Abstract
Given the modulatory role of neuropeptide Y (NPY) in the immune system, we investigated the effect of NPY on the production of NO and IL-1β in microglia. Upon LPS stimulation, NPY treatment inhibited NO production as well as the expression of inducible nitric-oxide synthase (iNOS). Pharmacological studies with a selective Y1 receptor agonist and selective antagonists for Y1, Y2, and Y5 receptors demonstrated that inhibition of NO production and iNOS expression was mediated exclusively through Y1 receptor activation. Microglial cells stimulated with LPS and ATP responded with a massive release of IL-1β, as measured by ELISA. NPY inhibited this effect, suggesting that it can strongly impair the release of IL-1β. Furthermore, we observed that IL-1β stimulation induced NO production and that the use of a selective IL-1 receptor antagonist prevented NO production upon LPS stimulation. Moreover, NPY acting through Y1 receptor inhibited LPS-stimulated release of IL-1β, inhibiting NO synthesis. IL-1β activation of NF-κB was inhibited by NPY treatment, as observed by confocal microscopy and Western blotting analysis of nuclear translocation of NF-κB p65 subunit, leading to the decrease of NO synthesis. Our results showed that upon LPS challenge, microglial cells release IL-1β, promoting the production of NO through a NF-κB-dependent pathway. Also, NPY was able to strongly inhibit NO synthesis through Y1 receptor activation, which prevents IL-1β release and thus inhibits nuclear translocation of NF-κB. The role of NPY in key inflammatory events may contribute to unravel novel gateways to modulate inflammation associated with brain pathology.
Keywords: Inflammation, Interleukin, Lipopolysaccharide (LPS), NF-kappaB, Nitric Oxide, Microglia, Neuropeptide Y
Introduction
Brain inflammation is characterized primarily by microglia activation (1). Several stimuli, such as ATP (2), blood-derived factors, or microbial signals (e.g. lipopolysaccharide (LPS)), induce significant morphological changes in microglial cells (3). They become undistinguishable from active macrophages and are able to migrate and proliferate at sites of neuronal injury, where they release both neurotrophic and neurotoxic factors, and inflammatory mediators, such as adhesion molecules, cytokines, and complement molecules among others (4–6). Consequently, microglia response remains controversial because it can either be beneficial or deleterious depending on the nature, concentration, and time of exposure to the activating stimulus, and the cellular interactions of the targeted tissue. Once the triggering stimulus wanes, microglia participate in the down-modulation of the immune response and in the regulation of their own apoptosis via secretion of anti-inflammatory cytokines (3).
One of the outcomes of microglia activation is the production of nitric oxide (NO) from the conversion of l-arginine to l-citrulline by Ca2+-independent inducible nitric oxide synthase (iNOS)2 (7–9). NO is produced by numerous cells, and it is of particular importance for blood flow regulation, sleep-wake cycle, food intake and thermal regulation, immune system function, and neuronal transmission (10). Particularly, in the central nervous system, NO regulation presents itself as an opportunity to intervene in human health. NO can grant neuroprotection through the following mechanisms: reduction of Ca2+ influx, due to S-nitrosylation of caspase 3 and NR1 and NR2 subunits of the N-methyl-d-aspartate receptors, which leads to a decrease of cell death; activation of cyclic AMP-responsive element-binding protein and Akt via stimulation of the soluble guanylate cyclase-cyclic GMP-protein kinase G pathway; and generation of biliverdin, a precursor of bilirubin, which acts as an antioxidant and antinitrosive molecule, through the induction of the activity of heme oxygenase 1 (10).
However, NO can act as a pathophysiological agent because it is associated with hypertension, diabetes, and heart failure among other pathologies (8). In the central nervous system, high amounts of NO inhibit mitochondrial cytochrome oxidase in neurons, causing them to depolarize and to release glutamate and ultimately to die by excitotoxicity via N-methyl-d-aspartate receptors (11, 12). NO can also react with superoxide anions and form peroxynitrite, which detains strong oxidant properties and can damage cellular components when protein nitration takes place (10).
In rodents, LPS and IL-1β are two well documented stimuli able to induce the synthesis of NO by glial cells (13, 14). LPS is a Gram-negative cell wall component that binds to Toll-like receptor 4, mimicking the development of an inflammatory reaction (15, 16). IL-1β is a well recognized proinflammatory cytokine involved in excitotoxicity, ischemia, brain trauma, inflammation, and cell death (17–19) but also in the synthesis of neurotrophic factors (20). The mature and biologically active form of IL-1β becomes ready to be released to the extracellular space after interleukin-converting enzyme (ICE or caspase-1) cleaves the precursor form pro-IL-1β. To exert its biological function, mature IL-1β must bind to IL-1 main receptor 1 (IL-1R1), which requires an accessory protein (IL-1RAcP) for signal transduction of the IL-1·IL-1R1 complex. A type 2 receptor for IL-1α/β exists, but it lacks a signaling-competent cytosolic part, acting solely as a decoy receptor (20–22).
In patients suffering from rheumatoid arthritis or osteoarthritis, IL-1β secretion by monocytes can be modulated by neuropeptide Y (NPY) (23). NPY is a highly abundant neuropeptide in the peripheral and central nervous system, and it is involved in anxiety and stress-related behaviors, food intake, memory and learning, and epileptogenesis (24, 25). In recent years, NPY has been also described to play a pivotal role in the immune system; it is released from sympathetic nerves innervating immune organs, and NPY receptors are present on the surface of various leukocyte subgroups modulating the release of different cytokines (26). NPY can also increase the number of blood CD-161-positive cells (a marker for activated monocytes) and IL-1β secretion by neutrophils and peripheral blood mononuclear cells (27, 28). Additionally, exogenous NPY decreases tissue immigration of blood monocytes (29). Indeed, all NPY receptors are involved in the functioning of the immune system. In particular, Y1 receptor appears to have a bimodal role (30). NPY signaling via this receptor acts as a key activator of antigen-presenting cell function. Consequently, antigen-presenting cells in Y1R receptor-deficient mice are functionally impaired to produce Th1-promoting cytokines and present antigens to T cells (31). On the other hand, in experimental autoimmune encephalomyelitis and in dextran sodium salt-induced colitis, loss of signaling through Y1 receptor on immune cells results in protection against disease (32, 33). In the present study, we revealed a novel signaling pathway through which NPY inhibits NO production induced by IL-1β; Y1R signaling prevents NF-κB activation triggered by IL-1β, in an inflammatory context occurring in a microglial cell line. Assessing the role of NPY in the regulation of microglia function could provide therapeutic targets for the prevention of neurological dysfunctions in several central nervous system injuries and chronic diseases, such as epilepsy, ischemia, stroke, Alzheimer disease, or multiple sclerosis (3, 34).
EXPERIMENTAL PROCEDURES
Cell Line Culture
The murine N9 microglia cell line (a kind gift from Prof. Claudia Verderio, CNR Institute of Neuroscience, Cellular and Molecular Pharmacology, Milan, Italy) was grown in RPMI medium supplemented with 30 mm glucose (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were kept at 37 °C in a 95% atmospheric air and 5% CO2 humidified atmosphere. The number of viable cells was evaluated, counting trypan blue-excluding cells. For immunocytochemistry studies, cells were plated at a density of 2 × 104 cells/well in 24-well trays or were plated at a density of 5 × 105 cells/well in 6-well trays (for the remaining experiments).
Cell treatment included the following incubation setup: NPY (human, rat/amidated sequence) (1 μm) (Bachem, Bubendorf, Switzerland) for 6 or 24 h, LPS (100 ng/ml) (Sigma) for 24 h, IL-1β (1.5 ng/ml) (R&D Systems, Minneapolis, MN) for 15 min or 6 h, ATP (1 mm) (Sigma) for 30 min, Y1 receptor agonist [Leu31,Pro34]NPY (porcine, amidated sequence) (1 μm) (Bachem) for 6 or 24 h, Y1 receptor antagonist BIBP3226 (1 μm, in water) (Bachem), Y2 receptor antagonist BIIE0246 (1 μm, in 30% DMSO) (Tocris, Bristol, UK), Y5 receptor antagonist L152-804 (1 μm, in 100% DMSO) (Tocris), and IL-1ra (150 ng/ml) (R&D Systems). All antagonists were added 30 min prior to cell treatment.
