Abstract
Processive stepping of myosin Va (myoV) has been tracked by monitoring either the tail position (center of mass) or the position of one or both heads. Here, we combine these two approaches by attaching a quantum dot to one of the motor domains and a bead to the tail. Using laser trapping and total internal reflection microscopy, the position of one head and the tail are observed simultaneously as myoV moves processively on an actin filament bundle against the resistive load of the laser trap. The head moves one step (73 ± 10 nm) for every two steps of the tail (35 ± 9 nm). One tail step occurs concurrently with quantum dot-labeled head movement, whereas the other occurs with movement of the unlabeled head, consistent with a hand-over-hand model. Load increases the probability of the motor taking a back step. The back step is triggered by the motor taking a shorter forward step (head step, 68 ± 11 nm; tail step, 32 ± 10 nm), likely one actin monomer short of its preferred binding site. During a back step, the motor reverses its hand-over-hand motion, with the leading head detaching and reattaching to one of multiple actin sites behind the trailing head. After a back step, the motor can correct its mistake and step processively forward at resistive loads <0.7 piconewton or stall or detach at higher loads. Back stepping may provide a mechanism to ensure efficient cargo delivery even when myoV encounters obstacles within the actin cytoskeletal meshwork or when other motors are attached to the same cargo.
Keywords: Actin, Microscopic Imaging, Molecular Motors, Myosin, Single Molecule Biophysics, TIRF Microscopy, Force, Laser Trap, Quantum Dot
Introduction
Myosin Va (myoV),3 a double-headed processive molecular motor, converts chemical energy derived from ATP hydrolysis into ∼36-nm steps as it transports intracellular cargo along actin filament tracks (1–4). Each motor domain binds actin and contains a catalytic site. It is followed by a long α-helical neck composed of six calmodulin-binding IQ motifs that serves as a lever arm to amplify small nucleotide-induced changes generated within the motor domain. Following the head is an α-helical coiled-coil tail that dimerizes the molecule, and a C-terminal globular tail that binds cargo and which is involved in regulating motor activity (for review, see Ref. 5).
For myoV to take multiple forward steps in a hand-over-hand fashion without detaching from actin, the mechanochemical cycles of each head must be out of phase and biased so that the rear or trailing head detaches first from actin and is then propelled forward by the lever arm rotation of the strongly bound leading head. This gating between heads probably results from intramolecular strain that occurs as the leading head attempts to complete its powerstroke and take a forward step but is resisted by the attached trailing head that has yet to complete its enzymatic cycle. This interhead coordination is so robust that under unloaded conditions in vitro, myoV is virtually guaranteed to step forward as evidenced by a low probability (0.3%) of backward steps (6). However, intracellular forces will oppose myoV cargo transport as the motor encounters obstacles within the actin cytoskeletal meshwork or when other motors are attached to the same cargo. Based on our own studies and those of others using single molecule laser trapping, resistive forces increase the probability of myoV taking a back step until the motor stalls (equal probability of a forward or backward step), or steps backward continuously in response to super stall forces (6, 7). Without knowing the spatial dynamics of the two heads and their relationship to the center of mass of the molecule (the tail) during such backward motions, one cannot distinguish between the motor slipping back along the actin filament versus the motor reversing its normal stepping pattern.
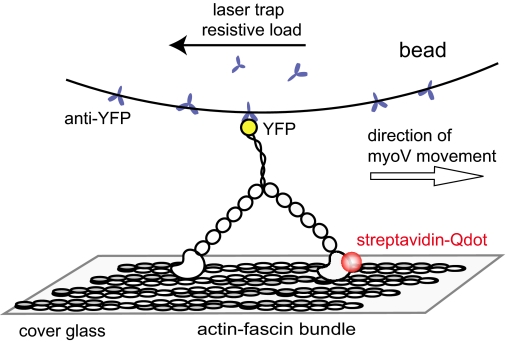
To define the stepping dynamics of the heads during both forward and backward steps and to understand the structural basis for triggering a back step under resistive load, we have simultaneously monitored the position of one of the two heads and the tail, in a combination laser trap and total internal reflectance fluorescence (TIRF) microscope. By attaching a quantum dot (Qdot) to the N terminus of one head, and a polystyrene bead to the C terminus of the motor, the head and tail positions were determined with nanometer resolution as load was applied to the tail (Fig. 1). Using this approach, backward motions of the motor in response to resistive load were characterized as true reversals of the hand-over-hand stepping mechanism. As myoV normally walks along the 36 nm pseudo-repeat of the actin filament with each head stepping on the 13th actin monomer, back steps are triggered when the leading head lands one actin monomer short of its preferred binding site. Following a back step, the motor will either continue its processive forward stepping or dynamically stall before terminating its run, depending on the resistive load. The ability of myoV to take a back step may allow the motor to continue transporting cargo even when physical challenges are encountered within the cell.
