Abstract
The mechanisms responsible for 17β-estradiol (E2)-stimulated breast cancer growth and development of resistance to tamoxifen and other estrogen receptor α (ERα) antagonists are not fully understood. We describe a new tool for dissecting ERα action in breast cancer, p-fluoro-4-(1,2,3,6,-tetrahydro-1,3-dimethyl-2-oxo-6-thionpurin-8-ylthio) (TPSF), a potent small-molecule inhibitor of estrogen receptor α that does not compete with estrogen for binding to ERα. TPSF noncompetitively inhibits estrogen-dependent ERα-mediated gene expression with little inhibition of transcriptional activity by NF-κB or the androgen or glucocorticoid receptor. TPSF inhibits E2-ERα-mediated induction of the proteinase inhibitor 9 gene, which is activated by ERα binding to estrogen response element DNA, and the cyclin D1 gene, which is induced by tethering ERα to other DNA-bound proteins. TPSF inhibits anchorage-dependent and anchorage-independent E2-ERα-stimulated growth of MCF-7 cells but does not inhibit growth of ER-negative MDA-MB-231 breast cancer cells. TPSF also inhibits ERα-dependent growth in three cellular models for tamoxifen resistance; that is, 4-hydroxytamoxifen-stimulated MCF7ERαHA cells that overexpress ERα, fully tamoxifen-resistant BT474 cells that have amplified HER-2 and AIB1, and partially tamoxifen-resistant ZR-75 cells. TPSF reduces ERα protein levels in MCF-7 cells and several other cell lines without altering ERα mRNA levels. The proteasome inhibitor MG132 abolished down-regulation of ERα by TPSF. Thus, TPSF affects receptor levels at least in part due to its ability to enhance proteasome-dependent degradation of ERα. TPSF represents a novel class of ER inhibitor with significant clinical potential.
Keywords: Breast Cancer, Estrogen, Ligand Binding Protein, Receptors, Steroid Hormone Receptor, ER Inhibitor, ER-dependent Cell Growth, Estrogen Receptor, TPSF
Introduction
Estrogen receptor α (ERα)3 is a well studied member of the steroid/nuclear receptor family of transcription regulators. ERα acts in the nucleus to regulate gene expression by binding to estrogen response elements (EREs) and related DNA sequences (1–4) and through association with transcription factors bound at SP1 and AP-1 DNA binding sites (4–7). In response to high affinity estrogen binding, ERα dimerizes, binds to ERE DNAs, and undergoes a conformational change in the ligand binding domain that facilitates the recruitment of coactivators (8). Bound coactivators promote assembly of a multiprotein complex that enables chromatin remodeling and stabilization of an active transcription complex (9–11). In contrast, antagonist-occupied ERα recruits corepressors (12).
At detection, growth of most human breast cancers depends on 17β-estradiol (E2) binding to ERα (13–16). Treatment strategies that inhibit estrogen-dependent breast cancer include selective ER modulators such as tamoxifen, which binds in the ERα ligand binding pocket, and aromatase inhibitors, which block estrogen production. Nearly half of patients treated with aromatase inhibitors develop resistance (17). The long-term effectiveness of tamoxifen is limited by the development of resistance in nearly all patients with metastatic breast cancer and in ∼40% of patients with primary breast cancers (18). The development of resistance to current therapies underscores the need to develop new small molecule antagonists that act outside the ligand binding pocket of ERα. We recently described an in vitro high throughput screening strategy to identify small molecule inhibitors of ERα binding to DNA. We identified 8-benzylsulfanylmethyl-1,3-dimethyl-3,7-dihydropurine-2,6-dione (TPBM) as a small molecule inhibitor of ERα binding to ERE DNA (19). Using a cell-based screen, we evaluated ∼200 small molecules structurally related to TPBM and identified butyrophenone, p-fluoro-4-(1,2,3,6, -tetrahydro-1,3-dimethyl-2-oxo-6-thionpurin-8-ylthio) (TPSF) as a novel inhibitor of ERα >15-fold more potent than TPBM. Although structurally related to TPBM, TPSF exhibits an entirely different mode of action. Although TPBM inhibits in vitro binding of E2-ERα to a labeled ERE, TPSF does not. TPSF strongly reduces ERα levels in breast cancer cells, whereas TPBM has little or no effect on the level of ERα. Here we demonstrate the selectivity of TPSF and its ability to inhibit expression of an endogenous ERα-regulated gene that contains EREs and a gene regulated by tethering of ERα through other proteins. We show that TPSF inhibits anchorage-dependent and anchorage-independent growth of tamoxifen-sensitive and tamoxifen-resistant ERα-containing breast cancer cells and demonstrate that TPSF enhances proteasome-dependent degradation of ERα.
EXPERIMENTAL PROCEDURES
Cell Culture
Unless otherwise indicated, cells were maintained at 37 °C in 5% CO2 in growth medium containing 1% penicillin and streptomycin and fetal bovine serum (FBS) (Atlanta Biological, Atlanta, GA) or calf serum and transferred to phenol red-free medium containing charcoal-dextran (CD)-stripped serum at least 2 days before treatment with E2, 4-hydroxytamoxifen (OHT), or TPSF. ERα-positive MCF-7 and ER-negative MDA-MB-231, human breast cancer cells, were cultured in MEM supplemented with 10% calf serum and switched to MEM containing 5% CD-treated calf serum 3 or 4 days before the experiment. The medium was changed on day 2. Tet-inducible MCF7ERαHA cells were maintained in DMEM supplemented with 1 mm sodium pyruvate, 0.5 μg/ml puromycin, and 10% FBS. Four days before the experiment, MCF7ERαHA cells were switched to the above medium without phenol red containing 10% 6× stripped CD-treated FBS without puromycin (20–23). ZR-75 human breast cancer cells were maintained in MEM containing 10% calf serum and transferred to medium containing 10% CD-CS 4 days before the experiment. BT474 human breast cancer cells were maintained in improved MEM (iMEM) containing 10% FBS and transferred to phenol red-free iMEM containing 10% CD-FBS 4 days before the experiment. T47D-KBluc breast cancer cells expressing an (ERE)3-luciferase reporter gene (24) were maintained in phenol red-free RPMI 1640 containing 2 mm l-glutamine, 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, 10 mm Hepes, pH 7.5, 1 mm sodium pyruvate, 10% FBS. Four days before induction with E2, cells were transferred to medium without phenol red containing 10% 2× CD calf serum. T47D/A1–2 cells that stably express the glucocorticoid receptor (GR) and contain a mouse mammary tumor virus (MMTV)-luciferase reporter (25) were maintained in MEM supplemented with 10 mm HEPES, pH 7.4, 2 mm glutamine, 5% FBS, and 0.2 mg/ml Geneticin (G418). Four days before the experiment the cells were transferred to the above phenol red-free medium (phenol red-free) containing 10% 2× CD-CS. HeLa-AR1C-PSA-Luc-A6 cells that stably express androgen receptor (AR) and a prostate-specific antigen (PSA)-Luc reporter were maintained in phenol-red free MEM supplemented with 2 mm l-glutamine, 1 mm sodium pyruvate, 10% FBS under selection with 0.1 mg/ml hygromycin B (Roche Applied Science), and 0.5 mg/ml G418. Four days before the experiment, cells were transferred to medium containing 10% 2× CD-CS.
Fluorescence Anisotropy Assays
The fluorescence anisotropy microplate assay for analyzing binding of ERα to the fluorescein-labeled consensus ERE was as described (19).
Competitive Radioligand Binding Assays
The relative binding affinity of TPSF for ERα and ERβ was determined in competitive radioligand binding assays using 2 nm [3H]E2 and a range of TPSF concentrations as described (26, 27).
Reporter Gene Assays
Reporter gene assays were performed to compare the ability of about 200 compounds structurally related to TPBM (19) to inhibit estrogen-dependent transcription in T47D-KBluc breast cancer cells stably transfected to express an (ERE)3-Luc reporter (24). The ability of TPSF to inhibit AR and GR transcriptional activity was assayed in HeLa AR1C-PSA-Luc-A6 cells that stably express human AR and a PSA-Luc reporter and in T47D/A1–2 cells that stably express GR and MMTV-Luc. Four days before each experiment, cells were switched to medium containing CD-treated serum as described above. HeLa AR-PSA-Luc cells (100,000 cells/well) and T47DA/1-2 and T47D-KBluc cells (200,000 cells/well) were plated in 1 ml of media in 24-well plates. After 24 h the indicated concentrations of E2, dihydrotestosterone, or dexamethasone in DMSO or DMSO vehicle alone with or without TPSF were added to each well. After 24 h, cells were washed once with phosphate-buffered saline and lysed in 100 μl of passive lysis buffer (Promega, Madison WI). Luciferase activity was determined using BrightGlo firefly luciferase reagent from Promega.
