Abstract
Prolyl 4-hydroxylases (P4Hs) catalyze the hydroxylation of collagens and hypoxia-inducible factor (HIF)-α subunits. We studied the zebrafish homologue of the recently characterized human transmembrane P4H (P4H-TM) that can hydroxylate HIF-α, but not collagens, in vitro and influence HIF-α levels in cellulo. The zebrafish P4H-TM mRNA had its highest expression in the eye and brain and lower levels in other tissues, including the kidney. Morpholino knockdown of P4H-TM in embryos resulted in a reduction in the size of the eye and head and morphological alterations in the head from 2 days postfertilization onward. In addition, pericardial edema, regarded as a sign of kidney dysfunction, developed from 3 days postfertilization onward. The phenotype was dependent on the P4H-TM catalytic activity because similar results were obtained with morpholinos targeting either translation initiation or catalytic residues of the enzyme. Structural and functional analyses of the morphant pronephric kidneys revealed fragmented glomerular basement membranes (BMs), disorganized podocyte foot processes, and severely compromised pronephric kidney function leading to proteinuria. The opacity of the eye lens was increased due to the presence of extra nuclei and deposits, and the structure of the lens capsule BM was altered. Our data suggest that P4H-TM catalytic activity is required for the proper development of the glomerular and lens capsule BMs. Many HIF target genes were induced in the P4H-TM-deficient morphants, but the observed phenotype is not likely to be mediated at least solely via the HIF pathway, and thus P4H-TM probably has additional, as yet unknown, substrates.
Keywords: Basement Membrane, Enzyme Catalysis, Eye, Hypoxia, Kidney, HIF, Prolyl 4-Hydroxylase, Zebrafish
Introduction
Prolyl 4-hydroxylases (P4Hs)4 catalyze the post-translational formation of 4-hydroxyproline (4-Hyp) in their substrate proteins. P4Hs belong to the group of Fe(II)- and 2-oxoglutarate-dependent dioxygenases and require Fe2+, 2-oxoglutarate, O2, and ascorbate (1, 2). At least two distinct groups of P4Hs exist: the collagen P4Hs (C-P4Hs) that reside in the lumen of the endoplasmic reticulum (1, 3) and the cytoplasmic and nuclear hypoxia-inducible factor (HIF) P4Hs (for reviews, see Refs. 4–6). The human C-P4Hs are α2β2 tetramers in which the α subunits are responsible for the P4H activity, whereas the β subunits are identical to protein-disulfide isomerase (1). Three C-P4H isoenzymes differing in their α subunits exist in humans and catalyze the synthesis of 4-Hyp in -X-Pro-Gly- sequences of collagens, the most abundant extracellular matrix (ECM) proteins, and in some other proteins with collagen-like motifs. The role of 4-Hyp in collagens is to promote their structural stability because, in the absence of 4-Hyp, the collagen polypeptides are not folded into the correct triple-helical conformation at human body temperature. Surprisingly, C-P4H isoenzyme I was also recently found to hydroxylate a proline in an -X-Pro-Gly- sequence in Ago2 (Argonaute 2), a cytosolic protein essential for RNA-induced silencing complexes (7). Inhibition of the prolyl 4-hydroxylation of Ago2 was found to alter its stability and, therefore, effective RNA interference (7). The three human HIF-P4Hs (HIF-P4Hs 1–3, also known as PHDs 1–3, and EglN2, -1, and -3, respectively) hydroxylate one or two prolines in the oxygen-dependent degradation domain (ODDD) of the HIF-α subunits in a -Leu-X-X-Leu-Ala-Pro- sequence (8–10). The 4-Hyp in HIF-α acts to enable the binding of an E3 ubiquitin ligase, the von Hippel Lindau tumor suppressor (pVHL), leading to subsequent polyubiqitinylation and rapid proteasomal degradation of HIF-α in normoxia. In hypoxia, the prolyl 4-hydroxylation is inhibited, and HIF-α is stabilized and translocates into the nucleus, where it dimerizes with HIF-β and initiates the transcription of numerous genes involved in the regulation of oxygen homeostasis. There are reports that link the activity of HIF-P4Hs to the regulation of the stability of some other targets in addition to HIF-α, such as HIF-P4H-3, which regulates the stability of the transcription factor ATF-4 (11) and the β2 adrenergic receptor (12), and HIF-P4H-1, which regulates cyclin D1 levels in a manner requiring its catalytic activity but remaining independent of HIF-α, thus indicating the hydroxylation of a so far unidentified polypeptide (13). Indirect evidence has also been reported for hydroxylation of the large subunit of RNA polymerase II and IκB kinase-β by HIF-P4Hs (14, 15).
We have recently cloned and characterized a novel human transmembrane P4H (P4H-TM) and shown that is located in the endoplasmic reticulum membranes, with its catalytic site inside the lumen (16). Its sequence resembles more closely those of the C-P4H α subunits than the HIF-P4Hs, but it lacks the peptide-substrate-binding domain of the C-P4Hs (17). Recombinant P4H-TM hydroxylated the two prolines in the HIF-1α ODDD in vitro, whereas it did not hydroxylate any prolines in recombinant full-length type I collagen polypeptides (16). Overexpression of P4H-TM in cultured neuroblastoma cells reduced HIF-1α ODDD reporter levels, whereas its knockdown increased the HIF-1α protein level, in the same way as observed with the HIF-P4Hs (16). P4H-TM also hydroxylated to a low extent a HIF-1α ODDD in which the two prolines targeted by the HIF-P4Hs were mutated, suggesting that, at least in vitro, it can hydroxylate some additional prolines besides those recognized by the HIF-P4Hs (16). P4H-TM is a vertebrate enzyme because the lowest organism in which its homologue is found is the zebrafish (16). This same protein was studied by Oehme et al. (18), who suggested that the polypeptide is oriented with its catalytic site facing cytosol and showed that its overexpression suppressed cellular HIF-1α and -2α reporter transactivation activity and reduced the HIF-2α protein level. We exploited here the morpholino knockdown technology to study the in vivo role of P4H-TM in zebrafish and found that P4H-TM deficiency led to basement membrane (BM) defects in several tissues and compromised pronephric kidney function. The deficiency induced many HIF target genes, but it seems highly unlikely that the phenotypic defects are mediated at least solely by the HIF pathway, and thus P4H-TM probably has additional, as yet unidentified, substrates.
EXPERIMENTAL PROCEDURES
Zebrafish Maintenance
Wild-type AB-line zebrafish obtained from the Zebrafish International Resource Center were maintained and raised as described earlier (19).
RT-PCR and Q-PCR Analysis
Whole zebrafish embryos, larvae, or dissected adult zebrafish tissues were immersed in RNAlater (Ambion) at room temperature immediately after collecting and stored either at room temperature or at 4 °C, according to the manufacturer's instructions. For RNA isolation, the tissues were removed from RNAlater, and total RNA was isolated using TriPure isolation reagent (Roche Applied Science) or an EZNA total RNA kit (OMEGA Bio-tek), and reverse transcription was carried out with the iScript cDNA Synthesis Kit (Bio-Rad). Q-PCR was performed with iTaq SYBR Green Supermix with ROX (Bio-Rad), SsoFast EvaGreen Supermix (Bio-Rad), and the Stratagene MX3005 thermocycler (Stratagene). The sequences of the primers used in Q-PCR are listed in supplemental Table S1. The mRNA expression levels were calculated relative to β-actin or 18S, the primer sets for which were as published earlier (see Refs. 20 and 21, respectively).