RNA Isolation from N9 Microglial Cells
The mRNA was isolated using the RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Briefly, cells were first lysed in a highly denaturing buffer containing guanidine thiocyanate, which ensured the inactivation of RNases. Samples were applied to spin columns, where total RNA bound to the membrane. Exclusion of contaminants and small size RNA allowed the purification of a high quality mRNA-enriched solution. RNA samples were stored in diethylpyrocarbonate-treated water (Sigma) at −80 °C prior to quantification by optical density measurement at 260 nm (RNA/DNA calculator GeneQuant II, Amersham Biosciences). The purity and integrity of the samples were determined using the ratio A260/A280 (only samples whose ratios were between 1.7 and 2.2 were transcribed) and by visual confirmation on the agarose gel.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis of NPY and NPY Receptor Expression in N9 Microglial Cells
A total of 2 μg of mRNA extracted was transcribed using 10 units/μl Reverase reverse transcriptase (Bioron GmbH, Ludwigshafen, Germany) and 0.05 μg/μl Oligo-p(dT)15 primers (Bioron GmbH). Amplification of NPY, NPY receptors, and β-actin was performed in a 50-μl reaction system (Bioron GmbH) containing 5 μl of template cDNA, 5 μl of 10× PCR reaction buffer, 10 mm deoxynucleotide mix, 0.2 μm upstream and downstream primers, 5000 units/ml TaqDNA polymerase (Amersham Biosciences), and RNase-free water.
Primer sequences were as follows: NPY forward, 5′-AGA GAT CCA GCC CTG AGA CA-3′; NPY reverse, 5′-AAC GAC AAC AAG GGA AAT GG-3′; Y1 receptor forward, 5′-AAC CTC TCC TTC TCA GAC TTG C-3′; Y1 receptor reverse, 5′-CAC AGT GTT GAA GAT GGT AAG G-3′; Y2 receptor forward, 5′-CTC CAA GCA AAT CAG CTT CC-3′; Y2 receptor reverse, 5′-GTT TTG TGC CTT CGC TGA TGG-3′; Y5 receptor forward, 5′-GTG TTC CCG AGG TGC TTC TA-3′; Y5 receptor reverse, 5′-ATT CCG AGC AGC AGC TGT AT-3′. Amplicons for NPY (236 bp), for Y1 receptor (615 bp), for Y2 receptor (318 bp), for Y5 receptor (524 bp), and for β-actin (428 bp) were run in a 1.5% agarose gel stained with ethidium bromide for visual confirmation. Densitometric analysis for the evaluation of mRNA expression of NPY and NPY receptors was performed on the Versa-Doc imaging system (model 3000, Bio-Rad).
Griess Assay
Production of NO was determined through the formation and accumulation of its stable metabolite product, nitrite (NO2−). Cells were incubated with lysis mixture solution (137 mm NaCl, 20 mm Tris-HCl, 1% Triton X-100, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 0.5 mm sodium vanadate (all from Sigma), pH 8.0). After gentle homogenization, the total amount of protein was quantified using the Bio-Rad method. A standard solution of 10 mm NaNO2 (Sigma) was diluted to concentrations ranging from 1 to 100 μm and applied in duplicates to 96-well EIA/RIA plates (Costar, Corning Glass). Griess reagents were added (1:1) to each well: 0.1% N-1-naphthylenediamine dihydrochloride and 1% sulfanilamide in 5% phosphoric acid (all from Sigma). Under acidic conditions, in the presence of nitrite, a pink chromophoric azo compound is produced (protocol adapted from Huygen (35), originally reported by Griess in 1879 (77)). Optical density was recorded at 540 nm in an ELISA plate reader (SPECTRA max 384 Plus, Molecular Devices).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (Sigma) and then placed for 20 min in permeabilizing solution (0.3% bovine serum albumin (BSA) and 3% Triton X-100 (all from Sigma)). Nonspecific binding was prevented by incubating cells in a 3% BSA and 0.3% Triton X-100 solution for 30 min at room temperature. Cells were kept overnight at 4 °C in a primary antibody solution and then washed with PBS and incubated for 1 h at room temperature with the corresponding secondary antibody.
Antibodies used were as follows: rabbit polyclonal anti-NPY (1:1000) (Sigma); sheep polyclonal anti-Y1R (1:1000) (AbD Serotec, Oxfordshire, UK); rabbit monoclonal anti-iNOS (1:250) (Millipore Corp., Bedford, MA); rat monoclonal anti-CD11b (1:1000) (AbD Serotec); rabbit monoclonal anti-NF-κB p65 (1:100) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in 0.1% Triton X-100, 0.3% BSA solution; and Alexa Fluor 594 goat anti-rabbit, Alexa Fluor 594 donkey anti-sheep, Alexa Fluor 594 goat anti-rat, Alexa Fluor 488 donkey anti-rabbit, and Alexa Fluor 488 goat anti-rat (all 1:200 in PBS, from Molecular Probes, Eugene, OR). For nuclear labeling, cell preparations were stained with Hoechst 33342 (2 μg/ml) (Molecular Probes) in 0.3% BSA for 5 min at room temperature and mounted in Dakocytomation fluorescent medium (Dakocytomation Inc.). Fluorescent images were acquired using a confocal microscope (LSM 510 Meta, Carl Zeiss, Göttingen, Germany).
Nuclear and Cytosolic Extracts for NF-κB p65 Labeling
After cell treatment with 1.5 ng/ml IL-1β and/or 1 μm NPY, cells were lysed and collected in 500 μl of buffer 1 (10 mm HEPES, 10 mm NaCl, 3 mm MgCl2, 0.1% Triton X-100, 1 mm EGTA, 0.1% chymostatin, 0.1% leupeptin, 0.1% antipain, and 0.1% pepstatin (all from Sigma), pH 7.5). Samples were centrifuged for 12 min at 2,300 × g at 4 °C, and the supernatant corresponding to the cytosolic extract was collected. Pellets were resuspended in 30 μl of buffer 2 (25 mm HEPES, 300 mm NaCl, 5 mm MgCl2, 20% glycerol, 1 mm EGTA, 0.1% chymostatin, 0.1% leupeptin, 0.1% antipain, and 0.1% pepstatin (all from Sigma), pH 7.5), centrifuged at 10,600 × g for 20 min at 4 °C, and the supernatant corresponding to the nuclear extract was collected. The protocol was adapted from Santos et al. (36). Total protein from each sample was quantified using the Bio-Rad method.
Western Blotting
Total protein from cell lysates (prepared as described under “Griess Assay”) was quantified using the Bio-Rad method. Afterward, samples were loaded onto 10% acrylamide/bisacrylamide gels (Bio-Rad) (for NPY and NF-κB p65 detection, the percentage of acrylamide/bisacrylamide used was 15%). Proteins were separated by SDS-PAGE using a Bicine/SDS (Sigma) electrophoresis buffer (pH 8.3) and then transferred to PVDF membranes (Millipore) with a 0.2-μm pore size for NPY and a 0.45-μm pore size for the remaining proteins under the following conditions: 300 mA, 90 min at 4 °C in a solution containing 10 mm CAPS (Sigma) and 10% methanol (VWR International S.A.S.), pH 11.0 (protocol adapted from Pinheiro et al. (37)). Membranes were blocked in Tris-buffered saline containing 5% low fat milk and 0.1% Tween 20 (Sigma) for 1 h at room temperature and then incubated overnight at 4 °C with the primary antibody solution diluted in 1% TBS-Tween, 0.5% low fat milk.
The following primary antibodies were used: rabbit monoclonal anti-iNOS (1:1,000) (BD Biosciences), rabbit monoclonal anti-NF-κB p65 (1:100) (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-NPY (1:100) (Sigma), and sheep polyclonal anti-Y1R (1:10,000) (AbD Serotec). After rinsing three times with TBS-T 0.5% low fat milk, membranes were incubated for 1 h at room temperature with an alkaline phosphatase-linked secondary antibody anti-rabbit IgG (1:20,000), and anti-sheep IgG (1:1,000), in 1% TBS-T, 0.5% low fat milk (GE Healthcare). For endogenous control immunolabeling, primary antibody solutions consisted of mouse monoclonal anti-α-tubulin (1:10,000) and rabbit monoclonal anti-histone (1:10,000) (Millipore). Protein-immunoreactive bands were visualized in a Versa-Doc imaging system (model 3000, Bio-Rad), after incubation of the membrane with ECFTM (GE Healthcare) for 5 min.