Endogenous Gene Expression
MCF-7 cells and MCF7ERαHA cells were maintained for 4 days in medium containing 5% 1× CD-CS (MCF-7 cells) or 10% 6× stripped CD FBS (MCF7ERαHA cells). For assays of TPSF inhibition of PI-9 induction in MCF-7 cells, cells were preincubated for 24 h with TPSF and then maintained for 4 h with and without E2 and TPSF or with vehicle alone. To induce ERα expression, MCF7ERαHA cells were maintained in medium containing 0.5 μg/ml doxycycline (Dox) for 24 h. E2 or OHT was added with or without TPSF and maintained for 24 h. For the induction of cyclin D1, 24 h after plating the cells, E2 with and without TPSF was added, and cells were maintained for 24 h. RNA was extracted, and mRNA levels were measured by quantitative RT-PCR as described (19, 28). Actin mRNA level is used as the qRT-PCR internal standard. Primers used in qRT-PCR were: ERα, forward (5′-GGAGACGGACCAAAGCCACT) and reverse (5′-TTCCCAACAGAAGACAGAAGATG); cyclin D1, forward (5′-TCATGGCTGAAGTCACCTCTTGGT) and reverse (5′-TCCACTGGATGGTTTGTCACTGGA); PI-9, forward (5′-TGGAATGAACCGTTTGACGAA) and reverse (5′-CATCTGCACTGGCCTTTGCT); IL-8 forward (5′-GAGGGTTGTGGAGAAGTTTTTG) and reverse (5′-CTGGCATCTTCACTGATTCTTG); β-actin forward (5′-AAGCCACCCCACTTCTCTCTAA) and reverse (5′-AATGCTATCACCTCCCCTGTGT).
Cell Growth and Viability Assays
Cells were maintained in CD-treated serum for at least 4 days before each experiment. To minimize cell aggregation, MCF-7 cells were harvested in 10 mm HEPES, pH 7.4, 1 mm EDTA. Other cell lines were harvested in trypsin-EDTA. To assay anchorage-dependent cell growth, 1000 cells/well were plated in a 96-well plate. For slow-growing ZR-75 cells, 2000 cells were plated/well. Cells were maintained in medium containing CD-treated serum for 24 h, and the medium was then changed, and E2 and DMSO vehicle or TPSF in DMSO was added. The medium was replaced after 2 days, except for BT474 cells, whose medium was not changed. After 4 days, cell viability was determined using Promega CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS) (Promega).
To assay anchorage-independent cell growth in soft agar, 1%, and 0.7% Select Agar (Invitrogen) was prepared in water and warmed at 40 °C before use. 1.5 ml of 0.5% bottom agar diluted in medium was added to each well of a 6-well cell culture plate and allowed to solidify at room temperature. Top agar was prepared by dilution in warm medium containing the various treatments. MCF-7 cells were resuspended in 1.5 ml of 0.35% top agar at 5000 cells/well and plated in 3 wells for each condition. The plate was kept at room temperature for 30 min until the top agar solidified, then 0.5 ml of medium containing the respective treatments was added on top of the agar. Culture medium on top of the agar was changed every 3–4 days. Colonies were visible by 1 week and counted at day 16 using a dissecting microscope. Photographs of colonies were taken using a Zeiss AxioImager2 imaging system at 6× magnification.
Western Blot
MCF-7 cells were plated at 200,000 cells/well in 6-well plates in MEM containing 5% 1× CD FBS. The medium was changed at day 2, and at day 4 the medium was replaced with fresh medium containing the indicated treatments. Whole cell extracts were prepared after 24 h in 1× radioimmune precipitation assay buffer (Millipore, CA) containing protease inhibitor mixture (Roche Applied Science). Extract (20 μg of protein/lane) was run on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. ER was detected using a 1:4000 dilution of ERα antibody ER6F11 (Bio Care Medical). The blot was stripped for 25 min and reprobed using a 1:10,000 dilution of β-actin monoclonal antibody (Sigma).
Statistical Analysis
Results are expressed as the mean ± S.E. of at least three independent experiments. Student's t test was used for comparison of the means between two groups. Significance was established when p < 0.05. The comparisons are described in the figure legends and are not shown in the body of the figures.
RESULTS
TPSF Is a Structure-specific Inhibitor That Acts Outside of the Ligand Binding Pocket of ERα
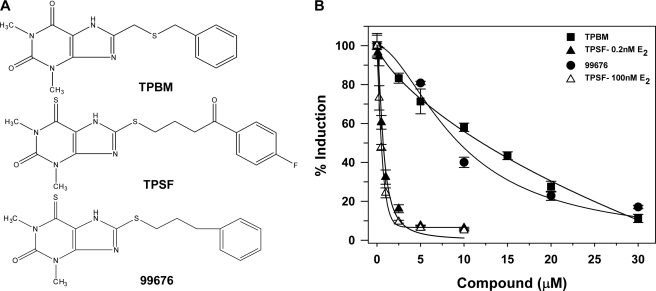
We evaluated the ability of ∼200 compounds structurally related to TPBM (Fig. 1A), a first generation ERα inhibitor (19), to inhibit E2-ERα-mediated gene expression in ERα-positive T47D-KBluc human breast cancer cells containing (ERE)3-Luc (24). Of the compounds tested, TPSF (Fig. 1A) was ∼16-fold more potent than TPBM in inhibiting ERα-mediated gene expression (Fig. 1B).
FIGURE 1.
Structure-specific inhibition of E2-ERα-mediated gene expression by TPSF. A, shown are structures of three ERα inhibitors. TPBM is a recently described ERα inhibitor (19). TPSF is butyrophenone, p-fluoro-4-(1,2,3,6,-tetrahydro-1,3-dimethyl-2-oxo-6-thionpurin-8-ylthio and is known also as theophylline, 8-(3-p-fluorobenzoylpropyl)thio-6-thio-). NSC-99676 is similar to TPSF except TPSF has C O and fluorine substitutions at the phenyl ring. B, shown are potency and efficacy of TPSF (triangles), TPBM (squares), and 99676 (circles). Inhibition of E2-ERα activation of ERE-Luc was evaluated in dose-response studies of T47D (ERE)3-Luc cells maintained in 0.2 nm E2 (filled triangles, squares, and circles) or 100 nm E2 (open triangles) with the indicated concentrations of TPBM, TPSF, or 99676 present for 24 h before assay. Activity of the reporter in the presence of the tested concentration of E2 with DMSO and no inhibitor was set to 100%. Data are the average of three experiments ± S.E. Some symbols overlap, and some error bars are smaller than the symbols. IC50 values were calculated by curve fitting using Sigma Plot.
O and fluorine substitutions at the phenyl ring. B, shown are potency and efficacy of TPSF (triangles), TPBM (squares), and 99676 (circles). Inhibition of E2-ERα activation of ERE-Luc was evaluated in dose-response studies of T47D (ERE)3-Luc cells maintained in 0.2 nm E2 (filled triangles, squares, and circles) or 100 nm E2 (open triangles) with the indicated concentrations of TPBM, TPSF, or 99676 present for 24 h before assay. Activity of the reporter in the presence of the tested concentration of E2 with DMSO and no inhibitor was set to 100%. Data are the average of three experiments ± S.E. Some symbols overlap, and some error bars are smaller than the symbols. IC50 values were calculated by curve fitting using Sigma Plot.
To test whether TPSF is a structure-specific inhibitor, we compared the ability of TPSF to inhibit ERα-mediated gene expression to the structurally similar small molecule 99676. TPSF differs from NSC 99676 by having hydrophilic C O and fluorine substitutions at the phenyl ring (Fig. 1A). The similar, but more hydrophobic, 99676 had an IC50 of 9 μm and was ∼13-fold less potent than TPSF (Fig. 1B). The results suggest that TPSF is a structure-specific inhibitor of ERα and is not simply acting by promiscuous inhibition due to micelle formation.
O and fluorine substitutions at the phenyl ring (Fig. 1A). The similar, but more hydrophobic, 99676 had an IC50 of 9 μm and was ∼13-fold less potent than TPSF (Fig. 1B). The results suggest that TPSF is a structure-specific inhibitor of ERα and is not simply acting by promiscuous inhibition due to micelle formation.
If TPSF inhibited ERα by competing with E2 for binding in the ligand binding pocket of the receptor, increasing the E2 concentration should reduce the ability of TPSF to bind ERα and block its action. To test this, we varied the E2 concentration by 500-fold and tested the ability of TPSF to inhibit (ERE)3-Luc in T47D cells. Increasing the concentration of E2 from 0.2 to 100 nm only slightly increased the IC50 for inhibiting E2-ERα-mediated transcription from 0.4 μm (Fig. 1B, filled triangles) to 0.7 μm (Fig. 1B, open triangles), suggesting TPSF acts outside the ERα ligand binding pocket.
If TPSF is a highly potent ERα ligand, it might retain the ability to inhibit ERα at 100 nm E2. To test whether TPSF binds in the ligand binding pocket of ERα, the ability of TPSF to compete with radiolabeled E2 for binding to ERα was evaluated. In competitive radiometric binding assays performed across a broad range of concentrations (26, 27), TPSF had virtually no ability to compete with E2 for binding to ERα. With E2 set at 100%, TPSF had a relative binding affinity for ERα of ∼0.001%, indicating that it is not a classical ligand that competes with E2 for binding in the ERα ligand binding pocket.