FIGURE 1.
Schematic of the experimental setup. MyoV HMM containing a C-terminal YFP was attached to a polystyrene bead coated with anti-YFP antibody. A fluorescently labeled actin-fascin bundle was bound to the surface of the cover glass by N-ethylmaleimide-myosin. A streptavidin-coated 655 nm Qdot was attached to the N terminus of one of the myoV motor domains through a biotin-streptavidin linkage. The Qdot was excited in the TIRF field, and fluorescence was collected with a 100× NA 1.49 objective.
EXPERIMENTAL PROCEDURES
Myosin V Preparation
To enable the simultaneous detection of the myoV head and tail, we used an expressed myoV HMM construct containing an N-terminal biotin tag for attachment of a streptavidin Qdot, and a C-terminal YFP for attachment to an anti-fluorescent protein antibody-coated polystyrene bead (8). In later experiments, we used a heterodimeric myoV HMM that had a biotin tag on only one of its two heads, thus facilitating Qdot labeling of only one head. For this construct, one heavy chain had YFP and FLAG tag at the C terminus and an N-terminal biotin motif, whereas the other heavy chain had YFP at the C terminus and an His8 motif at the N terminus. The heterodimeric construct was purified by a similar protocol described previously (9). The protein was passed first through a His-SELECT nickel affinity column (Sigma-Aldrich) and then through an anti-FLAG column (Sigma-Aldrich) with the final consisting of the heterodimeric motor.
Actin Bundle Preparation
Chicken skeletal actin was labeled with TRITC-phalloidin as described previously (10). Parallel actin bundles were formed by adding expressed fascin to actin filaments as described in Ref. 11. Briefly, 1 μm TRITC-labeled actin filaments were mixed with 1 μm fascin in buffer containing 25 mm KCl and 25 mm imidazole (pH 7.4) on ice for 1 h. Actin bundles were then perfused into a flow cell and allowed to bind to a cover glass surface coated with N-ethylmaleimide-modified chicken skeletal muscle myosin for 5 min.
The flow cell was then rinsed with buffer A (25 mm KCl, 1 mm EGTA, 4 mm MgCl2, and 25 mm imidazole (pH 7.4)) three times to remove unbound actin.
Simultaneous Detection of MyoV Head and Tail Displacements
Beads coated with an anti-YFP antibody (3E6; Invitrogen) for linkage to the myoV tail provide a well defined attachment strategy compared with nonspecific absorption. Beads were prepared as follows: 2.5 μl of 75 pm 1-μm polystyrene beads (Polysciences, Inc., Warrington, PA) were mixed with 1 μl of 50 mg/ml BSA, 4 μl of 0.5 mg/ml anti-fluorescent protein antibody (3E6; Invitrogen), 37.5 μl of buffer A and incubated on ice for 1 h. The mixture was briefly spun in a microcentrifuge, the supernatant removed, and the pellet resuspended in 50 μl of buffer B (buffer A plus 100 units/ml pyruvate kinase, 0.5 mm phosphoenol pyruvate, an oxygen scavenging system (0.1 mg/ml glucose oxidase, 0.18 mg/ml catalase, and 3 mg/ml glucose), 5 μm ATP, and 0.1 mg/ml CaMΔall, a calcium-insensitive calmodulin mutant (10)). To prepare Qdot-labeled myoV, 0.1 mg/ml myoV (homo- or heterodimeric, containing an N-terminal biotin tag and a C-terminal YFP) was diluted 1/50 in buffer A. 2 μl of this solution was mixed with 2 μl of 0.2 μm streptavidin Qdot (655-nm emission; Invitrogen) for 10 min at room temperature, giving a Qdot:myoV ratio of 53:1. This mixture was then further diluted 50–60-fold, with a final myoV concentration of 60–72 pm. To conjugate the Qdot-myoV to the bead, 2 μl of 60 pm myoV was mixed with 2 μl of 3.75 pm bead (precoated with anti-fluorescent protein antibody and BSA) and allowed to incubate for 10 min at room temperature. Finally, this mixture was diluted with 200 μl of buffer B and perfused into the flow cell, which had fascin-actin bundles adhered to the coverslip surface (see above).