ELISA for IL-1β
Cells were plated and treated with NPY as described above (see “Cell Line Culture”). For the quantification of IL-1β protein levels, a mouse IL-1β ELISA kit was used following the manufacturer's instructions (eBioscience, San Diego, CA). Cells were left at room temperature for 5 min in lysis buffer (137 mm NaCl, 20 mm Tris-HCl, 1% Triton X-100, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, 1 μg/ml leupeptin, 0.5 mm sodium orthovanadate (all from Sigma), pH 8.0). Total protein concentration was determined by the bicinchoninic acid method (BCA), and samples were stored at −80 °C.
Microtiter plates (MaxiSorp, Nunc A/S, Roskilde, Denmark) were coated with 100 μl/well capture antibody in coating buffer. Plates were sealed and left overnight at 4 °C. Wells were washed, blocked with 1× assay diluent, and left at room temperature for 1 h. After washing, 100 μl of each sample was added, as well as standard solutions, after performing 2-fold serial dilutions of the top standard. The plate was sealed and left incubating for 2 h at room temperature. Afterward, 100 μl/well detection antibody diluted in 1× assay diluent was added, and the plate was sealed and incubated at room temperature for 1 h. Washes were repeated, and 100 μl/well Avidin-HRP diluted in 1× assay diluent was added. Then the plate was sealed and kept at room temperature for 30 min. Wells were soaked in wash buffer for 5 min prior to aspiration, and 100 μl/well substrate solution was added to each well and incubated at room temperature for 15 min. Afterward, 50 μl/well stop solution was added. Optical density was recorded at 450 and 570 nm (values later subtracted from those obtained with 450 nm) in an ELISA plate (SPECTRA max 384 Plus, Molecular Devices).
Data Analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA). Statistical significance was considered relevant for p values of <0.05 using one-way analysis of variance followed by Bonferroni's post hoc test for comparison among experimental settings and Dunnett's post hoc test for comparison with control condition. In Western blotting studies for NPY and Y1R, statistical significance was determined using an unpaired one-tailed Student's t test. Data were presented as means ± S.E. For every immunocytochemistry analysis, five independent microscopy fields were acquired per coverslip with a ×40 objective (about 40 cells/field). Every experimental condition was tested in three sets of independent experiments, unless stated otherwise, and performed in duplicates.
RESULTS
Expression of NPY and Y1 Receptor Increase in Murine N9 Microglia Cell Line upon LPS-induced Inflammation
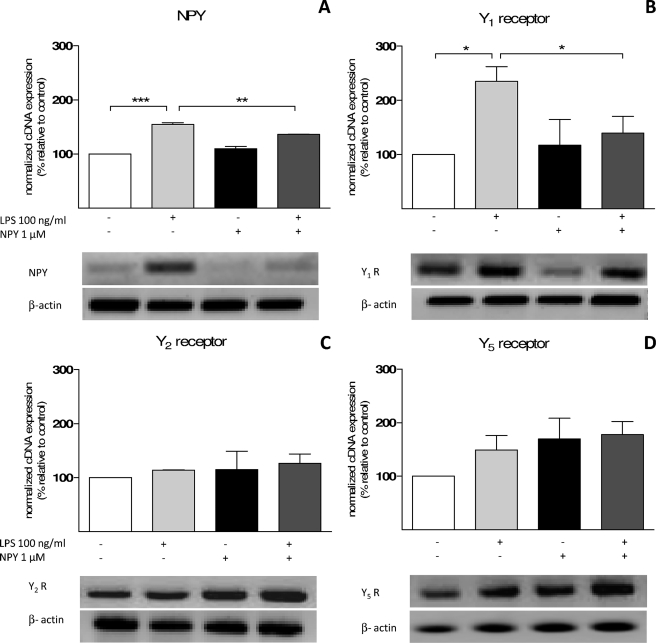
Murine N9 microglia cell line was used as a biological model to study endotoxin-induced inflammation. First, we performed conventional PCR as a qualitative approach to identify the expression of NPY and NPY receptors. We amplified cDNA coding for NPY, Y1 receptor (Y1R), Y2 receptor (Y2R), and Y5 receptor (Y5R). In order to have a semiquantitative analysis, β-actin was used as an endogenous control, given its stable expression in every experimental condition (38). Mouse hippocampal samples were used as a positive control because the hippocampus is a brain region known to highly express NPY and its receptors Y1, Y2, and Y5 (39–41). As a negative control, we used samples from negative transcription reactions (no template controls). Before RNA extraction, cells were treated with 1 μm NPY and challenged with 100 ng/ml LPS for 24 h. LPS is a key element of the outer membrane of Gram-negative bacteria, which binds to the CD14·TLR4·MD2 receptor complex, promoting the secretion of proinflammatory cytokines and the activation of several signaling cascades (42). The N9 microglial cell line did not abundantly express NPY in control conditions, although there was a significant increase of NPY cDNA when cells were treated with 100 ng/ml LPS (p < 0.001, n = 3) (Fig. 1A). Moreover, NPY treatment inhibited the described LPS effect (p < 0.01, n = 3) (Fig. 1A). Y1R, Y2R, and Y5R were detected in N9 microglia cell line, and a significant increase in Y1R expression was observed upon LPS challenge (p < 0.05, n = 3) (Fig. 1B), whereas no significant differences were obtained for Y2R (Fig. 1C) or for Y5R (Fig. 1D). Furthermore, NPY treatment caused a significant decrease in Y1R cDNA copies, when challenged with LPS, as compared with LPS alone (p < 0.05) (Fig. 1B). This decrease did not differ significantly from control levels.
FIGURE 1.
Murine N9 microglial cell line expresses NPY and receptors Y1, Y2, and Y5. Shown is RT-PCR detection of amplified products for NPY (236 bp) (A), Y1R (616 bp) (B), Y2R (318 bp) (C), and Y5R (524 bp) (D). Cells were treated with 1 μm NPY and challenged with 100 ng/ml LPS for 24 h. For semiquantitative analysis, results were normalized to β-actin (428 bp), an endogenous control. LPS-stimulated microglia significantly expressed higher levels of Y1R and NPY cDNA copies. Representative agarose gels for each amplified PCR product are depicted below the respective graph. Data are expressed as mean ± S.E. (error bars) (n = 3) and as a percentage of control (*, p < 0.05; **, p < 0.01; ***, p < 0.001, using Bonferroni's multiple comparison test).
To determine if the differences observed in cDNA levels translated into significant alterations of the pattern of protein expression, we performed immunocytochemistry for NPY and Y1R. To visualize microglia morphology, we labeled the α chain of αMβ2-integrin, CD11b, a well known surface marker, closely associated with microglial activation, and a mediator of the diapedesis process of leukocytes through the endothelium (43). As expected, LPS treatment led to an altered cell morphology shown by an increase of CD11b expression and a bloated cell body (Fig. 2A, LPS). Furthermore, we observed an increase in NPY labeling (Fig. 2A, top) and in both Y1R signal and distribution (Fig. 2A, bottom) induced by LPS. In addition, by Western blotting analysis, we could observe a significant increase in NPY (4 kDa) and Y1R (44 kDa) protein levels (meanNPY = 135.5 ± 12.25%; p < 0.05, n = 6; meanY1R = 150.20 ± 18.74%; n = 4) after LPS challenge (Fig. 2B).
FIGURE 2.
LPS induces NPY and Y1 receptor expression. A, confocal microscopy photomicrographs show NPY and Y1R localization (in red) on microglial cells (green) under basal conditions (control) and after 100 ng/ml LPS challenge for 24 h (LPS). Microglial cells stimulated with LPS exhibited an activated phenotype and expressed higher levels of both NPY and Y1R. Cell morphology was visualized with CD11b labeling (in green), and nuclear morphology is shown with Hoechst 33342 staining (in blue). Scale bar, 10 μm. B, Western blotting analysis of LPS stimulatory effect on NPY (4 kDa) and Y1R (44 kDa). After LPS challenge, an increase in NPY and Y1R protein levels was observed. A representative blot is shown below each graph. Data are expressed as mean ± S.E. (error bars) (n = 6 for NPY and n = 4 for Y1R) and as a percentage of control (*, p < 0.05; using Student's t test for comparison with control).