TPSF Is a Specific Inhibitor of ERα Transactivation
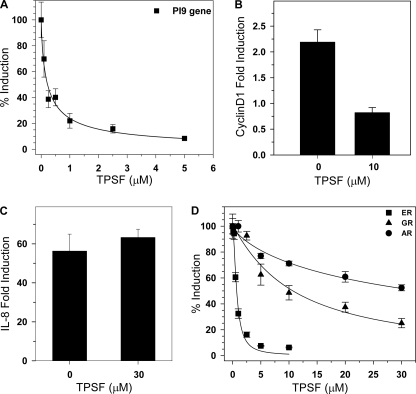
The ERα binding cleft for p160 coactivator LXXLL motifs has been a major target for development of peptide and small molecule inhibitors (29–33). Although these inhibitors are effective in reporter gene assays in transfected cells, in general they have not been shown to effectively inhibit expression of endogenous ER-regulated genes in breast cancer cells. We, therefore, tested the ability of TPSF to inhibit the expression of the endogenous E2-inducible PI-9 gene in MCF-7 cells. The serpin PI-9 is a tumor lethality factor (34–36) whose induction by estrogens enables breast cancer cells to evade apoptosis induced by the immune cells, cytotoxic T lymphocytes and natural killer cells (21, 23, 28). PI-9 also inhibits tumor necrosis factor-α (TNF-α) Fas and TRAIL (TNF-related apoptosis-inducing ligand)-mediated apoptosis (38, 39). Induction of PI-9 results from direct binding of E2-ERα to EREs and ERE half-sites (19, 40, 41). TPSF (IC50 = 0.2 μm) potently inhibited E2-ERα-stimulated induction of PI-9 mRNA (Fig. 2A).
FIGURE 2.
TPSF specifically inhibits expression of endogenous ER-regulated genes. A, TPSF inhibits E2 induction of PI-9 mRNA. For studies of PI-9 mRNA (filled squares), MCF-7 cells were incubated for 24 h with the indicated concentrations of TPSF and maintained for 4 h in 10 nm E2 and TPSF. PI-9 mRNA was quantitated by RT-PCR as described (23). B, TPSF inhibition of E2-ERα induction of cyclin D1 mRNA is shown. MCF-7 cells were plated and 24 h later treated with ethanol and DMSO vehicles, 10 nm E2, or 10 nm E2 and 10 μm TPSF. After 24 h, RNA was extracted, and cyclin D1 mRNA levels were measured by qRT-PCR. The level of cyclin D1 mRNA in the vehicle only sample was set to 1. -Fold induction of cyclin D1 in the presence of 10 μm TPSF was significantly different from the control (p < 0.05 using Student's t test). C, TPSF does not inhibit NF-κB induction of IL-8 mRNA. MCF-7 cells were maintained for 24 h in medium without TNF-α or with 10 ng/ml TNF-α with and without 30 μm TPSF and harvested, and IL-8 mRNA levels were determined by qRT-PCR. D, dose-response studies of inhibition of ERα, AR, and GR transactivation are shown. For each receptor, induction of luciferase reporter gene expression (AR and GR) or endogenous PI-9 mRNA (ER) in the presence of an appropriate ligand with DMSO minus TPSF was set to 100%. Cells were incubated for 24 h with 0.2 nm E2 for ERα (filled squares), 5 nm dexamethasone for GR (filled triangles), 1 μm dihydrotestosterone for AR (filled circles), and the indicated concentrations of TPSF. Data are the average ± S.E. for three experiments.
To examine the ability of TPSF to inhibit E2-ERα induction of a gene regulated by tethering of E2-ERα through DNA-bound transcriptional regulators, we tested the effect of TPSF on induction of cyclin D1 mRNA. Cyclin D1 is thought to contribute to the growth of MCF-7 and other breast cancer cells (42–44) and to tamoxifen-stimulated growth of breast cancer cells (45). As previously reported (43, 46), E2-ERα stimulated a 2–3-fold increase in cyclin D1 mRNA that was blocked by 10 μm TPSF (Fig. 2B). The data demonstrate that TPSF inhibits E2-dependent gene expression through mechanisms that include direct binding of ERα to EREs and through tethering of ERα to DNA-associated transcription regulators.
Specificity of TPSF inhibition of E2-ERα-mediated gene expression was evaluated by comparing the ability of TPSF to inhibit gene expression mediated by NF-κB (Fig. 2C) and by other steroid receptors (Fig. 2D). The NF-κB-regulated IL-8 gene was used to test specificity because many regulators and pathways including IκB and other kinases, the ubiquitin/proteasome pathway, and nuclear/cytoplasmic shuttling all influence NF-κB activity (47). Analyzing the effect of TPSF on NF-κB is a good way to test whether TPSF acts as a promiscuous inhibitor targeting diverse cell proteins and pathways. 30 μm TPSF had no effect on the NF-κB-mediated induction of IL-8 mRNA by TNF-α (Fig. 2C). In the same breast cancer cells where TPSF inhibited E2-ERα induction of PI-9 mRNA (Fig. 2A, IC50 = 0.2 μm), a >100-fold higher concentration of TPSF had no effect on NF-κB-mediated induction of IL-8 mRNA by TNF-α (Fig. 2C).
To further evaluate the specificity of TPSF, we compared the ability of TPSF to inhibit ERα to its effect on the AR- and GR-mediated activation of stably transfected reporter genes. AR was assayed in HeLa cells stably transfected to express AR and a PSA-luciferase reporter. TPSF only very weakly inhibited dihydrotestosterone-AR-mediated induction of the PSA-Luc reporter (IC50 = 33 μm, Fig. 2D). GR was assayed in T47D cells stably transfected to express GR and an MMTV-Luc reporter (25). TPSF weakly inhibited GR activation of the MMTV-luc reporter in T47D cells (IC50 = 10 μm). Although T47D cells contain substantial levels of the progesterone receptor (PR), cross-talk between ERα and PR makes them unsuitable for assaying inhibitor specificity using PR. The ER antagonist faslodex/fulvestrant/ICI 182,780 inhibited PR induction of the endogenous alkaline phosphatase gene (supplemental Fig. S1). TPSF did not inhibit NF-κB, and concentrations of TPSF required to inhibit AR (33 μm) and GR (10 μm) are far higher than the 0.2 and 0.4 μm TPSF required to inhibit the endogenous PI-9 gene and the stably transfected (ERE)3-Luc reporter. At low concentrations, TPSF is a relatively specific ERα inhibitor.
TPSF Inhibits E2 and OHT-induced Gene Expression in Tamoxifen-stimulated MCF7ERαHA Cells
Development of resistance to tamoxifen and other therapeutics that target ERα and estrogen production results in treatment failure in both primary and metastatic breast cancer. Recent studies show that tamoxifen-resistant breast cancer cells that retain dependence on ERα for growth lose their dependence on SRC3 and other p160 coactivators for E2-ERα-mediated gene transcription (22, 48). We explored the ability of TPSF to inhibit E2 and OHT-dependent gene expression in tamoxifen-resistant cells that are less dependent on p160 coactivators for transactivation.
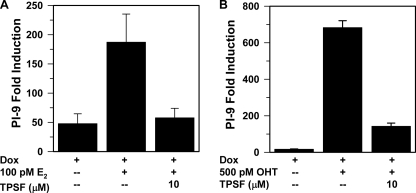
MCF7ERαHA cells are an MCF-7-breast cancer cell line engineered to increase ERα expression in response to Dox (20, 22). In Dox-induced MCF7ERαHA cells overexpressing ERα, tamoxifen and OHT are potent ERα agonists (22, 23) and increase ERα-mediated gene expression independent of SRC3 (22). Because OHT stabilizes ERα against degradation whereas E2 down-regulates ERα (21, 23), ERα levels are ∼4 times higher in OHT-treated MCF7ERαHA cells than in cells treated with E2. The elevated level of ERα in OHT-treated MCF7ERαHA cells compared with cells treated with E2 renders OHT more effective than E2 in inducing PI-9 gene expression and more difficult to inhibit. 10 μm TPSF inhibited E2-ERα (Fig. 3A) and OHT-ERα induction of PI-9 mRNA (Fig. 3B). This indicates that TPSF is an inhibitor of both E2-ERα and OHT-ERα-mediated gene expression in cells where tamoxifen is a full agonist.
FIGURE 3.
TPSF inhibits E2 and OHT-induced gene expression in a tamoxifen-stimulated cell line. A and B, MCF7ERαHA cells maintained in 6× CD-FBS (22, 37) were treated for 24 h with 0.5 μg/ml Dox to induce ERα expression (37) and 100 pm E2 and 10 μm TPSF (A) or 500 pm OHT and 10 μm TPSF (B) as indicated. PI-9 mRNA levels were measured by qRT-PCR. PI-9 mRNA in control MCF7ERαHA cells not treated with Dox, E2, or OHT was set equal to 1. The high level of ERα in Dox-treated cells results in ligand-independent transactivation of PI-9 (23). Data are the average, with the range shown, for two experiments for E2 and three experiments ± S.E. for OHT.