To image the myoV displacements associated with the Qdot and bead images, the flow cell was placed on the stage of a combined laser trap:TIRF microscope, based on a Nikon TE2000 inverted microscope equipped with a PlanApo 1.49 N.A. objective. The bead with its attached motor was captured by a custom-built laser trap (6). The bead's brightfield image was projected onto a quadrant photodiode detector, providing bead x and y positions that were filtered to 1 kHz and recorded following analog to digital (A/D) conversion. The trap stiffness was calibrated for each bead using the corner frequency from the bead position power spectrum (12, 13). The bead-motor complex was brought into contact with an actin bundle on the coverslip surface that was aligned to the x axis of the laser trap. The actin bundle and the myoV Qdot-labeled head were imaged simultaneously through the objective TIRF using a 488-nm argon ion laser for excitation (Spectra Physics, Santa Clara, CA). The fluorescent images of actin and the Qdot were separated by an image splitter (Optical Insights LLC, Santa Fe, NM) and projected onto an intensified CCD camera (XR-Mega S30; Stanford Photonics, Stanford, CA) and recorded as a TIFF stack using Piper Control software (Agile Automation, Inc., Union City, CA) at 10–30 frames/s. The effective pixel resolution at 2 × 2 binning was 117 nm/pixel. Because the Qdot image and bead position were recorded by separate computers, it was imperative that the two signals be precisely aligned in time. To accomplish this, an external timing pulse for each captured image frame was provided by the camera manufacturer and A/D converted and recorded by the computer that simultaneously recorded the bead position. This provided timing marks on the bead position trace that could be linked to a specific Qdot image frame.
Data Analysis
The TIFF stack of Qdot images was cropped to a smaller size and then analyzed by a custom-written MATLAB program to find the center of the Qdot intensity profile (i.e. point spread function) to a two-dimensional Gaussian distribution (14). This provided subpixel resolution of 6 nm (15). The bead traces were filtered with a 20-Hz low pass Gaussian filter in Clampfit (Axon Instruments, Inc, Foster City, CA). The Qdot and bead position traces were then analyzed by an objective step-finding algorithm to determine the step size and dwell time of a step (16).
RESULTS
Coordinated Head and Tail Movements of MyoV
The novel feature of this study is the simultaneous monitoring of the head and tail position of a single myoV, as it processively transports its bead cargo against the resistive load of the laser trap (Fig. 1). When a captured bead and its attached motor were brought into contact with an actin-fascin bundle, myoV stepped along the actin with the expected ∼72-nm steps, given that only one of the two motor domains is labeled with a Qdot (15). Steps were detected from the bead signal only after myoV took several steps, suggesting that an effective compliance exists between the motor and the bead. The compliance is due to the bead being held at its center by the laser trap while the motor is attached to the bead surface. Thus, as the motor begins to step, it applies a tangential force to the bead circumference, which will rotate the bead about its center in the trap, as we have shown previously (17). This results in no detectable linear displacement of the bead within the trap for the first few steps of the motor. This creates an effective compliance equivalent to ∼108 nm at maximum (i.e. three motor steps). Once this compliance has been taken up by the motor at loads of ∼0.1–0.2 pN, bead steps of ∼36 nm are detected, and there is a strict correlation between the head and cargo signals. The Qdot-labeled myoV head takes one step for every two steps that the cargo is moved, with one of the cargo steps being coincident with the Qdot-labeled head movement, within the 100-ms time resolution of the fluorescence camera system (Fig. 2a). This observation agrees well with a hand-over-hand mechanism for myoV processivity, with the second tail step originating from movement of the unlabeled head during the dwell period of the Qdot-labeled head. A total of 48 beads were captured, but only 24 beads showed a strict correlation between the bead and Qdot movement. By only analyzing data from these 24 beads, we eliminated the potential for multiple motors/bead so that the resulting 617-bead and 330-Qdot steps were generated by single motors that took seven steps on average during a run.