NPY Prevents the Production of NO and Decreases iNOS Expression after LPS Stimulation
Activation of microglia by inflammatory stimuli, such as pathogens, adhesion molecules, and cytokines, leads to the expression of high levels of nitric-oxide synthase (NOS), with consequent increasing levels of NO (44, 45). Using the Griess assay, we quantified NO production by microglial cells after 100 ng/ml LPS incubation for 24 h (Fig. 3). LPS-stimulated cells produced significantly more NO than control cells (meanCTR = 100%; meanLPS = 280.65 ± 50.38%), and this effect was reverted in the presence of NPY (1 μm) (meanNPY = 98.88 ± 7.41%; meanLPS + NPY = 101.85 ± 6.59%; p < 0.01, n = 3) (Fig. 3A). These results suggested that NO production stimulated by LPS was inhibited by NPY treatment. To discard any contribution from endogenous NPY, we treated cells with the monoclonal antibody NPY-05 (6 μg/ml), which acts as an NPY scavenger by binding to the carboxyl terminus of this peptide (46). As expected, nitrite levels after NPY-05 treatment were similar to control (meanNPY05 = 109.89%, n = 3) and significantly different from those obtained with LPS challenge alone (p < 0.001). In the presence of NPY-05 and LPS, nitrite production was similar to LPS alone (meanLPS = 264.52 ± 7.27%; meanLPS + NPY05 = 256.14 ± 10.13%; n = 3), indicating that, in our experimental conditions, endogenous NPY did not play any role in NO inhibition (Fig. 3B). The efficacy of the neutralizing antibody was determined by performing a concentration-response curve with increasing concentrations of NPY-05 (ranging from 60 ng/ml to 6 μg/ml) in the presence of LPS and NPY (data not shown).
FIGURE 3.
NPY inhibits the production of nitric oxide. A, LPS (100 ng/ml) significantly induced nitrite production, an indirect measure of the amount of NO, whereas NPY (1 μm) inhibited nitrite production upon LPS stimulation. B, preincubation with NPY-05 (6 μg/ml), a NPY scavenger, did not change the amount of NO when compared with control, indicating that, in our experimental conditions, endogenous NPY does not contribute to the inhibition of LPS-induced nitrite production. Data are expressed as mean ± S.E. (error bars) (n = 3) and as a percentage of control (**, p < 0.01; ***, p < 0.001, using Bonferroni's multiple comparison test).
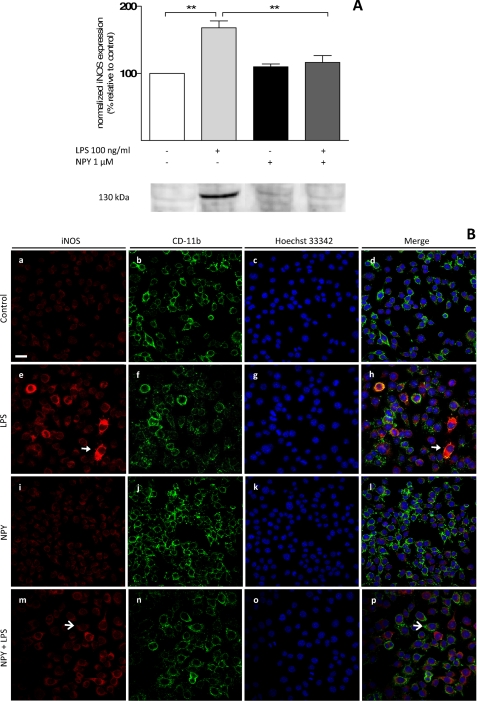
To determine how NPY exposure was involved in the inhibition of NO production, we tested whether NPY was affecting the synthesis of the converting enzyme iNOS, the isoform present in microglia. By Western blotting, we observed that LPS significantly induced an increase in iNOS protein levels (meanLPS = 167.86 ± 10.43%; n = 3, p < 0.001) and that this effect was abolished by NPY (meanLPS + NPY = 116.47 ± 10.13%; p < 0.01, n = 3) (Fig. 4A). Moreover, we also performed immunocytochemistry in the same experimental conditions (Fig. 4B), and we observed that cells treated with NPY alone (Fig. 4B, i–l) displayed a weak labeling signal for iNOS and CD11b that was similar to control conditions (Fig. 4B, a–d). The strongest fluorescent signal was observed when cells were challenged with LPS (Fig. 4B, e–h); furthermore, a moderate effect was visualized when cells were treated with both NPY and LPS (Fig. 4B, m–p).
FIGURE 4.
NPY inhibits inducible nitric-oxide synthase expression. Microglial cells were treated with 1 μm NPY and challenged with 100 ng/ml LPS for 6 h to assess the effect of NPY on iNOS (130 kDa) protein levels. A, NPY significantly inhibited LPS-stimulated iNOS protein levels. Below the graph, a representative blot illustrates this effect. Data are expressed as mean ± S.E. (error bars) (n = 3) and as a percentage of control (**, p < 0.01, using Bonferroni's multiple comparison test). B, immunolabeling against iNOS (in red) and CD11b (in green) shows a weaker fluorescent signal when cells were treated with NPY alone (i–l), similar to control conditions (a–d). The strongest fluorescent signal was observed when cells were challenged with LPS (e–h). A moderate effect was visualized when cells were treated with both NPY and LPS (m–p). Closed arrowheads point to iNOS-positive cells, whereas open arrowheads show negative labeling. Nuclear morphology is shown with Hoechst 33342 staining (in blue). Scale bar, 20 μm.
Y1 Receptor Activation Mimics the Effect of NPY on NO Production
To further characterize the action of NPY over the inhibition of NO production, we aimed at determining which NPY receptor(s) could be involved. For that purpose, we started by incubating cells with a selective agonist for Y1 receptor, [Leu31,Pro34]NPY (1 μm), for 24 h. Microglial cells treated with [Leu31,Pro34]NPY and LPS (meanLPS + Leu31, Pro34 = 109.14 ± 9.36%) produced NO levels similar to control (meanCTR = 99.65%), to NPY-treated cells (meanNPY = 98.88 ± 7.41%; Fig. 3A), and to cells exposed to LPS plus NPY (meanNPY + LPS = 116.30 ± 1.29%, n = 3) (Fig. 5A). Additionally, we used a selective antagonist for Y1R, BIBP3226 (1 μm), to further confirm that NPY-mediated inhibition of NO production was exclusively via Y1R. In fact, when Y1R was blocked, microglia stimulated with LPS or with LPS plus NPY significantly increase NO levels (meanLPS + BIBP3226 = 236.07 ± 5.32%; p < 0.001; meanLPS + NPY + BIBP3226 = 211.8 ± 26.70%; p < 0.001, n = 3) when compared with cells treated with LPS and NPY (meanNPY + LPS = 116.30 ± 1.29%) (Fig. 5A). To further exclude any contribution by other NPY receptors, cells were co-incubated with BIIE0246 and L152-804 (selective antagonists for Y2R and Y5R, respectively), and then treated with NPY and challenged with LPS. Blocking Y2R and Y5R did not affect the ability of NPY to inhibit NO production, even after LPS challenge (meanLPS + NPY + BIIE0246 + L152-804 = 105.98 ± 5.19%; p < 0.001, n = 4) (Fig. 5B).
FIGURE 5.
NPY inhibits nitric oxide production via Y1 receptor activation. Microglia cells were treated with a selective Y1 receptor agonist [Leu31,Pro34]NPY (1 μm) and a selective Y1 receptor antagonist BIBP3226 (1 μm) to determine the effect of Y1R activation on LPS-induced nitrite production. A, cells challenged with LPS and treated with [Leu31,Pro34]NPY display nitrite production similar to control levels. Accordingly, when cells were pretreated with BIBP3226, no NPY inhibitory effect was observed. B, the involvement of other receptors was excluded with the use of selective antagonists for Y2 receptor (BIIE0246; 1 μm) and for Y5 receptor (L152-804, 1 μm). When Y2R and Y5R were blocked, NPY inhibited NO production stimulated by LPS. Data are expressed as mean ± S.E. (error bars) (n = 3 for A, and n = 4 for B) and as a percentage of control (***, p < 0.001, using Bonferroni's multiple comparison test).