Whole Mount in Situ Hybridization
Whole mount in situ hybridization was performed as described previously (22) with minor modifications, using digoxigenin-11-UTP-labeled probes (Roche Applied Science) and an InsituPro robot (Intavis). For synthesis of the antisense P4H-TM probes, two sequences, one spanning 449 bp and the other 680 bp of the P4H-TM cDNA, were amplified by PCR with the primers 5′-GCGGAATTCGCTTCAGGTGGTCCGCTATGAGC-3′ and 5′-CTGGATCCACATTGATCCAATTATTGGC-3′ and primers 5′-GCGGAATTCATGTTGTCTGGCAAAATGATGGAACC-3′ and 5′-ATAGACTTTTGCGGAGGTCTTGAAGAACC-3′, respectively. The amplified fragments were cloned into the pSP72 plasmid (generating P4H-TM-1 and 2-pSP72, respectively), followed by linearization with BglII and XhoI, respectively. Sp6 RNA polymerase was used to synthesize the antisense probes. The sense probes used as negative controls were synthesized by linearizing the P4H-TM-pSP72 plasmid with XbaI and BglII, respectively, and using T7 RNA polymerase for their synthesis. Anti-digoxigenin antibody (1:2000 to 1:4000) (Roche Applied Science) and BM purple AP substrate (Roche Applied Science) were used to detect the probe after in situ hybridization. After washing, the embryos were equilibrated in glycerol and photographed using an Olympus SZX12 stereo microscope and an Olympus CC-12 CCD camera (Olympus).
For the cryosections, the embryos were transferred to 15% sucrose followed by 30% sucrose and embedding in OTC compound (Sakura). 10 μm sections were cut and stained with eosin for histology.
Antisense Morpholino Experiments
Three independent antisense morpholinos targeting either the translational start site (TM1) or an exon splice acceptor site of the zebrafish P4H-TM gene (TM2 and TM3) were obtained from GeneTools. In addition, a 5-bp mismatch morpholino (MM) was obtained for TM1. The sequences of the morpholinos used were as follows: TM1, 5′-GTTCCATCATTTTGCCAGACAACAT-3′; TM2, 5′-CATGGCCCACCTAAAAATGAAGAAA-3′; TM3, 5′-CCAACCTACAGAGGGCAGCATAGCA-3′; MM, 5′-GTTGCATGATTTTCCCAGAGAAGAT-3′ (mismatched base pairs indicated in boldface type); and a random control (RC), 5′-CCTCTTACCTCAGTTACAATTTATA-3′. The sequence for the p53 morpholino has been published earlier (23). The morpholinos were diluted in 0.2 m KCl (pH 7.2), and 0.2% rhodamine dextran tracer was added to the mixture to monitor for a uniform oligonucleotide distribution in the injected embryos. Microinjection of the newly fertilized eggs was performed at the 1–4-cell stage with 1 nl of morpholino solution (concentration varying from 125 to 750 μm). The embryos were screened under fluorescent light at 24 h postinjection to determine the success of the injection and were observed and photographed at 1–4 days postfertilization (dpf) in 2% methyl cellulose containing 0.17 mg/ml Tricaine using a Zeiss Lumar V.12 fluorescense stereomicroscope and a Zeiss AxioCam MRm camera (Zeiss). Embryos for morphological analysis were fixed in 4% paraformaldehyde overnight at 4 °C and processed to paraffin sections as described under “Histological Analysis.” RT-PCR of total RNA isolated from a pool of injected embryos was performed to verify the efficiency of the splice site-targeting morpholinos (TM2 and TM3). Gene-specific primers yielding a 599-bp amplicon from wild-type RNA and a 517-bp amplicon from the targeted P4H-TM mRNA were used to verify the deletion of exon 2 (TM2). Gene-specific primers from exons 1 and 9 generating 1428- and 1452-bp products from wild-type P4H-TM RNA were used to demonstrate the reduction in the amount of wild-type P4H-TM in TM3-targeted embryos. Primers amplifying β-actin were used as controls.
Histological Analysis
Whole zebrafish embryos/larvae were fixed in 4% paraformaldehyde at 4 °C overnight, embedded first in 2% agarose followed by embedding in paraffin. 5-μm sections were cut and stained with hematoxylin and eosin for general histological analysis and with alkaline phosphatase (Promega) to reveal endogenous tubulus activity.
o-Dianisidine Staining
For hemoglobin staining, anesthetized 48-h postfertilization embryos were incubated in 0.6 mg/ml o-dianisidine (Sigma-Aldrich) in 40% EtOH, 0.01 m NaAc, pH 4.5, and 0.65% H2O2 for 15 min covered from light. The samples were then fixed with 4% paraformaldehyde overnight and analyzed using an Olympus SZX16 inverted microscope. The amount of stained red blood cells in individual embryos was scored 1–3. Each experimental group consisted of 6–9 embryos.
Analysis of Heart Function
Heart function was determined from time lapse images taken with a Zeiss Lumar V.12 stereomicroscope linked to a Zeiss AxioCam MRm camera. Analyses were done using the Axiovision software. Heart rate was determined from 20-s time lapse videos. Ventricle performance was assayed by calculating the longitudinal fractional shortening (equal to (LDd − LDs)/LDd, where LDd represents longitudinal diameter at diastole and LDs is longitudinal diameter at systole).
Transmission Electron Microscopy (TEM)
Whole zebrafish larvae were fixed in a mixture of 1% glutaraldehyde and 4% formaldehyde in 0.1 m phosphate buffer, postfixed in 1% osmium tetroxide, dehydrated in acetone, and embedded in Epon LX 112 (Ladd Research Industries). Thin sections were cut with a Leica Ultracut UCT ultramicrotome, stained in uranyl acetate and lead citrate, and examined in a Philips CM100 transmission electron microscope. Images were captured using a Morada CCD camera (Olympus Soft Imaging Solutions GMBH).
Fluorescent Dye Injection
Lysine-fixable FITC-dextran (70 and 500 kDa; Molecular Probes) was dissolved in 0.2 m KCl (pH 7.2), and fluorescent tracers were injected into the cardinal veins of 72-h postfertilization larvae anesthetized with 0.17 mg/ml Tricaine (Sigma). After 5 h of incubation, the fish water was collected, and the larvae were fixed in 4% paraformaldehyde at 4 °C overnight, followed by processing to paraffin sections. Uptake of the filtered fluorescent dextran by the duct cells was evaluated in serial sections using an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus). Fluorescence in the fish water was determined using a 1420 Multilabel Counter Victor3 (PerkinElmer Life Sciences), excitation wave length 490 nm, emission wave length 535 nm, and measuring time 1 s.
