. 272, 28819–28822 [DOI] [PubMed] [Google Scholar]
- 48.Waldmann R., Bassilana F., de Weille J., Champigny G., Heurteaux C., Lazdunski M. (1997) J. Biol. Chem. 272, 20975–20978 [DOI] [PubMed] [Google Scholar]
- 49.Hesselager M., Timmermann D. B., Ahring P. K. (2004) J. Biol. Chem. 279, 11006–11015 [DOI] [PubMed] [Google Scholar]
- 50.Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., Lazdunski M. (2000) J. Biol. Chem. 275, 25116–25121 [DOI] [PubMed] [Google Scholar]
- 51.Chen X., Kalbacher H., Gründer S. (2005) J. Gen. Physiol. 126, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherwood T. W., Askwith C. C. (2009) J. Neurosci. 29, 14371–14380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez de la Rosa D., Canessa C. M., Fyfe G. K., Zhang P. (2000) Annu. Rev. Physiol. 62, 573–594 [DOI] [PubMed] [Google Scholar]
- 54.Mano I., Driscoll M. (1999) Bioessays 21, 568–578 [DOI] [PubMed] [Google Scholar]
- 55.Adams C. M., Snyder P. M., Price M. P., Welsh M. J. (1998) J. Biol. Chem. 273, 30204–30207 [DOI] [PubMed] [Google Scholar]
- 56.Springauf A., Gründer S. (2010) J. Physiol. 588, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babinski K., Lê K. T., Séguéla P. (1999) J. Neurochem. 72, 51–57 [DOI] [PubMed] [Google Scholar]
- 58.de Weille J. R., Bassilana F., Lazdunski M., Waldmann R. (1998) FEBS Lett. 433, 257–260 [DOI] [PubMed] [Google Scholar]
- 59.Salinas M., Lazdunski M., Lingueglia E. (2009) J. Biol. Chem. 284, 31851–31859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones N. G., Slater R., Cadiou H., McNaughton P., McMahon S. B. (2004) J. Neurosci. 24, 10974–10979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X., Paukert M., Kadurin I., Pusch M., Grunder S. (2006) Neuropharmacology 50, 964–974 [DOI] [PubMed] [Google Scholar]
- 62.Chen C. C., Zimmer A., Sun W. H., Hall J., Brownstein M. J., Zimmer A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8992–8997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sluka K. A., Price M. P., Breese N. M., Stucky C. L., Wemmie J. A., Welsh M. J. (2003) Pain 106, 229–239 [DOI] [PubMed] [Google Scholar]
- 64.Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price M. P., McIlwrath S. L., Xie J., Cheng C., Qiao J., Tarr D. E., Sluka K. A., Brennan T. J., Lewin G. R., Welsh M. J. (2001) Neuron 32, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 66.Voilley N. (2004) Curr. Drug Targets Inflamm. Allergy 3, 71–79 [DOI] [PubMed] [Google Scholar]
- 67.Xu T. L., Duan B. (2009) Prog. Neurobiol. 87, 171–180 [DOI] [PubMed] [Google Scholar]
- 68.Zha X. M., Wemmie J. A., Green S. H., Welsh M. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16556–16561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng J. K., Ji R. R. (2008) Neurochem. Res. 33, 1970–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tominaga M. (2007) Handb. Exp. Pharmacol. 179, 489–505 [DOI] [PubMed] [Google Scholar]
- 71.Baumann T. K., Chaudhary P., Martenson M. E. (2004) Eur. J. Neurosci. 19, 1343–1351 [DOI] [PubMed] [Google Scholar]
- 72.Baumann T. K., Burchiel K. J., Ingram S. L., Martenson M. E. (1996) Pain 65, 31–38 [DOI] [PubMed] [Google Scholar]
- 73.Chung W. S., Farley J. M., Swenson A., Barnard J. M., Hamilton G., Chiposi R., Drummond H. A. (2010) Am. J. Physiol. Cell Physiol. 298, C1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Celotto A. C., Capellini V. K., Baldo C. F., Dalio M. B., Rodrigues A. J., Evora P. R. (2008) Braz. J. Med. Biol. Res. 41, 439–445 [DOI] [PubMed] [Google Scholar]
- 75.Akata T. (2007) J. Anesth. 21, 220–231 [DOI] [PubMed] [Google Scholar]
- 76.Valeyev A. Y., Hackman J. C., Wood P. M., Davidoff R. A. (1996) J. Neurophysiol. 76, 3555–3558 [DOI] [PubMed] [Google Scholar]
- 77.Dib-Hajj S. D., Tyrrell L., Cummins T. R., Black J. A., Wood P. M., Waxman S. G. (1999) FEBS Lett. 462, 117–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.