NPY Modulates the Release of IL-1β
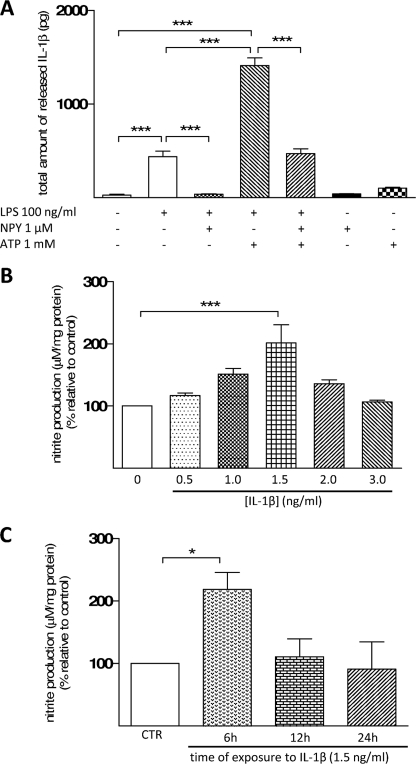
Another key feature of inflammation is the release of IL-1β by microglial cells. Using a quantitative method, such as ELISA, we observed that in the presence of LPS, there was a significant release of biologically active IL-1β (mature form) to the media (439.13 ± 58.90 pg; p < 0.001, n = 5) (Fig. 6A). When cells were simultaneously treated with NPY and LPS, the release of IL-1β was similar to control (control = 28.34 ± 9.06 pg; LPS + NPY = 36.94 ± 5.09 pg; p < 0.001, n = 5). To perceive the strength of this effect, cells were treated with ATP (1 mm). This nucleotide activates ICE in an inflammatory context (47) and, when co-administered with LPS, triggers a massive release of IL-1β (48–51). In the presence of these stimuli, we observed a highly significant release of IL-1β to the media (1412.69 ± 82.02 pg; p < 0.001, n = 5). Nevertheless, in microglia treated with NPY, this effect greatly diminished (41.21 ± 2.81 pg; p < 0.001, n = 5) (Fig. 6A). Moreover, because IL-1β has been described as a stimulator of NO production, a concentration-response curve was performed to determine which concentration of IL-1β would lead to a significant production of NO. We observed that 1.5 ng/ml IL-1β (201.67 ± 29.06%; p < 0.001, n = 3) (Fig. 6B) was the only concentration able to significantly increase NO production. Then we challenged cells with the selected concentration of IL-1β for 6, 12, and 24 h, and we found that 1.5 ng/ml IL-1β treatment for 6 h led to a significant production of NO (218.20 ± 32.85%; p < 0.05, n = 3) (Fig. 6C).
FIGURE 6.
NPY inhibits the release of interleukin-1β. A, microglia were stimulated with LPS (100 ng/ml) for 24 h to determine the effect of this endotoxin on IL-1β release. LPS induced the release of IL-1β, an effect inhibited by NPY treatment. Additionally, cells were challenged with both LPS and ATP (1 mm) for 30 min, which induced a massive release of IL-1β. NPY was also able to significantly reduce the amount of IL-1β released by microglial cells. B, a concentration-response curve was performed for IL-1β to observe which concentration induced a significant nitrite production. C, the selected concentration (1.5 ng/ml) was used to study which time of incubation is necessary to obtain a significant increase in NO production. Data are expressed as mean ± S.E. (error bars) (n = 5 for A; n = 3 for B and C) and as a percentage of control (A, ***, p < 0.001, using Bonferroni's multiple comparison test) (B and C, *, p < 0.05; ***, p < 0.001, using Dunnett's multiple comparison test).
NPY Blocks IL-1β-induced Production of NO through Y1 Receptor Activation
We sought to determine whether NPY could prevent the effect mediated by IL-1β on NO production. As previously shown, cells challenged with 1.5 ng/ml IL-1β for 6 h showed significant levels of NO (meanIL-1β = 209.57 ± 6.42%; p < 0.001, n = 3), whereas NPY co-exposure prevented this effect (meanNPY = 126.10 ± 1.77%; meanIL-1β + NPY = 111.28 ± 6.81%; p < 0.001, n = 3) (Fig. 7A). To assess how selective and robust was IL-1β-induced production of NO, we co-incubated microglial cells with a selective antagonist of IL-1β receptor (IL-1ra). To block the functional effects of IL-1β, a 102- to 103-fold higher dose of IL-1ra is needed (52); therefore, we used 150 ng/ml. We observed that microglial cells stimulated with IL-1β showed an increase of NO production (meanIL-1β = 204.66 ± 4.27%; p < 0.001, n = 3) and that IL-1ra inhibited this effect (meanIL-1β + IL-1ra = 112.96 ± 5.32%; p < 0.001, n = 3). Upon LPS challenge and in the presence of IL-1ra, NO production was inhibited (meanLPS = 236.07 ± 5.32%; meanIL-1ra + LPS = 114.94 ± 4.29%; p < 0.001, n = 3) (Fig. 7B). Moreover, we investigated if Y1R could be involved in this effect. In fact, when cells were exposed to IL-1β and treated with NPY and Y1R-selective antagonist, BIBP3226, the inhibitory effect of NPY was not observed (meanIL-1β + NPY = 112.98 ± 4.29%; meanIL-1β + NPY + BIBP3226 = 209.14 ± 9.36%; p < 0.001, n = 3). In contrast, in the presence of Y1R-selective agonist, [Leu31,Pro34]NPY, cells challenged with IL-1β produced NO levels similar to control (meanCTR = 99.65%; meanIL-1β + Leu31,Pro34 = 99.66 ± 0.67%; p < 0.001, n = 3) (Fig. 7C).
FIGURE 7.
NPY inhibits IL-1β-induced nitric oxide production through Y1 receptor activation. A, cells treated with 1.5 ng/ml IL-1β for 6 h produced significantly higher levels of NO; in the presence of NPY, IL-1β-induced NO production was prevented. B, selective IL-1β receptor antagonist IL-1ra (150 ng/ml) completely blocked the action of IL-1β over NO production. LPS-induced nitrite production is mediated by IL-1β because IL-1ra blocked this effect upon LPS challenge. C, cells were treated with a selective Y1 receptor agonist [Leu31,Pro34]NPY (1 μm) or with a selective Y1 receptor antagonist BIBP3226 (1 μm) to determine the effect of Y1 receptor activation on IL-1β-induced nitrite production. Activation of Y1R prevented IL-1β-induced NO production, whereas preincubation with BIBP3226 abolished this effect. Data are expressed as mean ± S.E. (error bars) (n = 3) and as a percentage of control (***, p < 0.001, using Bonferroni's multiple comparison test).
NPY Inhibits Nuclear Translocation of NF-κB p65 Induced by IL-1β Challenge
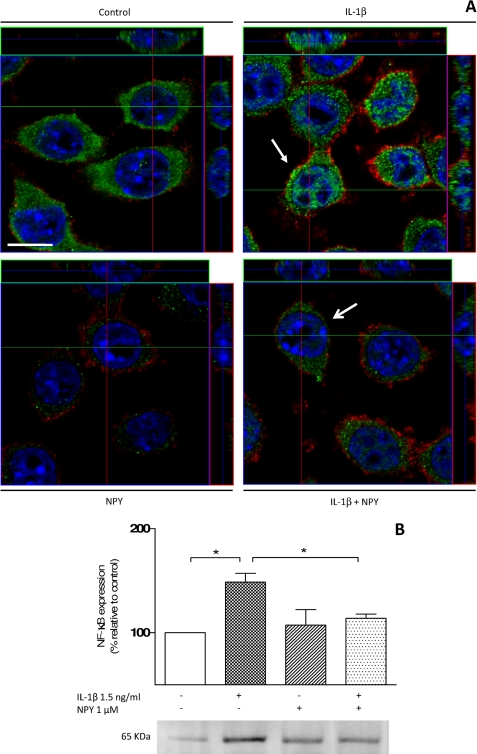
NF-κB is a well known transcription factor, which, upon microglia activation, is able to induce iNOS synthesis as well as proinflammatory cytokines, such as IL-1β (53). In the presence of IL-1β, we observed an increase in the signal of NF-κB p65-positive cells (Fig. 8A, top, right pictogram). When cells were treated with 1 μm NPY or with NPY plus IL-1β, little nuclear translocation of NF-κB p65 was detected (Fig. 8A, bottom), similar to control cells (Fig. 8A, top, left pictogram). Nuclear fraction extracts showed significantly increased NF-κB protein levels for cells challenged with IL-1β (meanIL-1β = 148.72 ± 8.37%; p < 0.05, n = 3), opposite to cells treated with NPY plus IL-1β (meanNPY + IL-1β = 107.31 ± 14.85%; p < 0.05, n = 3) (Fig. 8B).