Statistical Analyses
The statistical analyses were performed using Student's two-tailed t test, and the data are expressed as means ± S.D.
RESULTS
P4H-TM Is Conserved in Vertebrates and Has Its Highest Expression in the Zebrafish in the Eye and Brain
A sequence homology search in GenBankTM using the human P4H-TM cDNA as a bait identified P4H-TM homologues in many vertebrates but not in Caenorhabditis elegans or Drosophila melanogaster (16). The zebrafish Danio rerio was the lowest vertebrate with a P4H-TM homologue (GenBankTM accession number ENSDARP00000002179). Amino acid sequence alignment of the 487-amino acid zebrafish P4H-TM with the corresponding 503- and 502-amino acid mouse and human polypeptides, respectively, revealed 52 and 51% overall identity between the zebrafish and mouse P4H-TMs and between the zebrafish and human ones, respectively, whereas the identity between the human and mouse P4H-TMs is 91% (supplemental Fig. S1). The catalytically critical amino acids (two histidines (His344 and His457 in the zebrafish P4H-TM) and one aspartate (Asp346), which bind the iron atom, and the lysine (Lys467) that binds the C5 carboxyl group of 2-oxoglutarate) are conserved in the zebrafish P4H-TM (supplemental Fig. S1). The highest degree of sequence identity between the zebrafish and human P4H-TM polypeptides is found in the catalytic C terminus, the 176 C-terminal residues of the zebrafish P4H-TM being 76% identical to those of human P4H-TM (supplemental Fig. S1). Like the human and mouse P4H-TM polypeptides, the zebrafish P4H-TM has a putative transmembrane domain in its N-terminal region, spanning residues 57–79 (supplemental Fig. S1), and a predicted calcium-binding EF-hand motif in the middle part of the polypeptide (18). The short cytosolic N-terminal part is only 14% identical between the zebrafish and human P4H-TMs, whereas the transmembrane domain is 62% identical (supplemental Fig. S1). Amino acids 80–123 (i.e. those located immediately after the transmembrane domain) show a very low sequence identity, only 18%, between the zebrafish and human P4H-TMs (supplemental Fig. S1).
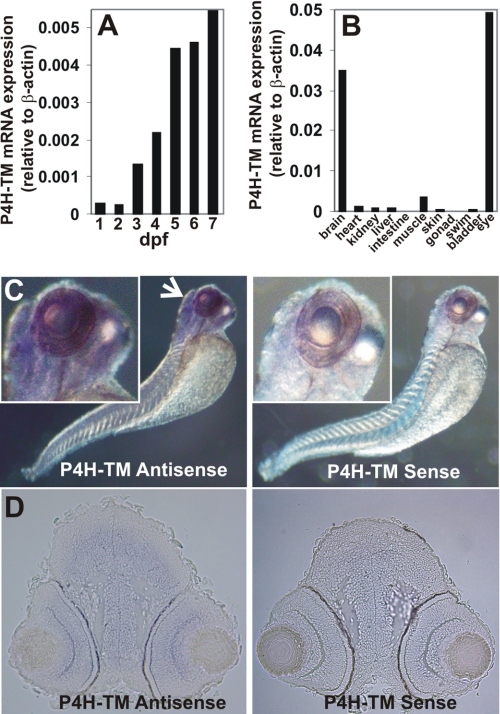
Quantitative real-time RT-PCR (Q-PCR) analysis during zebrafish development indicated that the P4H-TM mRNA expression level is low for the first 2 dpf, increases 5-fold at 3 dpf, and reaches a relatively steady level at 5 dpf, being about 20-fold relative to the levels at 1 and 2 dpf (Fig. 1A). We also studied the expression of P4H-TM mRNA by Q-PCR in several dissected adult female zebrafish tissues. The highest expression levels were found in the eye and brain, being 10- and 7-fold higher than that in the 7-dpf whole larvae, respectively, and 10–14-fold higher than in any other tissue studied (Fig. 1B). The P4H-TM mRNA level in the muscle was slightly lower than that in the 7-dpf whole larvae, whereas the expression levels in the heart, kidney, and liver were even lower and comparable with each other (Fig. 1B). The lowest expression was seen in the intestine and gonad (Fig. 1B). In situ hybridization of 3-dpf wild-type zebrafish larvae with an antisense probe coding for nucleotides 987–1435 of P4H-TM revealed strong staining in the brain and eye, whereas no staining was seen in the control (Fig. 1C). Cryosections from 3-dpf wild-type zebrafish larvae in situ hybridized with an antisense probe coding for nucleotides 1–680 of P4H-TM were prepared and counterstained with eosin (Fig. 1D). Strong staining was seen in telen- and diencephalon (Fig. 1D) and in mesen-, meten-, and myelencephalon (data not shown); thus, P4H-TM was expressed throughout the brain (Fig. 1D). In the eye, all retinal layers showed staining, whereas no staining in the brain or eye was seen in the control (Fig. 1D).
FIGURE 1.
Analysis of P4H-TM expression in zebrafish. A, Q-PCR analysis of P4H-TM mRNA expression in 1–7-dpf embryos (B) and various female adult tissues relative to β-actin. Each bar represents a value for a pool of 50 embryos (A) or for two adults (B). C, whole mount in situ hybridization analysis of P4H-TM expression with an antisense and sense (negative control) probe in 3-dpf larva. The arrow indicates intense staining in the head and eye. D, cryosections of the head of the 3-dpf larva show P4H-TM expression in telencephalon and diencephalon and in retina.