TPSF Inhibits Estrogen-dependent Growth of MCF-7 Cells and Exhibits Low Toxicity in ERα Negative MDA-MB-231 Cells
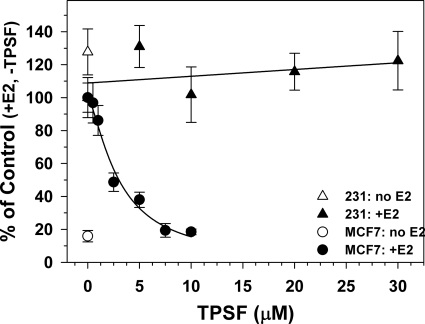
To determine whether TPSF specifically inhibits ERα-dependent growth of breast cancer cells with minimal nonspecific cell toxicity, we tested TPSF inhibition of cell growth in MCF-7 cells and ERα-negative MDA-MB-231 human breast cancer cells. Compared with MCF-7 cells in estrogen-depleted medium, both 1 and 10 pm E2 stimulated a 4–5-fold increase in cell number after 4 days (Fig. 4 and data not shown). TPSF elicited a dose-dependent inhibition of estrogen-dependent growth of MCF-7 cells (IC50 = 2 μm) and completely blocked E2-dependent growth at 7.5 μm (Fig. 4, filled circles). However, TPSF did not inhibit E2-independent cell growth (Fig. 4, compare 7.5 and 10 μm TPSF (filled circles) to no E2 or TPSF (open circle)). TPSF did not inhibit growth of ERα-negative MDA-MB-231 cells at all concentrations, including 30 μm (Fig. 4, filled triangles). To rule out the possibility that MDA-MB-231 cells are unusually resistant to TPSF or other ERα inhibitors, we compared the effects of TPSF and OHT on the growth of MDA-MB-231 cells. TPSF was less toxic to ER-negative MDA-MB-231 cells than OHT (supplemental Fig. 2). The results suggest that TPSF specifically inhibits ERα-mediated growth of breast cancer cells with low nonspecific toxicity in ERα-negative cells.
FIGURE 4.
Inhibition of E2-ERα-dependent breast cancer cell growth by TPSF. MCF-7 and MDA-MB-231 cells were maintained for 4 days in 5% CD-CS, and 1000 MCF-7 cells (circles) or MDA-MB-231 cells (triangles) were plated per well in 96-wellplates. After 24 h the medium was changed to 5% CD-CS with 1 pm E2 (filled circles or triangles) or without E2 (open circle and open triangle) and DMSO vehicle and the indicated concentrations of TPSF. Medium was replaced after 2 days, and cells were assayed with MTS after a total of 4 days. Cell number was determined using a standard curve of cell number versus absorbance based on plating a known number of cells and assaying using MTS. Each data point is the average of 8 wells ± S.E. The percentage of cells present after 4 days with E2 and without TPSF was set equal to 100. By curve-fitting in Sigma Plot, the IC50 for inhibition of E2-dependent growth of MCF-7 cells by TPSF was 2 μm.
TPSF Inhibits Anchorage-independent Growth of MCF-7 Cells
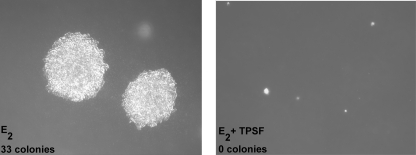
The capacity for anchorage-independent growth is a hallmark of cancer cells. Growth in soft agar is often used to evaluate anchorage-independent growth of human breast cancer cells. We tested the ability of TPSF to inhibit E2-stimulated growth of MCF-7 cells in soft agar. MCF-7 cells grown in medium containing E2 formed large colonies after 16 days (Fig. 5, E2). The addition of 10 μm TPSF completely inhibited growth of MCF-7 cells in soft agar (Fig. 5, E2+TPSF). When colonies in equal areas of the soft agar plate were counted, the E2-treated plate had 33 colonies >0.5 mm in diameter, whereas there were no colonies >0.5 mm in diameter in the E2 and TPSF-treated plate. The data indicate that TPSF inhibits estrogen stimulation of anchorage-dependent (Fig. 4) and anchorage-independent (Fig. 5) growth of breast cancer cells.
FIGURE 5.
TPSF inhibits growth of MCF-7 cells in soft agar. 5000 MCF-7 cells were plated into top agar containing 1 pm E2 (left) or E2 + 10 μm TPSF (right) as described under “Experimental Procedures.” After 16 days colonies were photographed at 5× magnification and counted. Photographs are representative of the entire plate and of duplicate experiments.
TPSF Inhibits E2-ERα-dependent Growth of Tamoxifen-resistant Breast Cancer Cells
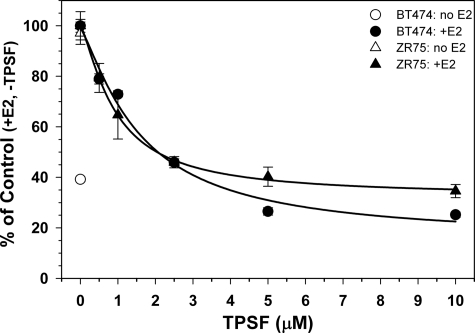
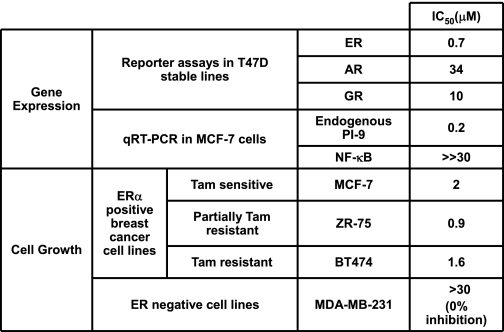
The ability of TPSF to inhibit E2-ERα-dependent cell growth was tested using human cell models of tamoxifen-resistant breast cancer. ZR-75 cells are usually reported as partially tamoxifen and OHT-resistant (49–51), whereas BT474 cells are fully tamoxifen-resistant and contain amplified expression of HER2 and AIB1 (52, 53). TPSF inhibited E2-ERα-dependent growth of BT474 and ZR-75 cells with near maximal inhibition at 5 μm TPSF (Fig. 6). Because TPSF has minimal nonspecific toxic effects, cell numbers after TPSF treatment were not 0 and represented cells plated at day 0 plus E2-ERα-independent cell growth over the 4 days. TPSF IC50 values were 0.9 μm for slow-growing ZR-75 cells and 1.6 μm for BT474 cells. The lower levels of ERα in ZR-75 compared with MCF-7 cells (54) may be responsible for the greater potency of TPSF in ZR-75 cells. Some tamoxifen-resistant breast cancers regress after tamoxifen withdrawal, suggesting that tamoxifen stimulates tumor growth (45, 55–57). The MCF7ERαHA cell line is a model for tamoxifen-stimulated breast cancer, where tamoxifen and OHT act as full agonists (Fig. 3) (21, 23). In MCF7ERαHA cells treated with Dox, overexpression of ERα increased E2-independent ERα-mediated cell growth, which was modestly increased by 1 pm E2 with and without 5 μm OHT and was inhibited by 5 μm TPSF (supplemental Fig. 3). Table 1 summarizes the effect of TPSF on gene expression and cell growth.
FIGURE 6.
TPSF inhibits E2-ERα-dependent growth of tamoxifen-resistant BT474 and ZR-75 cells. Cells were maintained in medium containing 10% CD-FBS (ZR-75) (triangles) or 10% CD-CS (BT474) (circles) with or without 100 pm E2 and the indicated concentrations of TPSF. Viable cells were measured by comparison to a standard curve of cell number versus absorbance using the MTS assay. Data represent the average of at least 4 wells. IC50 values for TPSF inhibition of cell growth were calculated by curve-fitting using Sigma Plot. Although some portion of ZR-75 cell growth is likely ERα-independent, to calculate the IC50 using Sigma Plot, we used the conservative assumption that all cell growth beyond the 2000 ZR-75 cells plated was E2-ERα-dependent growth and, thus, subject to inhibition by TPSF.
TABLE 1.
TPSF and TPBM Have Different Modes of Action
Our data show that TPSF is a potent and selective inhibitor of ER-stimulated gene expression and breast cancer cell growth. We, therefore, began to assess how TPSF might exert its actions. We used our fluorescence anisotropy microplate assay (19, 58, 59) to compare the ability of TPSF and TPBM to inhibit binding of purified ERα to a fluorescein-labeled consensus ERE (flcERE). When polarized light excites the flcERE, most of the emitted light is depolarized because of rapid rotational diffusion of the flcERE that results in its position being largely randomized at the time of emission. Binding of the larger ERα protein to the flcERE slows rotation of the flcERE, increasing the likelihood that the complex is in the same plane at emission and excitation. Interaction of ERα with the flcERE increases fluorescence polarization/fluorescence anisotropy.