FIGURE 8.
NPY inhibits nuclear translocation of NF-κB after IL-1β challenge. A, confocal microscopy photomicrographs of microglial cells treated with 1 μm NPY and 1.5 ng/ml IL-1β for 15 min were taken to assess the role of NPY and IL-1β in the NF-κB signaling pathway. Cells were stained for NF-κB (in green), for CD11b (in red), and with Hoechst 33342 (nuclei in blue). Nuclear translocation of NF-κB was promoted by IL-1β and inhibited when cells were treated with NPY. Orthogonal sections show nuclear localization of NF-κB (in green). Scale bar, 10 μm. B, Western blotting analysis was performed to study the inhibitory effect of NPY on NF-κB (65 kDa) nuclear translocation upon IL-1β stimulation. After IL-1β challenge, a significant increase in NF-κB protein levels was observed. When cells were treated with NPY, the amount NF-κB was reduced to values comparable with control. A representative blot is shown below the graph. Data are expressed as mean ± S.E. (n = 3) and as a percentage of control (*, p < 0.05, using Bonferroni's multiple comparison test).
NPY Blocks IL-1β-induced iNOS Expression
IL-1β induces activation of the NF-κB pathway, ultimately leading to the synthesis of iNOS mRNA and NO production (13, 14, 54). Therefore, we aimed at uncovering whether NPY would inhibit IL-1β-induced iNOS expression. Cells that were stimulated with 1.5 ng/ml IL-1β showed an increased expression of iNOS when compared with control cells or with cells treated with NPY, as observed by immunocytochemistry (Fig. 9). To quantify this effect, we performed a Western blotting analysis of iNOS (130 kDa) protein levels under the same experimental conditions (Fig. 9B). We observed that IL-1β significantly induced an increase in iNOS protein levels (meanIL-1β = 129.52 ± 5.05%; p < 0.01, n = 4) and that this effect was abolished by NPY (meanIL-1β + NPY = 107.76 ± 2.44%; p < 0.01, n = 4) (Fig. 9B).
FIGURE 9.
NPY inhibits IL-1β-induced iNOS protein levels. A, confocal microscopy photomicrographs illustrate microglial cells treated with 1 μm NPY and 1.5 ng/ml IL-1β for 6 h to assess the role of NPY and IL-1β in iNOS synthesis. To determine whether NPY was blocking the synthesis of NO induced by IL-1β treatment, cells were stained for NF-κB (in green), for CD11b (in red), and with Hoechst 33342 (nuclei in blue). An increase of iNOS labeling was induced by IL-1β administration and inhibited to an intensity comparable with fluorescence control values when treated with NPY. Scale bar, 10 μm. B, to provide a quantitative analysis, iNOS protein values were measured by Western blot. NPY inhibited IL-1β-induced iNOS levels. A representative blot is shown below the graph. Data are expressed as mean ± S.E. (n = 4) and as a percentage of control (**, p < 0.01; ***, p < 0.001 using Bonferroni's multiple comparison test).
DISCUSSION
Activated microglia respond to brain injury or infection, acting as immunocompetent cells capable of phagocytosis and able to release a diversity of chemical mediators of inflammation, including chemokines, cytokines, reactive oxygen, and nitrogen intermediates (55, 56). In the immune system, increasing evidence has implicated NPY as a key modulator (27–29, 57, 58). In fact, NPY has been shown to play a major role in important functional properties of the central nervous system, such as neural stem cell proliferation and differentiation, modulation of neurotransmission, neuroprotection, response to brain injury, and epilepsy (59–62). These findings suggest that NPY could work as a modulator of the inflammatory reaction of the brain immune system, eventually acting as a microglial activation repressor.
In order to address this hypothesis, we used an endotoxin-mediated model of inflammation to unravel the role of NPY in inflammatory mediators, such as IL-1β and NO, produced by a microglial cell line. Our results showed that NPY was able to prevent NO production by microglia after LPS challenge. Additionally, NPY inhibited the release of IL-1β and also prevented IL-1β-induced production of NO via activation of Y1 receptor. This effect was mediated through the NF-κB p65 signaling pathway because NPY was able to block nuclear translocation of this transcriptional factor and associated synthesis of iNOS.
Using conventional reverse transcription PCR, immunocytochemistry analysis, and Western blotting, we characterized the murine N9 microglial cell line with regard to the expression of NPY and its receptors Y1, Y2, and Y5. Our results clearly showed an increase of Y1R and NPY labeling and protein levels across cell membrane and cytoplasm, respectively, when microglia were challenged with LPS. Additionally, NPY treatment inhibited this effect, suggesting that it could act as a negative regulator of Y1 receptor expression. In accordance with this observation, Teixeira et al. (63) also demonstrated that treatment with NPY resulted in a significant decrease of Y1 receptor transcript in differentiating osteoblasts.
In the present work, we identified an inhibitory role for NPY in LPS-induced NO production. It has been previously reported that N9 murine microglial cells produce and release NO following exposure to LPS (64). Activation of macrophages by bacterial cell wall components can lead to the expression of high levels of NOS, with the most expressed isoform being iNOS, which oxidizes l-arginine to yield l-citrulline and NO. For that reason, we performed Western blotting and immunocytochemistry to determine possible differences in iNOS expression levels attributable to NPY. Our results showed that iNOS expression was significantly reduced when NPY was present, implying that NPY could be preventing de novo synthesis of this enzyme. To discover which receptor NPY was acting upon, we treated cells with selective Y1R agonist [Leu31,Pro34]NPY and selective antagonists for Y1R, Y2R, and Y5R (BIBP3226, BIIE0246, and L152-804, respectively), and as previously reported in the olfactory mucosa (65), the inhibitory effect of NPY on NO production involved the activation of Y1 receptor. In a study conducted in healthy human volunteers to determine dose-dependent effects of NPY on nasal mucosal blood flow, NPY was able to inhibit intranasal NO production (65). Moreover, RT-PCR analysis performed on nasal mucosa biopsies revealed only Y1 receptor mRNA detection, leading to the suggestion that NPY-evoked vasoconstriction was mediated via Y1 receptors.
Upon brain insult, IL-1α/β is synthesized and proteolytically processed to mature IL-1β by caspase-1 (21). As part of the repertoire of inflammation, excessive IL-1β synthesis and release from microglia can be detrimental to the injured brain. Accordingly, Bernardino and colleagues (51) showed that 100 μm VX-765, a selective ICE/caspase-1 inhibitor, or 1 μm IL-1ra (IL-1 receptor antagonist) blocked exacerbation of AMPA-induced neuronal damage during transient exposure to LPS and ATP. It has been described that LPS activates Toll-like receptor 4 and, during co-activation of P2X7 receptors by ATP, causes the release of IL-1β from microglial cells (51, 66). Moreover, Ohtani et al. (67) had shown that, in rat cultured microglia, ATP induced iNOS expression and NO production, presumably in cooperation with macrophage colony-stimulating factor present in the culture media. Also, Schroeder et al. (68) had shown that inhibition of NO synthesis leads to an increase of IL-1β protein expression in ANA-1 murine macrophages. The authors suggested a negative feedback mechanism through which NO production inhibited the synthesis of IL-1β by S-nitrosation of NF-κB, a transcription factor implicated in immune and inflammatory reactions. Our findings can further provide an insightful understanding of the liaison between IL-1β and NO, suggesting NPY as a key modulator of their interplay. Our results showed that microglia significantly released IL-1β (the biologically active form) to the medium when cells were treated with LPS and that this effect was potentiated when cells were treated simultaneously with LPS and ATP. In the presence of NPY, the release of IL-1β was significantly reduced, and this effect was mimicked using a selective Y1R agonist, implying that NPY acted via Y1 receptor. Moreover, cells treated with IL-1β significantly increased NO production, an effect abolished in the presence of NPY or Y1R agonist. Furthermore, LPS challenge, together with IL-1ra treatment, led to the inhibition of NO production. Hence, blockage of IL-1β receptor with IL-1ra inhibited NO production, suggesting that LPS action on NO production is mediated through this cytokine. In fact, some reports have also shown that IL-1ra inhibited iNOS in astrocytes (69–71).