Knockdown of Zebrafish P4H-TM during Embryogenesis Leads to Morphological Alterations and Pericardial Edema
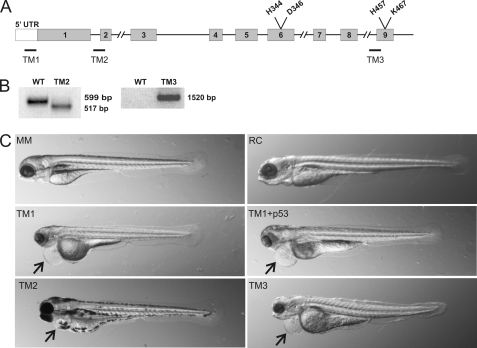
To study the role of zebrafish P4H-TM in vivo, we employed the antisense morpholino oligonucleotide technology (24, 25) to knock down P4H-TM expression during embryogenesis. Fertilized zebrafish eggs were injected with various concentrations of one of the three P4H-TM-specific morpholino-oligonucleotides TM1–TM3 (Fig. 2A) or a 5-bp mismatch morpholino for TM1 (MM) or an RC morpholino. To avoid the nonspecific p53-mediated effects on neuronal apoptosis during the first few days of development, a morpholino targeting p53 was co-injected with the P4H-TM-targeting morpholinos in some experiments. The TM1 morpholino prevents translation initiation and is therefore expected to markedly reduce the amount of the P4H-TM protein (Fig. 2A). Unfortunately, the efficiency of the TM1 knock-down could not be analyzed because of the lack of a functional antibody against zebrafish P4H-TM. The TM2 morpholino targets the mRNA splicing site at the intron 1-exon 2 boundary (Fig. 2A). As expected, the majority of the P4H-TM mRNAs of embryos injected with the TM2 morpholino lacked the 82-bp exon 2 (Fig. 2B), which causes a shift in the reading frame and a premature stop codon and results in the generation of a 143-amino acid truncated protein in which the amino acids encoded by exon 1 are followed by 12 scrambled amino acids verified by sequencing of the TM2 transcript. The TM3 morpholino targets the intron 8-exon 9 boundary (Fig. 2A), which is the last intron-exon boundary of the P4H-TM gene. Targeting of the last intron-exon boundary in pre-mRNA usually results in a final messenger with the last intron inserted (26). Therefore, TM3 led to the inclusion of intron 8 in the P4H-TM mRNA, resulting in a premature stop codon and generation of a P4H-TM polypeptide in which the amino acids encoded by exons 1–8 were followed by 5 scrambled amino acids verified by sequencing of the TM3 transcript (Fig. 2B). Because exon 9 codes for the catalytically critical amino acids His457 and Lys467, the majority of the P4H-TM mRNA expressed in the TM3-injected embryos codes for a truncated inactive polypeptide. Because we were unable to measure the P4H-TM protein levels, one must bear in mind that the truncation may have altered the stability of the mutant protein. However, point mutations of the corresponding histidine and lysine residues in the human collagen P4H have been reported to destroy the activity of the enzyme without affecting the stability of the enzyme tetramer (1).
FIGURE 2.
Knock-down of zebrafish P4H-TM by antisense morpholino oligonucleotides. A, schematic representation of the organization of the zebrafish P4H-TM gene. Gray boxes, exons; black lines, introns. Target sites of the sequence-specific morpholinos TM1–TM3 are indicated by black bars. B, RT-PCR of 4-dpf wild-type zebrafish embryos and embryos targeted with TM2 and TM3. A primer pair amplifying a 599-bp product from wild-type embryos and a 517-bp product from TM2-injected embryos lacking exon 2 is shown. Another primer pair from exon 1 and intron 8 was used to amplify P4H-TM cDNA from wild-type and TM3-injected embryos, yielding a product of 1520 bp from TM3. C, whole mounts of 4 dpf larvae injected with a 5-bp mismatch control for TM1 (MM; 500 μm), random control (RC; 750 μm), TM1 (500 μm), TM1 (500 μm) + p53 (750 μm), TM2 (125 μm), and TM3 (750 μm) are shown. All except for TM2 larvae were kept in 1-phenyl-2-thiourea water, preventing pigment formation. The arrows indicate pericardial edema.
The phenotypes of the morpholino-injected embryos were evaluated daily until 4 dpf, starting with the day of the injection. The embryos injected with TM1 showed a reduction in the sizes of the head and eye and morphological alterations in the head relative to the MM control starting at 2 dpf and persisting until 4 dpf (Fig. 2C) (i.e. throughout the effective time of morpholino knockdown). The phenotype of the embryos co-injected with the TM1 and p53 morpholinos was identical to that obtained with TM1 alone, except for the attenuation of the reduction in the size of the head (Fig. 2C). No obvious differences in the cerebral structures were detected between the TM1 + p53 and MM controls at 4 dpf, however (supplemental Fig. S2). The phenotypes of the embryos injected with TM2 and TM3 (Fig. 2C) were similar to that obtained with TM1, except that the morphological alteration in the head of the TM3-treated embryos was milder (Fig. 2C). Statistical analysis of the phenotypes of over 800 TM1- and TM2-injected embryos was performed. About 22% of the TM1-injected embryos died during the first 24 h, whereas this percentage was markedly higher, 59%, for TM2-injected embryos. No marked reduction in the vitality of the embryos after 24 h was observed because 70% of the TM1 and 30% of the TM2 larvae were alive at 3 dpf. Of the 3 dpf TM1 and TM2 live larvae, 83% showed a phenotype(s). In general, the phenotype of the TM2 larvae was more severe because about 35% of them expressed a development cessation phenotype at 3 dpf, whereas the same was observed for only 3% of the TM1 larvae. All three independent P4H-TM-targeting morpholinos resulted in pericardial edema, starting at 3 dpf and becoming more severe at 4 dpf (Fig. 2C). About 55% of the TM1 and 25% of the TM2 larvae had pericardial edema at 3 dpf. To determine whether the pericardial edema was of cardiac origin, the cardiac function of the TM1 and TM3 morphants and the controls was analyzed from 20-s time lapse movies recorded at 2 dpf (supplemental Movies S1–S4). The values for longitudinal fractional shortening were 0.28 ± 0.06 for RC (n = 5), 0.32 ± 0.07 for MM (n = 8), 0.23 ± 0.07 (n = 7) for TM1, and 0.24 ± 0.07 for TM3 (n = 7) embryos (p > 0.2 for all when compared with RC); therefore, no significant differences were found between TM1 and TM3 morphants and the controls in their ventricle performance. The measured heart rates, expressed as average beats/min (bpm) ± S.D., were 100 ± 4 bpm for RC (n = 8), 88 ± 6 bpm for MM (n = 8), 108 ± 5 bpm (n = 8) for TM1, and 86 ± 10 bpm for TM3 (n = 8) embryos; thus, the values for TM1 and TM3 were in the same range with the two controls. Normal cardiac function of the TM1 and TM3 morphants suggests that, as usually is the case at this stage of development, the observed pericardial edema is caused by a dysfunction of the pronephros, the primary function of which in zebrafish is osmoregulation (27).
P4H-TM Deficiency Alters mRNA Expression of Select Genes
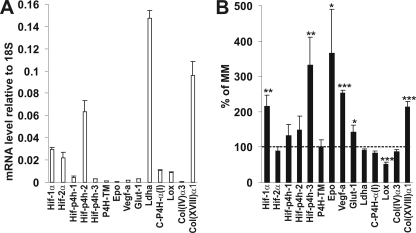
We have shown earlier that human P4H-TM is able to hydroxylate HIF-1α in vitro and that its overexpression reduces HIF-1α protein levels in cultured cells, whereas RNAi induces these levels (16). Because no antibodies against zebrafish HIF-α subunits were available, we used Q-PCR to study whether the knockdown of zebrafish P4H-TM leads to altered expression of selected known HIF target genes. In the wild-type 3-dpf embryos, the level of HIF-1α mRNA expression was higher than that of HIF-2α expression, and the HIF-P4H-2 mRNA was the most abundantly expressed HIF-P4H isoenzyme mRNA, as is also found in human cells (Fig. 3A) (28). The level of the P4H-TM mRNA expression was lower than that of any of the HIF-P4H mRNAs (Fig. 3A).
FIGURE 3.