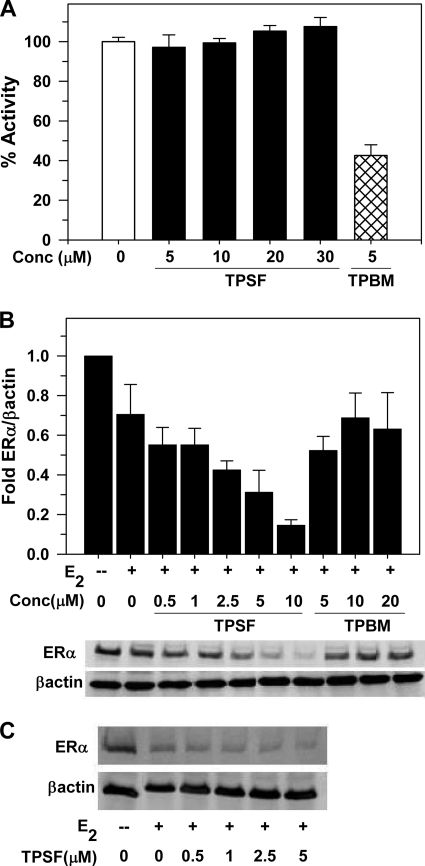
We compared the ability of TPBM and TPSF to inhibit binding of ERα to the flcERE. Consistent with our recent report (19), TPBM inhibited binding of E2-ERα to the ERE (Fig. 7A). Surprisingly, even at 30 μm, TPSF had no effect on binding of E2-ERα to the flcERE (Fig. 7A). Thus, in a direct in vitro assay containing only E2-ERα, the flcERE, TPSF did not inhibit binding of ERα to an ERE.
FIGURE 7.
Different modes of action of TPSF and TPBM. A, TPSF does not inhibit binding of E2-ERα to the flcERE. Fluorescence anisotropy microplate assay was performed as described (19) in the presence of increasing concentrations of TPSF (solid bars) and 5 μm TPBM (hatched bar). Consistent with our detailed dose-response study (19), 5 μm TPBM inhibited binding of TPBM to the flcERE by ∼60%. Data were plotted with the change in anisotropy for binding of E2-ERα to the flcERE in the absence of small molecule inhibitors (open bar) set to 100% (actual anisotropy: flcERE, 44 mA units; E2-ERα-flcERE, 81 mA units). Data are the average + S.E. of four experiments. The difference between 5 μm TPSF and the control (no inhibitor) was not significant (p > 0.05). The data for 5 μm TPBM were significantly different from both the control and from 5 μm TPSF (p < 0.01 using Student's t test) B, TPSF decreases ERα levels. MCF-7 cells were cultured in 5% CD calf serum for at least 2 days and maintained in the absence or presence of E2 and the indicated concentrations of TPSF or TPBM for 24 h and analyzed for ERα by Western blot using 8 μg of protein/lane with actin as internal standard. Data are from the Western blot shown and two additional Western blots from independent experiments and are presented as the mean ± S.E. Quantitation of ERα and actin was by PhosphorImager analysis. The value for ERα/actin in the absence of E2 was set equal to 1. C, T47D cells were maintained as described under “Experimental Procedures,” maintained in the absence or presence of E2 and the indicated concentrations of TPSF, harvested, and analyzed by Western blot as described for panel B.
We next compared the effects of TPSF and TPBM on the intracellular levels of E2-ERα in MCF-7 cells. TPBM at 5–20 μm had little or no effect on the level of E2-ERα. In contrast, TPSF elicited a concentration-dependent reduction in E2-ERα levels, with 10 μm TPSF decreasing the level of E2-ERα by ∼4-fold (Fig. 7B). TPSF also reduced E2-ERα levels in T47D breast cancer cells (Fig. 7C). Because TPSF had very little or no effect on the levels of AR and GR (supplemental Fig. 4), TPSF selectively down-regulates the level of ERα. The results indicate that TPBM and TPSF have distinct modes of action and that TPSF is not simply a more potent version of TPBM.
TPSF Does Not Alter the Level of ER mRNA
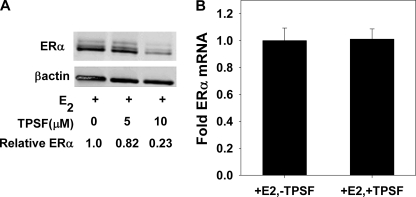
TPSF might reduce ERα levels by decreasing transcription or by destabilizing ERα mRNA. To test for the effects of TPSF at the mRNA level, we examined the effect of TPSF on ERα levels in HeLa cells that stably express ERα mRNA from a CMV promoter. TPSF retained the ability to down-regulate ERα protein from the CMV promoter and from the 2-kb ERα mRNA coding region that lacks ∼4 kb of 5′- and 3′-untranslated region (Fig. 8A). TPSF had no effect ERα mRNA levels in MCF-7 cells (Fig. 8B). Taken together the results suggest that TPSF down-regulates ERα protein levels through mechanisms that are independent of the level of ERα mRNA.
FIGURE 8.
TPSF does not alter the level of ERα mRNA. A, shown is a Western blot of HeLa-ER cell extract. HeLa cells stably transfected to express functional wild-type ERα (76) were maintained in MEM + 10% FBS. Four days before, the cells were plated in 6-well plates at 50,000 cells/well in MEM + 10% 1× CD-FBS. The medium was changed after 2 days and on day 4 replaced with fresh medium containing 10 nm E2 in DMSO or DMSO and the indicated concentration of TPSF. After 24 h, the cells were harvested, and extracts were prepared as described under “Experimental Procedures.” B, effect of TPSF on ERα mRNA levels in MCF-7 cells. Cells were maintained 4 days in MEM + 5% 1× CD-FBS as described under “Experimental Procedures.” Then cells were then maintained for 24 h in medium containing 10 nm E2 in DMSO or DMSO with or without 10 μm TPSF and ER mRNA levels determined by qRT-PCR as described under “Experimental Procedures.” Data were the average of three experiments ±S.E.
The Proteasome Inhibitor MG132 Blocks the Down-regulation of ERα by TPSF
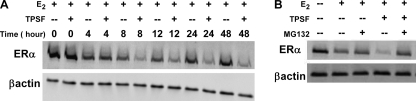
To further examine the effects of TPSF, we determined the time course of TPSF down-regulation of ERα. Consistent with a TPSF-induced increase in proteasome-dependent degradation of ERα, there was a progressive decrease in ERα protein levels after 6–8 h (Fig. 9A). To examine this further we tested the ability of the proteasome inhibitor MG132 to block the effects of TPSF. Compared with E2 alone, TPSF reduced ER levels, and the TPSF-mediated reduction in ERα levels was completely blocked by MG132 (Fig. 9B). Efforts to examine the effect of TPSF on ubiquitination of ERα in MCF-7 cells were complicated by the use of endogenous untagged endogenous ubiquitin and because E2 down-regulates ER and influences its degradation. The data suggests that much of TPSFs effectiveness as an ER inhibitor resides in its ability to enhance proteasome-mediated degradation of ERα.
FIGURE 9.
The proteasome inhibitor MG132 blocks degradation of ERα by TPSF. A, shown is the time course of the effect of TPSF on ER levels. MCF-7 cells were plated as described under “Experimental Procedures.” After 4 days in MEM + 5% 1× CD-FBS, the medium was replaced with medium containing 10 nm E2 with or without 10 μm TPSF. Cells were harvested at the indicated times, extracts prepared, and ERα protein levels were determined by Western blot as described under “Experimental Procedures.” B, MG132 reverses the down-regulation of ER by TPSF. Cells were treated as in panel A and maintained for 24 h in medium containing 10 nm E2 with or without 10 μm and 10 μm MG132. Preparation of cell extracts and Western blotting were as described under “Experimental Procedures.”
TPSF Does Not Enhance ERα Degradation by Reducing the Level of the Muc1 Oncoprotein
Kufe and coworkers (60) reported that the cytoplasmic domain of the Muc1 oncoprotein binds ERα and stabilizes ERα against degradation, which contributes to enhanced ERα transactivation and the estrogen-dependent growth of breast cancer cells. Although the mechanism by which Muc1 influences ER degradation is unknown, knockdown of Muc1 with RNAi enhanced degradation of ERα and inhibited ERα-mediated transactivation and growth of ER-positive breast cancer cells (60). Because the effects of TPSF and RNAi knockdown of Muc1 protein are similar, we tested whether TPSF influenced the level of the Muc1 cytoplasmic domain. Using the same antibody used by Kufe and coworkers (60), TPSF did not alter the level of the Muc1 cytoplasmic domain (supplemental Fig. 5), indicating that the reduction in ERα levels elicited by TPSF was likely independent of the level of Muc1.