In activated microglia, induction of iNOS and consequently NO production is likely to involve NF-κB (53). In broad terms, Toll-like receptors are activated by pathogen-associated molecular patterns and trigger a cascade of cellular signals leading to the activation of NF-κB. The Toll-like receptor superfamily includes IL-R1, through which IL-1β leads to NF-κB activation via a serine/threonine kinase called interleukin receptor-associated kinase (72). In relation to this, competitive inhibitors of serine/threonine protein kinases, such as calmodulin-regulated protein kinases, can modulate iNOS expression. Watterson et al. (73) screened the action of low molecular weight cell-permeable compounds described as calmodulin-regulated protein kinase inhibitors and found them to block the induction of both iNOS and IL-1β in primary cortical glial cultures and the microglial BV-2 cell line. Also, in rat aortic smooth muscle cells, NF-κB and C/EBP mediated IL-1β-induced iNOS expression (74). Hitherto, our data support a role for the NF-κB signaling pathway in the inflammation model used because this transcriptional factor was not able to translocate to the nucleus upon NPY treatment, even after IL-1β challenge. Interestingly, early in 1995, Ball et al. (75) reported the existence of a binding site for NF-κB in a promoter region of the human and murine Y1 receptor gene. Later, Musso et al. (76) showed that the murine Y1 receptor promoter region contained consensus sites for members of the κB-Rel family of transcription factors, which were able to bind κB-related nuclear complexes in a specific manner. The authors speculated on whether Y1 receptor could represent one of the κB site-containing genes regulated by κB-related factors responding to inflammatory stimuli.
In summary, our work established a novel role for NPY in the regulation of key events occurring during inflammation, converging with relevant evidence from the literature. Upon an endotoxin challenge, microglia respond with increased IL-1β and NO production, an effect inhibited by NPY via Y1 receptor activation, showing the involvement of the NF-κB signaling pathway in this process. Microglia are the smallest members of the glia family but greatly responsible for vital physiological responses to brain injury. Taken together, our data indicate a new integrated functional response of microglia cells and a key modulatory role for NPY. These findings may be valuable in revealing new drug targets to modulate the inflammatory reaction upon brain injury.
Acknowledgments
We thank Prof. Claudia Verderio (National Research Council, Institute of Neuroscience and Department of Medical Pharmacology, Milan, Italy) for the generous gift of the murine N9 microglial cell line, Prof. Paulo Santos (Center for Neuroscience and Cell Biology, Department of Zoology, University of Coimbra, Portugal), and Prof. Eric Grouzmann (Division of Clinical Pharmacology and Toxicology, Centre Hospitalier Universitaire Vaudois, Switzerland) for providing monoclonal antibody NPY-05, and Prof. Cláudia Cavadas (Faculty of Pharmacy, University of Coimbra, Portugal) for kind and constructive scientific suggestions.
This work was supported by the Fundação para a Ciência e a Tecnologia Portugal and Fundo-Europeu-De-Desenvolvimento-Regional Grants PTDC/SAU-NEU/68465/2006, PTDC/SAU-OSM/101469/2008 and SFRH/BD/23595/2005.
- iNOS
- inducible nitric-oxide synthase
- NOS
- nitric-oxide synthase
- ICE
- interleukin-converting enzyme
- IL-1ra
- interleukin-1 receptor antagonist
- NPY
- neuropeptide Y
- TBS-T
- Tris-buffered saline Tween 20®
- Y1R
- Y2R, and Y5R, Y1, Y2, and Y5 receptor, respectively
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
REFERENCES
- 1.Streit W. J., Walter S. A., Pennell N. A. (1999) Prog. Neurobiol. 57, 563–581 [DOI] [PubMed] [Google Scholar]
- 2.Inoue K. (1998) Pharmacol. Res. 38, 323–331 [DOI] [PubMed] [Google Scholar]
- 3.Garden G. A., Möller T. (2006) J. Neuroimmune Pharmacol. 1, 127–137 [DOI] [PubMed] [Google Scholar]
- 4.Minghetti L., Ajmone-Cat M. A., De Berardinis M. A., De Simone R. (2005) Brain Res. Rev. 48, 251–256 [DOI] [PubMed] [Google Scholar]
- 5.Block M. L., Zecca L., Hong J. S. (2007) Nat. Rev. Neurosci. 8, 57–69 [DOI] [PubMed] [Google Scholar]
- 6.Allan S. M., Rothwell N. J. (2003) Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer R. M., Ashton D. S., Moncada S. (1988) Nature 333, 664–666 [DOI] [PubMed] [Google Scholar]
- 8.Moncada S., Bolaños J. P. (2006) J. Neurochem. 97, 1676–1689 [DOI] [PubMed] [Google Scholar]
- 9.Knowles R. G., Moncada S. (1994) Biochem. J. 298, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calabrese V., Mancuso C., Calvani M., Rizzarelli E., Butterfield D. A., Stella A. M. (2007) Nat. Rev. Neurosci. 8, 766–775 [DOI] [PubMed] [Google Scholar]
- 11.Bal-Price A., Brown G. C. (2001) J. Neurosci. 21, 6480–6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown G. C. (2007) Biochem. Soc. Trans. 35, 1119–1121 [DOI] [PubMed] [Google Scholar]
- 13.Murphy S., Grzybicki D. (1996) Neuroscientist 2, 90–99 [Google Scholar]
- 14.Saha R. N., Pahan K. (2006) Antioxid. Redox Signal. 8, 929–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivest S. (2003) Brain Behav. Immun. 17, 13–19 [DOI] [PubMed] [Google Scholar]
- 16.Hagberg H., Mallard C. (2005) Curr. Opin. Neurol. 18, 117–123 [DOI] [PubMed] [Google Scholar]
- 17.Allan S. M. (2005) Methods Mol. Med. 104, 333–346 [DOI] [PubMed] [Google Scholar]
- 18.Bernardino L., Xapelli S., Silva A. P., Jakobsen B., Poulsen F. R., Oliveira C. R., Vezzani A., Malva J. O., Zimmer J. (2005) J. Neurosci. 25, 6734–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vezzani A., Granata T. (2005) Epilepsia 46, 1724–1743 [DOI] [PubMed] [Google Scholar]
- 20.Pinteaux E., Trotter P., Simi A. (2009) Cytokine 45, 1–7 [DOI] [PubMed] [Google Scholar]
- 21.Dinarello C. A. (2009) Annu. Rev. Immunol. 27, 519–550 [DOI] [PubMed] [Google Scholar]
- 22.Fantuzzi G., Dinarello C. A. (1999) J. Clin. Immunol. 19, 1–11 [DOI] [PubMed] [Google Scholar]
- 23.Hernanz A., Medina S., de Miguel E., Martín-Mola E. (2003) Regul. Pept. 115, 19–24 [DOI] [PubMed] [Google Scholar]
- 24.Heilig M. (2004) Neuropeptides 38, 213–224 [DOI] [PubMed] [Google Scholar]
- 25.Thorsell A., Heilig M. (2002) Neuropeptides 36, 182–193 [DOI] [PubMed] [Google Scholar]
- 26.Petitto J. M., Huang Z., McCarthy D. B. (1994) J. Neuroimmunol. 54, 81–86 [DOI] [PubMed] [Google Scholar]
- 27.Bedoui S., Kawamura N., Straub R. H., Pabst R., Yamamura T., von Hörsten S. (2003) J. Neuroimmunol. 134, 1–11 [DOI] [PubMed] [Google Scholar]