Q-PCR analysis of the mRNA levels of selected HIF target and ECM genes relative to 18S in controls (A) and TM1-deficient larvae compared with controls (MM) (B). A, mRNA levels of 3-dpf control larvae injected with MM morpholino are shown. The results are means ± S.D. (error bars) of three 30-larva pools. B, mRNA levels of 3-dpf TM1-injected larvae were compared with MM-injected controls. The results are shown as percentage of MM ± S.D. of three 30-larva pools. The dashed line indicates 100% of MM. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Epo, erythropoietin; Vegf-a, vascular endothelial growth factor-a; Glut-1, glucose transporter-1; Ldha, lactate dehydrogenase-a; C-P4H-α(I), collagen prolyl 4-hydroxylase α(I) subunit; Col(IV)α3, the α3 chain of type IV collagen; Col(XVIII)α1, the α1 chain of type XVIII collagen.
HIF-1α mRNA in the whole 3-dpf TM1-injected larvae was induced to about 2.2-fold compared with the control, whereas no change was seen in the level of HIF-2α mRNA (Fig. 3B). Stabilization of HIF-1α in mammalian cells may not increase its mRNA levels, although such increases have also been found (29), whereas increased mRNA levels have been reported to be associated with hypoxic stabilization of HIF-1α in zebrafish tissues (30), as was also observed here. The level of P4H-TM mRNA expression was not changed in the TM1-injected embryos, whereas the P4H-TM protein level is increased by the hypoxia response in several human cell lines but not in all of those studied (Fig. 3B) (16). The levels of expression of the well characterized HIF target genes hif-p4h-3, epo (erythropoietin), vegf-a (vascular endothelial growth factor-a), and glut-1 (glucose transporter-1) were all significantly increased in the TM1 embryos relative to MM (Fig. 3B). Despite the increase in the Epo mRNA level in TM1 morphants, no signs of polycythemia or of anemia, on the other hand, were found in the hemoglobin stainings of the embryos compared with the RC control (supplemental Fig. S3). The ECM enzyme gene lox (lysyl oxidase), which has been found to be highly hypoxia-inducible in several mammalian microarrays (31, 32), was, surprisingly, significantly down-regulated in the TM1 morphants (Fig. 3B). Of the other ECM genes analyzed, the level of type XVIII collagen α(1) chain mRNA expression was induced about 2.1-fold in the TM1 morphants, having also been found to be induced about 1.8-fold in Vhl mutant zebrafishes in which degradation of HIF-1α is prevented (33), whereas no alterations were found in the mRNA levels of the α(3) chain of type IV collagen or the α subunit of C-P4H isoenzyme I (Fig. 3B).
Deficiency of P4H-TM Disturbs the Normal Lens Development and Leads to Alteration of the Basement Membrane Structure in the Eye
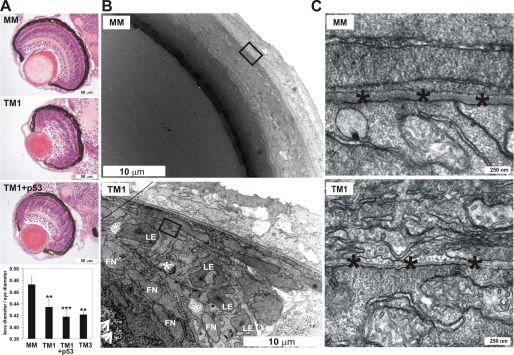
Given that our Q-PCR results obtained with adult zebrafish tissues indicated that the eye had the highest level of P4H-TM mRNA expression, and analysis of the whole mount zebrafish embryos/larvae deficient in P4H-TM revealed a reduction in the size of the eye at 2–4-dpf (Fig. 1B and 2C), we examined the morphology of the eye of the P4H-TM morphants in more detail. Hematoxylin and eosin-stained sections of 4-dpf eyes showed that the size of the eye and the transparency of the lens were reduced in the TM1 and TM3 morphants compared with MM-injected larvae (Fig. 4A) (data not shown). The ratio of the lens to eye diameters was significantly reduced in TM1, TM1 + p53, and TM3 morphants compared with controls (MM) (Fig. 4A). Moreover, blue staining spots, most likely representing nuclei, were present in the middle of the lens of the TM1 eyes but were not observed in the MM morphants (Fig. 4A). The retinal layers of the TM1 larvae were poorly organized compared with the controls (MM) (Fig. 4A), but the poor organization of the retinal layers in the TM1 eyes was corrected to some extent in the TM1- and p53-co-injected eyes (Fig. 4A), suggesting that this may have been a nonspecific p53-mediated alteration, whereas co-injection of p53 did not correct the lens phenotype (Fig. 4A). TEM revealed numerous extra fiber cell nuclei, cytoplasmic degeneration areas, and fiber cell deposits in the lens of the TM1 eyes compared with the control (Fig. 4B). Because all of the cell organelles, including nuclei, should have disappeared by 4 dpf, epithelial cell differentiation appears to be disturbed in the TM1 morphants (Fig. 4B). The morphology of the lens capsule BM was altered in the TM1 morphants compared with the control (Fig. 4C), being very thin and fragmented.
FIGURE 4.
Analysis of the eye phenotype of the P4H-TM-deficient larvae. A, light microscopy of hematoxylin and eosin-stained sections of eyes of TM1-injected and TM1 + p53-injected 4-dpf larvae reveals reduction in the size of the eye, opacity of the lens, and disorganization of the retinal layers in TM1 eyes compared with controls (MM). The lens diameter/eye diameter ratio was significantly reduced in TM1, TM1 + p53, and TM3 morphants compared with controls (MM). Averages ± S.D. (error bars) of 5–11 larvae were analyzed. B, TEM demonstrates the presence of numerous fiber cell nuclei (FN), degeneration areas (white asterisk), and deposits (arrowheads) in the lens of the TM1 morphants but not in controls (MM). LE, lens epithelial cells. C, a higher magnification TEM image shows a thin and fragmented BM (black asterisks) in TM1 morphants compared with controls (MM).
P4H-TM Is Essential for the Normal Structure and Function of the Pronephric Zebrafish Kidney and Its Deficiency Causes Proteinuria
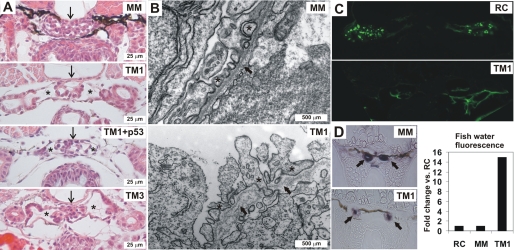
Because we had observed that the injection of embryos with any of the three P4H-TM-targeting morpholinos resulted in pericardial edema at 3–4 dpf, and cardiac dysfunction of the TM1 morphants seemed highly unlikely, we next focused our attention on studying the pronephric kidneys of the P4H-TM-deficient larvae in more detail. For histological analysis of the pronephros, 4-dpf morpholino-injected larvae were processed to paraffin sections and stained with hematoxylin and eosin (Fig. 5A). A clear reduction was observed in the number of podocytes, accompanied by dilatation of Bowman's capsule and the presence of bare capillary loops in the glomeruli of the TM1-injected larvae relative to the control (MM) (Fig. 5A). These changes were independent of p53 but dependent on the catalytic activity of P4H-TM because the phenotype was unchanged in larvae co-injected with the TM1 and p53 morpholinos and in TM3-injected larvae (Fig. 5A). TEM revealed fragmented glomerular BMs (GBMs), swelling of podocytes, and abnormal shaping and disorganization of the podocyte foot processes in the TM1-injected larvae compared with the controls (MM) (Fig. 5B). Some foot processes even protruded into the BM (Fig. 5B). Although intact slit diaphragms were seen between the foot processes in the glomeruli of the TM1 morphants, the possibility cannot be excluded that the alterations in foot process alignment may affect their function (Fig. 5B). Furthermore, the glomerular endothelial cells were swollen and detached from the GBM in the TM1 morphants (Fig. 5B). No gross abnormalities were seen in the structure of the pronephric tubuli (Fig. 5A).