DISCUSSION
Specificity and Toxicity of TPSF
An optimal small molecule inhibitor of E2-ERα action and growth of breast cancer cells will exhibit specificity for ERα and low nonspecific toxicity. Independent testing of TPSF at concentrations up to 10 μm against a panel of 60 cancer cell lines at the National Cancer Institute Developmental Therapeutics Program demonstrated that TPSF is generally not toxic to cancer cells (testing was terminated because <6 of the 60 cell lines showed 50% inhibition of cell growth at 10 μm TPSF). In agreement with this, we provide evidence that TPSF selectivity targets E2-ERα-dependent cell growth, with little effect on ERα-independent cell growth. After 4 days of treatment, E2 increased MCF-7 cell numbers by ∼4-fold, which corresponds to a doubling time of ∼1 day with E2 and ∼2 days without E2. The number of cells treated with 7.5 and 10 μm TPSF was similar to that seen without E2. In addition, studies using ER-negative MDA-MB-231 cells showed 30 μm TPSF was not toxic. The ability of 10–20 μm OHT to induce apoptosis of MDA-MB-231 cells suggests that these cells are not especially resistant to nonspecific toxic effects. Over several decades tamoxifen has displayed an excellent safety profile in humans. The toxicity of OHT is used only to demonstrate that MDA-MB-231 cells remain susceptible to cell death and that the failure of TPSF to damage the cells is therefore due to low toxicity rather than resistance of these cells to cell death.
Several lines of evidence support the specificity of TPSF for ERα. For example, NF-κB is regulated by a variety of signaling mechanisms that include the ubiquitin/proteasome pathway, nuclear/cytoplasmic shuttling, IκB, and other kinases and acetylases (47). In MCF-7 cells, TNF-α activation of NF-κB in MCF-7 cells increases IL-8 mRNA levels by ∼50-fold. The absence of an effect of 30 μm TPSF on TNF-α induction of IL-8 mRNA suggests that TPSF does not exhibit nonspecific effects on these diverse cell pathways.
TPSF specificity for ERα was also demonstrated relative to other steroid receptors. TPSF strongly down-regulated the level of ERα but had very little or no effect on the levels of AR and GR. TPSF was a more potent inhibitor of transactivation by ERα than by AR or GR. We have identified other compounds that inhibit AR and GR under the same assay conditions (data not shown), suggesting that the failure of low concentrations of TPSF to inhibit transactivation by AR and GR was not due to assay conditions. The partial inhibition of GR and AR by higher concentrations of TPSF will not impede future animal or human studies. Two recently described AR inhibitors being tested for prostate cancer therapy, harmol and pyrvinium (61), were strong inhibitors of GR but were used with some success as AR inhibitors in studies in mice (61). Mifepristone (RU-486), a classical PR antagonist that also inhibits GR, has been used in long-term clinical studies without significant GR-related pathology (62, 63).
Our initial ER inhibitor, TPBM, has proven useful as a selective inhibitor of the binding of E2-ERα to cellular genes (64, 65). The identification of a new coactivator binding surface on AR using moderate potency (IC50 ∼50 μm) small molecule inhibitors of AR selected by screening (66) that are unrelated to TPSF also supports the utility of small molecule inhibitors as probes to understand the mechanisms of steroid receptor action.
Inhibition of Gene Expression by TPSF
ERα activates gene expression by direct binding to ERE-related DNA sequences and by tethering to DNA-associated transcription factors. Our studies indicate that both of these mechanisms are inhibited by TPSF. TPSF inhibited the induction of PI-9 mRNA by E2-ERα and by OHT-ERα. PI-9 gene expression induced by E2-ERα results from binding to two adjacent ER binding sites in the PI-9 promoter region (40, 41). Induction of PI-9 may be a mechanism by which estrogens enable breast cancers to evade immune surveillance and apoptosis (21, 23). PI-9 inhibits granzyme B and cytotoxic T lymphocyte and natural killer and cell-mediated apoptosis of target cancer cells (21, 23, 28) and caspase 8-dependent apoptosis induced by TNF-α family members (38, 39). Expression of PI-9 is associated with a poor prognosis in some cancers (34–36, 67).
Cyclin D1 plays a key role in cell cycle progression and is induced by tethering E2-ERα to transcription factors bound at SP1 sites (46). Cyclin D1 induction is proposed to play a role in estrogen-dependent growth of breast cancer cells (42, 43, 45). Consistent with TPSF as a broad spectrum ERα inhibitor, 10 μm TPSF abolished E2 induction of cyclin D1 mRNA but did not reduce the level of cyclin D1 mRNA much below the basal (−E2) level. Inhibition of E2-ERα-dependent MCF-7 cell growth by 10 μm TPSF is consistent with a role for cyclin D1 in estrogen-stimulated growth of breast cancer cells. Our work extends earlier studies demonstrating that nearly complete loss of cyclin D1 after RNAi knockdown reduces growth of MCF-7 cells in medium containing estrogen (68). Because TPSF specifically targets the ERα-regulated component of target gene expression and did not influence basal gene expression, TPSF may be a promising new probe to help clarify the role of ERα regulation of specific genes in growth of breast cancer cells.
TPSF Is Effective in Tamoxifen-stimulated and Tamoxifen-resistant Breast Cancer Cells
Development of resistance to tamoxifen and other endocrine therapies is a multifactorial clinical challenge in the treatment of breast cancer. A therapeutically useful small molecule inhibitor of ERα should inhibit the growth of a primary tumor as do tamoxifen and its active metabolite OHT and also inhibit growth of tumor cells with acquired resistance to tamoxifen. Tamoxifen-resistant tumors can be grouped into three broad classes. Some tumors become independent of ERα for growth and may be unaffected by therapies that target ERα. Others tumors remain dependent on E2 and ERα for growth. A third group loses estrogen dependence but requires ERα for growth. The mechanisms involved in resistance to endocrine therapy are diverse. For example, ERα transactivation in tamoxifen-resistant cell lines may depend on as yet unidentified coregulators or may be independent of the p160 coactivators (22, 48).
One proposed mechanism for tumor resistance to antagonists is the overexpression of steroid receptors. Overexpression of AR was suggested as an important mechanism of resistance to endocrine therapy in castration-recurrent prostate cancer (69). A subset of breast cancers that contain high levels of ERα are often refractory to tamoxifen therapy (70–72). In MCF7ERαHA cells that overexpress ERα, tamoxifen and OHT are full agonists and induce PI-9 expression. In MCF7ERαHA cells maintained in the presence of OHT, levels of ERα are >10 times higher than in wild-type MCF-7 cells maintained in the presence of E2 (21). In cells expressing high levels of ERα, 10 μm TPSF inhibited both E2-ERα and OHT-ERα induction of PI-9 mRNA.
Clinical specimens of tamoxifen-resistant metastatic breast cancer can be difficult to obtain (73). We and others (49) have, therefore, evaluated ERα inhibitors using stable breast cancer cell lines resistant to tamoxifen. TPSF inhibited E2-ERα-dependent growth of ZR-75 human breast cancer cells (IC50 = 0.9 μm), whose slow growth is only weakly stimulated by E2 and are partially resistant to tamoxifen and OHT (49, 74). In contrast to OHT, TPSF blocked the growth of MCF7ERαHA cells that are tamoxifen-resistant because they overexpress ERα. TPSF inhibited E2-ERα-dependent growth of BT474 cells, which contain amplified HER2 and AIB1 and are fully tamoxifen-resistant in cell culture (49) and in xenograft studies (52). Thus, TPSF is effective in cells that become tamoxifen-resistant through different mechanisms.
Small Molecules Inhibitors of ERα
TPSF is structurally distinct from disulfide benzamide, a zinc chelator that acts outside the ERα ligand binding pocket. Disulfide benzamide promotes an ERα conformation conducive to the antagonist activity of OHT in tamoxifen-resistant cell lines. However, 5 μm disulfide benzamide inhibited the growth of ZR-75 cells by ∼20% but did not inhibit the growth of tamoxifen-resistant BT474 cells (49). In contrast, growth of both ZR-75 cells and BT474 cells was inhibited by TPSF (IC50 values = 0.9 and 1.6 μm, respectively). Because TPSF does not compete with E2 for binding to ER in a direct binding assay or in transactivation, TPSF is not a classical antagonist ligand and is distinct from known ERα inhibitors.
Fulvestrant is a high affinity ER ligand with nearly pure antagonist activity. Although fulvestrant is used therapeutically to treat advanced breast cancer, its use is limited by the fact that it can require several months for fulvestrant to reach a therapeutic level in serum (75). It has been known for many years that fulvestrant and related compounds, such as ICI 162,380, enhance the degradation of ERα (37, 76), although the mechanisms are not known. Recent solution of the structure of fulvestrant bound to the ligand binding domain of ERα suggests that fulvestrant binding may distort ERα structure so that a few hydrophobic amino acids are exposed near the surface, perhaps triggering recognition of ERα as a misfolded protein and rapid degradation (8). Although this is an attractive hypothesis, this idea has not been tested in experiments.