- 28.Bedoui S., von Hörsten S., Gebhardt T. (2007) Peptides 28, 373–376 [DOI] [PubMed] [Google Scholar]
- 29.Nave H., Bedoui S., Moenter F., Steffens J., Felies M., Gebhardt T., Straub R. H., Pabst R., Dimitrijevic M., Stanojevic S., von Hörsten S. (2004) J. Neuroimmunol. 155, 1–12 [DOI] [PubMed] [Google Scholar]
- 30.Wheway J., Mackay C. R., Newton R. A., Sainsbury A., Boey D., Herzog H., Mackay F. (2005) J. Exp. Med. 202, 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheway J., Herzog H., Mackay F. (2007) Peptides 28, 453–458 [DOI] [PubMed] [Google Scholar]
- 32.Bedoui S., Miyake S., Lin Y., Miyamoto K., Oki S., Kawamura N., Beck-Sickinger A., von Hörsten S., Yamamura T. (2003) J. Immunol. 171, 3451–3458 [DOI] [PubMed] [Google Scholar]
- 33.Hassani H., Lucas G., Rozell B., Ernfors P. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 288, G550–G556 [DOI] [PubMed] [Google Scholar]
- 34.Choi J., Koh S. (2008) Yonsei Med. J. 49, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huygen I. C. (1970) Anal. Chem. 42, 3. [DOI] [PubMed] [Google Scholar]
- 36.Santos A. E., Carvalho A. L., Lopes M. C., Carvalho A. P. (2001) J. Neurosci. Res. 66, 643–655 [DOI] [PubMed] [Google Scholar]
- 37.Pinheiro P. S., Rodrigues R. J., Rebola N., Xapelli S., Oliveira C. R., Malva J. O. (2005) Neurochem. Int. 47, 309–316 [DOI] [PubMed] [Google Scholar]
- 38.Bustin S. A. (2000) J. Mol. Endocrinol. 25, 169–193 [DOI] [PubMed] [Google Scholar]
- 39.de Quidt M. E., Emson P. C. (1986) Neuroscience 18, 527–543 [DOI] [PubMed] [Google Scholar]
- 40.de Quidt M. E., Emson P. C. (1986) Neuroscience 18, 545–618 [DOI] [PubMed] [Google Scholar]
- 41.Naveilhan P., Neveu I., Arenas E., Ernfors P. (1998) Neuroscience 87, 289–302 [DOI] [PubMed] [Google Scholar]
- 42.Cohen J. (2002) Nature 420, 885–891 [DOI] [PubMed] [Google Scholar]
- 43.V̆tvicka V., Hanikýrová M., V̆tvicková J., Ross G. D. (1999) Clin. Exp. Immunol. 115, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moss D. W., Bates T. E. (2001) Eur. J. Neurosci. 13, 529–538 [DOI] [PubMed] [Google Scholar]
- 45.Liu B., Gao H. M., Wang J. Y., Jeohn G. H., Cooper C. L., Hong J. S. (2002) Ann. N.Y. Acad. Sci. 962, 318–331 [DOI] [PubMed] [Google Scholar]
- 46.Brakch N., Allemandou F., Cavadas C., Grouzmann E., Brunner H. R. (2002) J. Neurochem. 81, 1166–1175 [DOI] [PubMed] [Google Scholar]
- 47.Abreu M. T., Arditi M. (2004) J. Pediatr. 144, 421–429 [DOI] [PubMed] [Google Scholar]
- 48.Griffiths R. J., Stam E. J., Downs J. T., Otterness I. G. (1995) J. Immunol. 154, 2821–2828 [PubMed] [Google Scholar]
- 49.Grahames C. B., Michel A. D., Chessell I. P., Humphrey P. P. (1999) Br. J. Pharmacol. 127, 1915–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrari D., Chiozzi P., Falzoni S., Dal Susino M., Melchiorri L., Baricordi O. R., Di Virgilio F. (1997) J. Immunol. 159, 1451–1458 [PubMed] [Google Scholar]
- 51.Bernardino L., Balosso S., Ravizza T., Marchi N., Ku G., Randle J. C., Malva J. O., Vezzani A. (2008) J. Neurochem. 106, 271–280 [DOI] [PubMed] [Google Scholar]
- 52.Arend W. P., Welgus H. G., Thompson R. C., Eisenberg S. P. (1990) J. Clin. Invest. 85, 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukaida N., Ishikawa Y., Ikeda N., Fujioka N., Watanabe S., Kuno K., Matsushima K. (1996) J. Leukoc. Biol. 59, 145–151 [DOI] [PubMed] [Google Scholar]
- 54.Weber A., Wasiliew P., Kracht M. (2010) Sci. Signal. 3, cm1. [DOI] [PubMed] [Google Scholar]
- 55.Turrin N. P., Rivest S. (2006) Mol. Neurobiol. 34, 221–242 [DOI] [PubMed] [Google Scholar]
- 56.Bernardino L., Malva J. O. (2007) in Interaction between Neurons and Glia in Aging and Disease (Malva J. O. ed) pp. 3–35, Springer, New York [Google Scholar]
- 57.Bedoui S., Miyake S., Straub R. H., von Hörsten S., Yamamura T. (2004) Trends Immunol. 25, 508–512 [DOI] [PubMed] [Google Scholar]
- 58.De la Fuente M., Del Río M., Medina S. (2001) J. Neuroimmunol. 116, 156–167 [DOI] [PubMed] [Google Scholar]
- 59.Agasse F., Bernardino L., Kristiansen H., Christiansen S. H., Ferreira R., Silva B., Grade S., Woldbye D. P., Malva J. O. (2008) Stem Cells 26, 1636–1645 [DOI] [PubMed] [Google Scholar]
- 60.Xapelli S., Bernardino L., Ferreira R., Grade S., Silva A. P., Salgado J. R., Cavadas C., Grouzmann E., Poulsen F. R., Jakobsen B., Oliveira C. R., Zimmer J., Malva J. O. (2008) Eur. J. Neurosci. 27, 2089–2102 [DOI] [PubMed] [Google Scholar]
- 61.Silva A. P., Pinheiro P. S., Carvalho A. P., Carvalho C. M., Jakobsen B., Zimmer J., Malva J. O. (2003) FASEB J. 17, 1118–1120 [DOI] [PubMed] [Google Scholar]
- 62.Howell O. W., Scharfman H. E., Herzog H., Sundstrom L. E., Beck-Sickinger A., Gray W. P. (2003) J. Neurochem. 86, 646–659 [DOI] [PubMed] [Google Scholar]
- 63.Teixeira L., Sousa D. M., Nunes A. F., Sousa M. M., Herzog H., Lamghari M. (2009) J. Cell. Biochem. 107, 908–916 [DOI] [PubMed] [Google Scholar]
- 64.Dimayuga F. O., Wang C., Clark J. M., Dimayuga E. R., Dimayuga V. M., Bruce-Keller A. J. (2007) J. Neuroimmunol. 182, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cervin A., Onnerfält J., Edvinsson L., Grundemar L. (1999) Am. J. Respir. Crit. Care Med. 160, 1724–1728 [DOI] [PubMed] [Google Scholar]
- 66.Bianco F., Ceruti S., Colombo A., Fumagalli M., Ferrari D., Pizzirani C., Matteoli M., Di Virgilio F., Abbracchio M. P., Verderio C. (2006) J. Neurochem. 99, 745–758 [DOI] [PubMed] [Google Scholar]
- 67.Ohtani Y., Minami M., Satoh M. (2000) Neurosci. Lett. 293, 72–74 [DOI] [PubMed] [Google Scholar]
- 68.Schroeder R. A., Cai C., Kuo P. C. (1999) Am. J. Physiol. 277, C523–C530 [DOI] [PubMed] [Google Scholar]
- 69.Mollace V., Muscoli C., Rotiroti D., Nisticó G. (1997) Biochem. Biophys. Res. Commun. 238, 916–919 [DOI] [PubMed] [Google Scholar]
- 70.Hu S., Ali H., Sheng W. S., Ehrlich L. C., Peterson P. K., Chao C. C. (1999) J. Neurosci. 19, 6468–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akama K. T., Van Eldik L. J. (2000) J. Biol. Chem. 275, 7918–7924 [DOI] [PubMed] [Google Scholar]
- 72.Doyle S. L., O'Neill L. A. J. (2006) Biochem. Pharmacol. 72, 1102–1113 [DOI] [PubMed] [Google Scholar]
- 73.Watterson D. M., Mirzoeva S., Guo L., Whyte A., Bourguignon J. J., Hibert M., Haiech J., Van Eldik L. J. (2001) Neurochem. Int. 39, 459–468 [DOI] [PubMed] [Google Scholar]
- 74.Teng X., Zhang H., Snead C., Catravas J. D. (2002) Am. J. Physiol. Cell Physiol 282, C144–C152 [DOI] [PubMed] [Google Scholar]
- 75.Ball H. J., Shine J., Herzog H. (1995) J. Biol. Chem. 270, 27272–27276 [DOI] [PubMed] [Google Scholar]
- 76.Musso R., Grilli M., Oberto A., Gamalero S. R., Eva C. (1997) Mol. Pharmacol. 51, 27–35 [DOI] [PubMed] [Google Scholar]
- 77.Griess P. (1879) Chem. Ber. 12, 426–428 [Google Scholar]