FIGURE 5.
Analysis of the kidney phenotype of the P4H-TM-deficient larvae. A, light microscopy of hematoxylin and eosin-stained sections shows a reduction in the number of podocytes in the glomeruli (arrow) and a dilatation of Bowman's capsule (asterisks) in the 4-dpf TM1-injected, TM1 + p53-injected, and TM3-injected larvae as compared with the MM-injected controls. All except MM larvae were kept in 1-phenyl-2-thiourea water. B, TEM reveals fragmentation of the GBM (arrows) in 4-dpf TM1 morphants compared with controls (MM). In addition, endothelial cells have become detached from the GBM, and the podocyte foot processes (asterisks) are abnormally shaped and poorly organized in the TM1 morphants relative to the controls (MM). C, RC-injected embryos display glomerular passage of 70-kDa FITC-dextran, as evidenced by uptake into apical endosomes of the pronephric duct cells, whereas no endocytosis is seen in the tubuli of the TM1-injected embryos. D, reduced endogenous alkaline phosphatase activity is seen in the TM1 pronephric tubuli relative to MM. The fluorescence emitted by the fish water in TM1 morphants was 15-fold compared with the RC and MM control morphants. A representative experiment is shown.
To assess the in vivo function of the pronephric zebrafish glomerulus, a functional assay to detect proteinuria, a symptom of compromised kidney filtration was employed. Different molecular weight fluorescent dextrans were injected into the general blood circulation of the larvae, followed by monitoring their passage through the pronephric glomerular filter and subsequent endocytic uptake into the apical cytoplasm of the tubular epithelial cells (34, 35). TM1, TM3, and RC-injected 3-dpf larvae were anaesthesized and intravenously injected with either 70- or 500-kDa FITC-labeled dextran followed by fixation and processing to paraffin sections 5 h after injection. The 500-kDa dextran can be used as an indicator of glomerular filtration barrier discrimination because it should be completely retained within the vascular system following intravenous injection in the wild-type larvae, whereas the 70-kDa dextran readily passes into the glomerular filtrate and can thus be used as a positive control (36). Confocal microscopy of the paraffin sections surprisingly revealed that no FITC-positive endosomes could be seen in the TM1 morphant tubuli after injection of either the 70- or 500-kDa dextran, whereas the RC control embryos showed FITC-containing endosomes after injection of the 70-kDa dextran but not its 500-kDa counterpart (Fig. 5C). To study whether the apparent lack of signs of proteinuria in the TM1 morphants was in fact caused by disturbed tubular endocytic function, endogenous alkaline phosphatase activity was determined by staining of the tubular histological sections, the fish water was collected after injection of the 500-kDa FITC-dextran, and its fluorescence was determined. The assumption was that if the 500-kDa dextran passes through the glomerulus but is not reabsorbed in the tubuli, it must be secreted into the water. The TM1 morphants displayed reduced endogenous alkaline phosphatase activity in their pronephric tubuli (Fig. 5D), and the fluorescence emitted by the TM1 fish water was about 15-fold compared with the controls (Fig. 5D), therefore supporting the hypothesis of the TM1 morphants possessing tubular defects in addition to a glomerular defect. Likewise, fluorescence was detected in the TM3 morphant fish water, its amount being about half of that in the TM1 fish water, however. This result demonstrates that, even expressed at a level detectable by Q-PCR but not with in situ hybridization, deficiency of P4H-TM leads to not only severe structural alterations in the pronephric zebrafish kidney but also to proteinuria and tubular defects, thus compromising the pronephros function.
DISCUSSION
Our data indicate that the expression pattern of the zebrafish homologue of vertebrate P4H-TM resembles that of the human enzyme (16, 18). The expression level was the highest in the eye and brain, whereas lower levels were observed in several other tissues, including skeletal muscle, the kidney, and the heart. Morpholino knockdown technology allowed us to investigate the in vivo role of P4H-TM during zebrafish development. Surprisingly, the most obvious phenotypic alteration observed with all three independent morpholinos targeting different sites in the P4H-TM mRNA was pericardial edema, which became visible at 3 dpf and more severe at 4 dpf. This phenotype was independent of p53 but dependent on the catalytic activity of P4H-TM. A similar phenotype has been reported by others for zebrafish morphants lacking the coxsackie and adenovirus receptor CAR, which was found to be required for renal epithelial differentiation in the zebrafish pronephric kidney (37). At this stage of development (3–4 dpf), pericardial edema most likely results from dysfunction of the pronephric kidney because the primary function of the fish pronephros is osmoregulation (27). We therefore analyzed the morphology and function of the pronephros in more detail. Severe alterations in the structure of the glomerulus were observed in the TM1 morphants at 3 dpf even at the light microscopic level, whereas TEM revealed changes in the ultrastructure of the glomerular endothelial and epithelial cells and fragmentation of the GBM, suggesting that the filtration function might consequently be altered. Surprisingly, we could not detect any uptake of either low or high molecular weight fluorescent dextrans into the pronephric tubular endosomes in the P4H-TM-deficient larvae, whereas in the controls, the low molecular weight fluorescent dextran passed through the glomerular filter and was taken up in the tubuli, and the high molecular weight dextran stayed in the circulation as expected. Importantly, however, we were able to show that the apparent lack of the signs of proteinuria in the serial sections analyzed by confocal microscopy was due to the fact that the 500-kDa FITC-dextran was not reabsorbed in the tubules but was secreted into the fish water by the P4H-TM morphants. Although no gross abnormalities were seen in the structure of the P4H-TM deficient tubuli at the light microscopic level, reduced endogenous alkaline phosphatase staining suggested that their function was compromised to some extent. Lack of P4H-TM was thus shown to lead to a severe leakage of high molecular weight dextran into the filtrate in P4H-TM-deficient 3-dpf larvae, suggesting that P4H-TM is critical to the normal development and function of the pronephric zebrafish kidney because its knockdown during development leads to kidney failure and proteinuria.