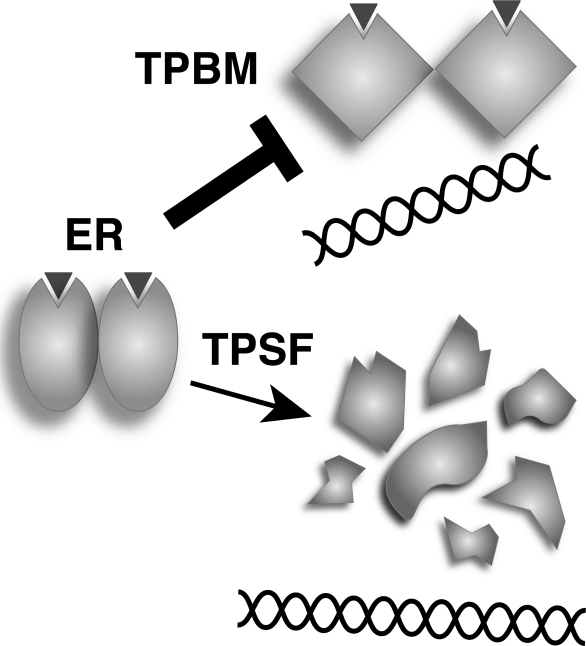
Structurally Related TPBM and TPSF Elicit Different Effects
Because ERα and other steroid receptors exhibit a high level of conformational flexibility, small molecules can elicit quite different conformations when they interact with ERα. For example, binding of E2 or OHT in the ERα ligand binding pocket resulted in functionally distinct agonist and antagonist conformations (8). Thus, binding of structurally related ERα inhibitors TPBM (19) and the more potent TPSF may cause distinct ERα conformations that are associated with different modes of action. The different actions of TPBM and TPSF are illustrated in Fig. 10. TPBM inhibited E2-ERα binding to ERE DNA in vitro but had no effect on the intracellular level of ERα. In contrast, TPSF had no effect on binding of E2-ERα to ERE DNA but elicited a concentration-dependent reduction in ERα protein levels. TPSF also reduced the ERα protein level in our HeLa-ERα cells that stably express FLAG-tagged ERα from a cytomegalovirus promoter that drives transcription of the ∼2000 nucleotide ERα cDNA (76) and did not reduce the level of ERα mRNA. Thus, at least part of the inhibitory effect of TPSF appears to reflect its ability to down-regulate ERα protein. E2-ERα induction of PI-9 mRNA and of the stably transfected (ERE)3-Luc reporter is inhibited by TPSF with IC50 values of 0.2 and 0.7 μm, respectively, with only a modest effect on ERα levels. It is possible that regulation of some genes is more sensitive to small changes ERα levels. Another possibility is that low concentrations of TPSF did not saturate ERα. Under these conditions, ERα may assume a transient TPSF-induced conformation sufficient to alter ERα function and inhibit E2-ERα-mediated transactivation at PI-9 and (ERE)3-Luc, whereas higher concentrations of TPSF may be required for an effect on levels of ERα.
FIGURE 10.
Schematic representation of the different modes of action of TPSF and TPBM.
In conclusion, TPSF is a potent and specific small molecule inhibitor of ERα that blocks ERα-mediated gene expression and estrogen-dependent growth of tamoxifen-sensitive and tamoxifen-resistant human breast cancer cells. TPSF inhibition of ERα is consistent with a role for estrogen induction of cyclinD1 in triggering the growth of breast cancer cells. TPSF represents a novel compound with potential for treating breast cancer and for probing the mechanisms of ER action.
Supplementary Material
Acknowledgments
We are most grateful to K. Carlson and Prof. J. Katzenellenbogen who performed the determination of the relative binding affinity of TPSF, Z. Erdogan and Prof. B. Katzenellenbogen for much helpful advice on MCF-7 cell growth assays, Dr. B. Huang and Prof. L.-F. Chen for important advice on growth of MCF-7 cells in soft agar, Prof. F. Wang for use of his microphotography system, Prof. E. Alarid who provided the MCF7ERαHA cells, and Prof. A. Nardulli who provided the MDA-MB-231 cells.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK 071909 (to D. J. S.) and RO1 HD 16910 and PO1-CA77739 (to E. M. W.). This work was also supported by a Bridge grant from the Endocrine Society (to D. J. S.). The University of Illinois at Urbana-Champaign has filed a novel use patent that claims TPSF. D. J. Shapiro, N. M. Patterson, M. Cherian, and C. Mao are co-inventors on the patent.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ERα
- estrogen receptor α
- E2
- 17β-estradiol
- TPSF
- butyrophenone, p-fluoro-4-(1,2,3,6,-tetrahydro-1,3-dimethyl-2-oxo-6-thionpurin-8-ylthio)
- TPBM
- 8-benzylsulfanylmethyl-1,3-dimethyl-3,7-dihydropurine-2,6-dione
- OHT
- 4-hydroxytamoxifen
- Luc
- luciferase
- MMTV
- mouse mammary tumor virus
- PSA
- prostate-specific antigen
- AR
- androgen receptor
- GR
- glucocorticoid receptor
- ERE
- estrogen response element
- flcERE
- fluorescein-labeled consensus ERE
- AIB1
- amplified in breast cancer 1
- Dox
- doxycycline
- qRT-PCR
- quantitative reverse transcriptase-PCR
- CD
- charcoal dextran
- PI-9
- proteinase inhibitor 9
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- PR
- progesterone receptor
- MEM
- minimum Eagle's medium
- CS
- calf serum.
REFERENCES
- 1.O'Lone R., Frith M. C., Karlsson E. K., Hansen U. (2004) Mol. Endocrinol. 18, 1859–1875 [DOI] [PubMed] [Google Scholar]
- 2.Carroll J. S., Brown M. (2006) Mol. Endocrinol. 20, 1707–1714 [DOI] [PubMed] [Google Scholar]
- 3.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 4.Jakacka M., Ito M., Weiss J., Chien P. Y., Gehm B. D., Jameson J. L. (2001) J. Biol. Chem. 276, 13615–13621 [DOI] [PubMed] [Google Scholar]
- 5.Safe S. (2001) Vitam. Horm. 62, 231–252 [DOI] [PubMed] [Google Scholar]
- 6.Kushner P. J., Agard D. A., Greene G. L., Scanlan T. S., Shiau A. K., Uht R. M., Webb P. (2000) J. Steroid Biochem. Mol. Biol. 74, 311–317 [DOI] [PubMed] [Google Scholar]
- 7.Qin C., Singh P., Safe S. (1999) Endocrinology 140, 2501–2508 [DOI] [PubMed] [Google Scholar]
- 8.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 9.Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 10.McKenna N. J., O'Malley B. W. (2002) Endocrinology 143, 2461–2465 [DOI] [PubMed] [Google Scholar]
- 11.McKenna N. J., O'Malley B. W. (2002) Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 12.Shang Y., Brown M. (2002) Science 295, 2465–2468 [DOI] [PubMed] [Google Scholar]
- 13.Henderson B. E., Feigelson H. S. (2000) Carcinogenesis 21, 427–433 [DOI] [PubMed] [Google Scholar]
- 14.Deroo B. J., Korach K. S. (2006) J. Clin. Invest. 116, 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabian C. J., Kimler B. F. (2005) J Clin. Oncol. 23, 1644–1655 [DOI] [PubMed] [Google Scholar]
- 16.Katzenellenbogen B. S., Montano M. M., Ekena K., Herman M. E., McInerney E. M. (1997) Breast Cancer Res. Treat. 44, 23–38 [DOI] [PubMed] [Google Scholar]
- 17.Anderson H., Bulun S., Smith I., Dowsett M. (2007) J. Steroid Biochem. Mol. Biol. 106, 49–54 [DOI] [PubMed] [Google Scholar]
- 18.Jordan V. C. (2001) Ann. N.Y. Acad. Sci. 949, 72–79 [DOI] [PubMed] [Google Scholar]
- 19.Mao C., Patterson N. M., Cherian M. T., Aninye I. O., Zhang C., Montoya J. B., Cheng J., Putt K. S., Hergenrother P. J., Wilson E. M., Nardulli A. M., Nordeen S. K., Shapiro D. J. (2008) J. Biol. Chem. 283, 12819–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler A. M., Solodin N., Preisler-Mashek M. T., Zhang P., Lee A. V., Alarid E. T. (2004) FASEB J. 18, 81–93 [DOI] [PubMed] [Google Scholar]
- 21.Jiang X., Patterson N. M., Ling Y., Xie J., Helferich W. G., Shapiro D. J. (2008) Endocrinology 149, 5366–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowler A. M., Solodin N. M., Valley C. C., Alarid E. T. (2006) Mol. Endocrinol. 20, 291–301 [DOI] [PubMed] [Google Scholar]
- 23.Jiang X., Ellison S. J., Alarid E. T., Shapiro D. J. (2007) Oncogene 26, 4106–4114 [DOI] [PubMed] [Google Scholar]
- 24.Wilson V. S., Bobseine K., Gray L. E., Jr. (2004) Toxicol Sci. 81, 69–77 [DOI] [PubMed] [Google Scholar]
- 25.Nordeen S. K., Kühnel B., Lawler-Heavner J., Barber D. A., Edwards D. P. (1989) Mol. Endocrinol. 3, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 26.Carlson K. E., Choi I., Gee A., Katzenellenbogen B. S., Katzenellenbogen J. A. (1997) Biochemistry 36, 14897–14905 [DOI] [PubMed] [Google Scholar]