Leakage of high molecular weight proteins (i.e. proteinuria) is typical for many human nephropathies and also a common complication of diabetes (38). Proteinuria can result from a failure of the formation of the slit diaphragm, an adherens junction formed between podocyte foot processes (39), but TEM analysis revealed the presence of normal slit diaphragms in the glomeruli of the larvae lacking P4H-TM. Structural integration of glomerular podocytes and blood vessels is required for the normal development of the pronephric zebrafish kidney (27). Podocytes are believed to organize vessel growth by expressing Vegf and thereby attracting and assembling the glomerular tuft (35), but vascular shear force has also been suggested to be required for proper glomerular capillary tuft formation (40). Also, degradation and remodeling of the GBM have been reported to be a key step in capillary tuft formation (40). The structural alterations observed in the pronephric kidney of the morphants lacking P4H-TM may therefore be a result of several dysregulated functions (i.e. failure of GBM remodeling, evidenced by fragmented GBMs, altered Vegf-a expression detected by Q-PCR and compromised blood flow caused by alterations in the endothelial cells seen in TEM). In terms of gene expression, we observed significant alterations in the levels of two ECM components in the TM1 morphants relative to the controls. Expression of the mRNA for lox was reduced, whereas that of the α(1) chain of type XVIII collagen was increased, both of which may have contributed to the alteration in GBM remodeling. Human lysyl oxidase, which was earlier thought to cross-link mainly elastin and fibrillar collagens, has been reported to be a critical mediator of bone marrow cell recruitment by cross-linking type IV collagen of the BMs of premetastatic sites (41). Down-regulation of lox may therefore have contributed to the altered BM structures in P4H-TM-deficient larvae. The expression pattern of zebrafish lox overlaps with that of P4H-TM being expressed in the central nervous system and eyes at 3 dpf, and its morpholino knockdown phenocopies some aspects of the P4H-TM-deficient larvae by showing reduced size of the head and eyes and pericardial edema (42). In addition, these larvae have defects in the organization of their notochord BM, muscle development, and neurogenesis (42). Collagen XVIII-deficient mice have been reported to have alterations in their kidney BMs and kidney filtering capacity (43), and collagen XVIII is required for preservation of the integrity of ECM and capillaries in the kidney (44). It is not clear at this point whether the increased expression of type XVIII collagen mRNA in P4H-TM deficient larvae contributes directly to the kidney phenotype or whether it is a feedback effect to compensate for the altered kidney structure and function.
In addition to the kidney, the eye contains highly specialized BMs (i.e. the lens capsule, inner limiting membrane, and Bruch's membrane). Abnormalities were also observed in the BM structure of the lens capsule in the P4H-TM-deficient morphants. The lens phenotype began to become visible at 2 dpf, whereas at 4 dpf, when the lens should have been completely enucleated, we observed numerous nuclei accompanied by degeneration particles and deposits in the lenses of the P4H-TM-deficient embryos. Because gene knockdown by antisense morpholino-oligonucleotides is transient, we were unable to follow the development of the eyes any longer in order to learn whether the P4H-TM deficiency merely led to a delay in development. It is possible, however, that the morphologically altered lens capsule BM may have contributed to the severely compromised epithelial cell differentiation. Interestingly, mice overexpressing endostatin, a proteolytic fragment of type XVIII collagen, under the K14 keratin promoter resulted in lens opacity and cataract and ultrastructural alterations in the skin BMs (45), suggesting that altered type XVIII collagen expression may have contributed to the lens phenotype in P4H-TM-deficient larvae. Also, others have reported that laminin γ1-deficient zebrafish larvae have little or no lens capsule BM at 3 dpf, but the process of lens enucleation is normal (46).
Although no data are currently available on the in vivo role of any of the human C-P4H or HIF-P4H homologues in zebrafish, two zebrafish lines carrying germ line mutations in the vhl homologue have been found in a Hubrecht target-selected ENU-mutagenized zebrafish library (33). The vhl mutant embryos display a hypoxic response and recapitulate key aspects of Chuvash polycythemia, which is caused by a deficiency in the binding of hydroxylated HIF-α to pVHL and its subsequent targeting for proteasomal degradation (33). Knockdown of P4H-TM did not result in any signs of polycythemia observed in the vhl mutants, and vice versa, no alterations in kidney or lens morphology were reported in the vhl mutants (33). Nevertheless, the P4H-TM-deficient morphants, like the vhl mutants, showed up-regulation of several HIF-regulated genes, although direct comparison of changes in gene expression is not possible because the vhl mutants were studied at 7 dpf, whereas the P4H-TM morphants were analyzed at 3 dpf. Zebrafish embryos injected with the pan-2-oxoglutarate-dependent dioxygenase inhibitor dimethyloxalylglycine mimicked the phenotype of the vhl mutants but also displayed additional alterations not seen in these. For example, strong Vegf-a mRNA expression was seen in the proximal renal tubule of the dimethyloxalylglycine-treated embryos but not in the vhl mutants, suggesting that a protein other than HIF-α regulated by 2-oxoglutarate may be responsible for controlling vegf-a expression there (33). Careful expression studies focused on zebrafish HIF-1α and -2α during development show that both forms are expressed in all tissues during early development (47), and their dysregulation may therefore have contributed to the phenotype observed in the P4H-TM-deficient morphants. It seems highly unlikely, however, that the phenotypic abnormalities found here could be mediated at least solely by changes in the HIF pathway, and thus P4H-TM probably has additional, as yet unknown, substrate(s), the reduced hydroxylation of which results in the observed phenotype.
Supplementary Material
Acknowledgments
We thank T. Aatsinki, R. Juntunen, E. Lehtimäki, R. Polojärvi, and M. Siurua and the Biocenter Oulu EM Core Facility for excellent technical assistance.
This work was supported by Academy of Finland Grants 120156 (to P. K.), 200471 and 202469 (to J. M.), and 118431 (to M. R.); the S. Juselius Foundation (to P. K., J. M., and M. R.); FibroGen Inc. (to J. M.); the Knut and Alice Wallenberg Foundation; the Swedish Research Council; the Swedish Foundation for Strategic Research; and Söderberg's Foundation (to K. T.). The Tampere Zebrafish facility is supported by the Biocenter Finland. K. I. K. is a scientific founder and consultant of FibroGen Inc., which is developing HIF-P4H inhibitors as potential therapeutic agents. J. M. and K. I. K. own equity in this company, and the company itself has supported research in the laboratory headed by K. I. K. in the past and currently supports research in the laboratory of J. M. K. T. is co-founder of NephroGenex, Inc., and BioLamina AB Inc. but receives no research support from those companies.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Movies S1–S4, and Table S1.
- P4H
- prolyl 4-hydroxylase
- 4-Hyp
- 4-hydroxyproline
- C-P4H
- collagen P4H
- HIF
- hypoxia-inducible factor
- ECM
- extracellular matrix
- ODDD
- oxygen-dependent degradation domain
- P4H-TM
- transmembrane P4H
- dpf
- days postfertilization
- BM
- basement membrane
- TEM
- transmission electron microscopy
- GBM
- glomerular basement membrane
- Q-PCR
- quantitative PCR
- MM
- mismatch morpholino
- RC
- random control
- bpm
- beats/min.