- 27.Katzenellenbogen J. A., Johnson H. J., Jr., Myers H. N. (1973) Biochemistry 12, 4085–4092 [DOI] [PubMed] [Google Scholar]
- 28.Jiang X., Orr B. A., Kranz D. M., Shapiro D. J. (2006) Endocrinology 147, 1419–1426 [DOI] [PubMed] [Google Scholar]
- 29.Chang C., Norris J. D., Grøn H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. (1999) Mol. Cell. Biol. 19, 8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C. Y., Norris J. D., Jansen M., Huang H. J., McDonnell D. P. (2003) Methods Enzymol. 364, 118–142 [DOI] [PubMed] [Google Scholar]
- 31.Gunther J. R., Moore T. W., Collins M. L., Katzenellenbogen J. A. (2008) ACS Chem. Biol. 3, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaFrate A. L., Gunther J. R., Carlson K. E., Katzenellenbogen J. A. (2008) Bioorg. Med. Chem. 16, 10075–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norris J. D., Paige L. A., Christensen D. J., Chang C. Y., Huacani M. R., Fan D., Hamilton P. T., Fowlkes D. M., McDonnell D. P. (1999) Science 285, 744–746 [DOI] [PubMed] [Google Scholar]
- 34.ten Berge R. L., de Bruin P. C., Oudejans J. J., Ossenkoppele G. J., van der Valk P., Meijer C. J. (2003) Histopathology 43, 462–469 [DOI] [PubMed] [Google Scholar]
- 35.ten Berge R. L., Meijer C. J., Dukers D. F., Kummer J. A., Bladergroen B. A., Vos W., Hack C. E., Ossenkoppele G. J., Oudejans J. J. (2002) Blood 99, 4540–4546 [DOI] [PubMed] [Google Scholar]
- 36.van Houdt I. S., Oudejans J. J., van den Eertwegh A. J., Baars A., Vos W., Bladergroen B. A., Rimoldi D., Muris J. J., Hooijberg E., Gundy C. M., Meijer C. J., Kummer J. A. (2005) Clin. Cancer Res. 11, 6400–6407 [DOI] [PubMed] [Google Scholar]
- 37.Reese J. C., Katzenellenbogen B. S. (1991) Nucleic Acids Res. 19, 6595–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kummer J. A., Micheau O., Schneider P., Bovenschen N., Broekhuizen R., Quadir R., Strik M. C., Hack C. E., Tschopp J. (2007) Cell Death Differ. 14, 1486–1496 [DOI] [PubMed] [Google Scholar]
- 39.Cunningham T. D., Jiang X., Shapiro D. J. (2007) Cell. Immunol. 245, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krieg A. J., Krieg S. A., Ahn B. S., Shapiro D. J. (2004) J. Biol. Chem. 279, 5025–5034 [DOI] [PubMed] [Google Scholar]
- 41.Krieg S. A., Krieg A. J., Shapiro D. J. (2001) Mol. Endocrinol. 15, 1971–1982 [DOI] [PubMed] [Google Scholar]
- 42.Musgrove E. A., Lee C. S., Buckley M. F., Sutherland R. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8022–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuman E., Ladha M. H., Lin N., Upton T. M., Miller S. J., DiRenzo J., Pestell R. G., Hinds P. W., Dowdy S. F., Brown M., Ewen M. E. (1997) Mol. Cell. Biol. 17, 5338–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabbah M., Courilleau D., Mester J., Redeuilh G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilker R. L., Planas-Silva M. D. (2006) Cancer Res. 66, 11478–11484 [DOI] [PubMed] [Google Scholar]
- 46.Castro-Rivera E., Samudio I., Safe S. (2001) J. Biol. Chem. 276, 30853–30861 [DOI] [PubMed] [Google Scholar]
- 47.Chen Lf., Fischle W., Verdin E., Greene W. C. (2001) Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 48.Naughton C., MacLeod K., Kuske B., Clarke R., Cameron D. A., Langdon S. P. (2007) Mol. Endocrinol. 21, 2615–2626 [DOI] [PubMed] [Google Scholar]
- 49.Wang L. H., Yang X. Y., Zhang X., An P., Kim H. J., Huang J., Clarke R., Osborne C. K., Inman J. K., Appella E., Farrar W. L. (2006) Cancer Cell 10, 487–499 [DOI] [PubMed] [Google Scholar]
- 50.Coradini D., Biffi A., Cappelletti V., Di Fronzo G. (1995) Cancer Detect. Prev. 19, 348–354 [PubMed] [Google Scholar]
- 51.Arteaga C. L., Koli K. M., Dugger T. C., Clarke R. (1999) J. Natl. Cancer Inst. 91, 46–53 [DOI] [PubMed] [Google Scholar]
- 52.Schiff R., Massarweh S. A., Shou J., Bharwani L., Mohsin S. K., Osborne C. K. (2004) Clin. Cancer Res. 10, 331S–336S [DOI] [PubMed] [Google Scholar]
- 53.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 54.Reese J. C., Katzenellenbogen B. S. (1992) Mol. Cell. Biol. 12, 4531–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howell A., Dodwell D. J., Anderson H., Redford J. (1992) Ann. Oncol. 3, 611–617 [DOI] [PubMed] [Google Scholar]
- 56.Canney P. A., Griffiths T., Latief T. N., Priestman T. J. (1987) Lancet 1, 36. [DOI] [PubMed] [Google Scholar]
- 57.Ishii Y., Waxman S., Germain D. (2008) Cancer Res. 68, 852–860 [DOI] [PubMed] [Google Scholar]
- 58.Wang S., Zhang C., Nordeen S. K., Shapiro D. J. (2007) J. Biol. Chem. 282, 2765–2775 [DOI] [PubMed] [Google Scholar]
- 59.Wang S. Y., Ahn B. S., Harris R., Nordeen S. K., Shapiro D. J. (2004) Biotechniques 37, 807–817 [DOI] [PubMed] [Google Scholar]
- 60.Wei X., Xu H., Kufe D. (2006) Mol. Cell 21, 295–305 [DOI] [PubMed] [Google Scholar]
- 61.Jones J. O., Bolton E. C., Huang Y., Feau C., Guy R. K., Yamamoto K. R., Hann B., Diamond M. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7233–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramondetta L. M., Johnson A. J., Sun C. C., Atkinson N., Smith J. A., Jung M. S., Broaddus R., Iyer R. B., Burke T. (2009) Cancer 115, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 63.Grunberg S. M., Weiss M. H., Russell C. A., Spitz I. M., Ahmadi J., Sadun A., Sitruk-Ware R. (2006) Cancer Invest. 24, 727–733 [DOI] [PubMed] [Google Scholar]
- 64.Pan Y. F., Wansa K. D., Liu M. H., Zhao B., Hong S. Z., Tan P. Y., Lim K. S., Bourque G., Liu E. T., Cheung E. (2008) J. Biol. Chem. 283, 32977–32988 [DOI] [PubMed] [Google Scholar]
- 65.Powell E., Wang Y., Shapiro D. J., Xu W. (2010) J. Biol. Chem. 285, 16125–16134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Estébanez-Perpiñá E., Arnold L. A., Arnold A. A., Nguyen P., Rodrigues E. D., Mar E., Bateman R., Pallai P., Shokat K. M., Baxter J. D., Guy R. K., Webb P., Fletterick R. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16074–16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.ten Berge R. L., Oudejans J. J., Ossenkoppele G. J., Meijer C. J. (2003) J. Pathol. 200, 4–15 [DOI] [PubMed] [Google Scholar]
- 68.Grillo M., Bott M. J., Khandke N., McGinnis J. P., Miranda M., Meyyappan M., Rosfjord E. C., Rabindran S. K. (2006) Breast Cancer Res. Treat. 95, 185–194 [DOI] [PubMed] [Google Scholar]
- 69.Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 70.Lewis J. S., Jordan V. C. (2005) Mutat. Res. 591, 247–263 [DOI] [PubMed] [Google Scholar]
- 71.Thorpe S. M., Christensen I. J., Rasmussen B. B., Rose C. (1993) Eur. J. Cancer 29A, 971–977 [DOI] [PubMed] [Google Scholar]
- 72.Romain S., Chinot O., Guirou O., Soullière M., Martin P. M. (1994) Int. J. Cancer 59, 17–19 [DOI] [PubMed] [Google Scholar]
- 73.Abukhdeir A. M., Vitolo M. I., Argani P., De Marzo A. M., Karakas B., Konishi H., Gustin J. P., Lauring J., Garay J. P., Pendleton C., Konishi Y., Blair B. G., Brenner K., Garrett-Mayer E., Carraway H., Bachman K. E., Park B. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffmann J., Bohlmann R., Heinrich N., Hofmeister H., Kroll J., Künzer H., Lichtner R. B., Nishino Y., Parczyk K., Sauer G., Gieschen H., Ulbrich H. F., Schneider M. R. (2004) J. Natl. Cancer Inst. 96, 210–218 [DOI] [PubMed] [Google Scholar]
- 75.Robertson J. F. (2007) Oncologist 12, 774–784 [DOI] [PubMed] [Google Scholar]
- 76.Cheng J., Zhang C., Shapiro D. J. (2007) Endocrinology 148, 4634–4641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.