REFERENCES
- 1.Myllyharju J. (2008) Ann. Med. 40, 402–417 [DOI] [PubMed] [Google Scholar]
- 2.Ozer A., Bruick R. K. (2007) Nat. Chem. Biol. 3, 144–153 [DOI] [PubMed] [Google Scholar]
- 3.Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 4.Fraisl P., Aragonés J., Carmeliet P. (2009) Nat. Rev. Drug Discov. 8, 139–152 [DOI] [PubMed] [Google Scholar]
- 5.Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell. 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 6.Semenza G. L. (2009) Physiology 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 7.Qi H. H., Ongusaha P. P., Myllyharju J., Cheng D., Pakkanen O., Shi Y., Lee S. W., Peng J., Shi Y. (2008) Nature 455, 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 9.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 10.Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Günzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., Kaelin W. G., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13459–13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köditz J., Nesper J., Wottawa M., Stiehl D. P., Camenisch G., Franke C., Myllyharju J., Wenger R. H., Katschinski D. M. (2007) Blood 110, 3610–3617 [DOI] [PubMed] [Google Scholar]
- 12.Xie L., Xiao K., Whalen E. J., Forrester M. T., Freeman R. S., Fong G., Gygi S. P., Lefkowitz R. J., Stamler J. S. (2009) Sci. Signal. 2, ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q., Gu J., Li L., Liu J., Luo B., Cheung H. W., Boehm J. S., Ni M., Geisen C., Root D. E., Polyak K., Brown M., Richardson A. L., Hahn W. C., Kaelin W. G., Jr., Bommi-Reddy A. (2009) Cancer Cell. 16, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cummins E. P., Berra E., Comerford K. M., Ginouves A., Fitzgerald K. T., Seeballuck F., Godson C., Nielsen J. E., Moynagh P., Pouyssegur J., Taylor C. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18154–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuznetsova A. V., Meller J., Schnell P. O., Nash J. A., Ignacak M. L., Sanchez Y., Conaway J. W., Conaway R. C., Czyzyk-Krzeska M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2706–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koivunen P., Tiainen P., Hyvärinen J., Williams K. E., Sormunen R., Klaus S. J., Kivirikko K. I., Myllyharju J. (2007) J. Biol. Chem. 282, 30544–30552 [DOI] [PubMed] [Google Scholar]
- 17.Myllyharju J., Kivirikko K. I. (1999) EMBO J. 18, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oehme F., Ellinghaus P., Kolkhof P., Smith T. J., Ramakrishnan S., Hütter J., Schramm M., Flamme I. (2002) Biochem. Biophys. Res. Commun. 296, 343–349 [DOI] [PubMed] [Google Scholar]
- 19.Westerfield M. (ed) (2000) The Zebrafish Book, University of Oregon Press, Eugene, OR [Google Scholar]
- 20.McCurley A. T., Callard G. V. (2008) BMC Mol. Biol. 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pressley M. E., Phelan P. E., 3rd, Witten P. E., Mellon M. T., Kim C. H. (2005) Dev. Comp. Immunol. 29, 501–513 [DOI] [PubMed] [Google Scholar]
- 22.Thisse C., Thisse B. (2008) Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 23.Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corey D. R., Abrams J. M. (2001) Genome Biol. 2, REVIEWS1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heasman J. (2002) Dev. Biol. 243, 209–214 [DOI] [PubMed] [Google Scholar]
- 26.Morcos P. A. (2007) Biochem. Biophys. Res. Commun. 358, 521–527 [DOI] [PubMed] [Google Scholar]
- 27.Drummond I. A. (2005) J. Am. Soc. Nephrol. 16, 299–304 [DOI] [PubMed] [Google Scholar]
- 28.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor C. T. (2008) J. Physiol. 586, 4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ton C., Stamatiou D., Liew C. C. (2003) Physiol. Genomics. 13, 97–106 [DOI] [PubMed] [Google Scholar]
- 31.Elvidge G. P., Glenny L., Appelhoff R. J., Ratcliffe P. J., Ragoussis J., Gleadle J. M. (2006) J. Biol. Chem. 281, 15215–15226 [DOI] [PubMed] [Google Scholar]
- 32.Manalo D. J., Rowan A., Lavoie T., Natarajan L., Kelly B. D., Ye S. Q., Garcia J. G., Semenza G. L. (2005) Blood 105, 659–669 [DOI] [PubMed] [Google Scholar]
- 33.van Rooijen E., Voest E. E., Logister I., Korving J., Schwerte T., Schulte-Merker S., Giles R. H., van Eeden F. J. (2009) Blood 113, 6449–6460 [DOI] [PubMed] [Google Scholar]
- 34.Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Stemple D. L., Zwartkruis F., Rangini Z., Driever W., Fishman M. C. (1998) Development 125, 4655–4667 [DOI] [PubMed] [Google Scholar]
- 35.Majumdar A., Drummond I. A. (2000) Dev. Biol. 222, 147–157 [DOI] [PubMed] [Google Scholar]
- 36.Kramer-Zucker A. G., Wiessner S., Jensen A. M., Drummond I. A. (2005) Dev. Biol. 285, 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raschperger E., Neve E. P., Wernerson A., Hultenby K., Pettersson R. F., Majumdar A. (2008) Dev. Biol. 313, 455–464 [DOI] [PubMed] [Google Scholar]
- 38.Cooper M. E., Mundel P., Boner G. (2002) Semin. Nephrol. 22, 393–398 [DOI] [PubMed] [Google Scholar]
- 39.Patrakka J., Tryggvason K. (2009) Nat. Rev. Nephrol. 8, 463–468 [DOI] [PubMed] [Google Scholar]
- 40.Serluca F. C., Drummond I. A., Fishman M. C. (2002) Curr. Biol. 12, 492–497 [DOI] [PubMed] [Google Scholar]
- 41.Erler J. T., Bennewith K. L., Cox T. R., Lang G., Bird D., Koong A., Le Q. T., Giaccia A. J. (2009) Cancer Cell 15, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynaud C., Baas D., Gleyzal C., Le Guellec D., Sommer P. (2008) Matrix Biol. 27, 547–560 [DOI] [PubMed] [Google Scholar]
- 43.Utriainen A., Sormunen R., Kettunen M., Carvalhaes L. S., Sajanti E., Eklund L., Kauppinen R., Kitten G. T., Pihlajaniemi T. (2004) Hum. Mol. Genet. 13, 2089–2099 [DOI] [PubMed] [Google Scholar]
- 44.Hamano Y., Okude T., Shirai R., Sato I., Kimura R., Ogawa M., Ueda Y., Yokosuka O., Kalluri R., Ueda S. (2010) J. Am. Soc. Nephrol. 21, 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elamaa H., Sormunen R., Rehn M., Soininen R., Pihlajaniemi T. (2005) Am. J. Pathol. 166, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J., Gross J. M. (2007) Invest. Ophthalmol. Vis. Sci. 48, 2483–2490 [DOI] [PubMed] [Google Scholar]
- 47.Rojas D. A., Perez-Munizaga D. A., Centanin L., Antonelli M., Wappner P., Allende M. L., Reyes A. E. (2007) Gene Expr. Patterns 7, 339–345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.