FIGURE 2.
Simultaneous monitoring of head and tail position. The blue circles are the x positions of the Qdot as determined by a two-dimensional Gaussian fit. The pink line is the x positions of the bead from the quadrant detector, filtered with a 20-Hz low pass Gaussian filter. The solid red and green lines are the steps found by the automatic step-finding program (see “Experimental Procedures”). Pink shaded and white areas alternate to show whenever a step is taken. The light blue background highlights periods when myoV takes a back step. The resistive load applied to the bead (right y axis) is indicated for the beginning and end of the bead position trace. a, raw data trace showing the simultaneous movement of the bead (tail) and head (Qdot) during processive motion. For every one ∼72-nm step of the head, there are two ∼36-nm steps of the tail. b, example of a back step event in which both the bead and Qdot-labeled head move backward (first blue stripe). The second blue stripe in b and the blue stripe in c show examples of a back step where the tail position changes, but the Qdot-labeled head position remains stationary. These are events where the unlabeled head and the tail move backward, but the position of the Qdot-labeled head remains fixed. d, example of a run that terminates during the back step.
The step displacements of the head (73 ± 10 nm) and cargo (35 ± 9 nm) were well fit by Gaussian distributions (Fig. 3a and Table 1). The size of each cargo displacement was half that of the head, confirming that the 73-nm step of one head moves the center of mass of the molecule (equivalent to the position of the neck-tail junction), by half that distance. The effect of any compliance is minimal given the good correlation between head and cargo movement both temporally and spatially. The observed step amplitudes agree with literature values obtained using single actin filaments, suggesting that our use of actin-fascin bundles to facilitate interaction of the motor with the track did not affect the inherent stepping pattern of the motor. Furthermore, we did not observe any appreciable motion perpendicular to the direction of travel, either in the bead or Qdot signal (data not shown), indicating that the motor traveled along a single actin filament within the bundle.
FIGURE 3.
Analysis of various head and tail step sizes. a, Gaussian fits of head and tail normal forward steps and the smaller forward step taken before the back step. Normal steps are shown in red, and the shorter steps before the back step are in blue. The Gaussian fits to the various head and tail steps were all normalized to 100%, with the peak of the fit set to 100 and with the total number of steps, and the number of steps at the highest recorded data point for each condition as follows: normal head steps (299, 109), normal tail steps (598, 136), head steps before a back step (30, 10), tail steps before a back step (52, 11). All fits had r2 values >0.96. b, Gaussian fits of tail back steps (r2 = 0.87) and forward recovery steps (r2 = 0.92). c, Gaussian fits of head back steps (r2 = 0.55) and forward recovery steps (r2 = 0.77). The black dashed line is the fit of one broad Gaussian peak to the head back steps. The green dashed line is the fit of one Gaussian peak to the head recovery step. The Gaussian fit, although poor, is only meant to illustrate that the data are considerably broader than the head steps shown in a.
TABLE 1.
Head and tail step sizes
Head position was determined from Qdot signal with tail position determined from bead signal. Mean ± S.D. are from Gaussian fits to the histograms in Fig. 3.
| Signal | Step sizea |
|||
|---|---|---|---|---|
| Normal step | Step before a back step | Back step | Recovery step | |
| Head | 73 ± 10.3 | 68.0 ± 11 | 48.5 ± 26.9 | 60.6 ± 21.8 |
| Tail | 34.7 ± 8.6 | 31.9 ± 9.7 | 28.6 ± 13.7 | 30.8 ± 10.4 |
a Step size is in nm.
Backward Steps Are Reversals of Normal Steps
As the myoV stepped forward against the load of the laser trap, backward steps in both the Qdot and bead signals were observed more frequently (Figs. 2, b–d, and 4a). Although previous studies have described backward steps under resistive loads, the experimental setups could only monitor the center of mass of the motor (6, 7). Backward motor displacements could therefore result from a slip, with the two heads forcibly detached from actin, and then reattached behind their previous position on actin. Alternatively, the motor could effectively walk backward by reversing its normal mode of stepping. With positional data for both the head and tail, we can distinguish between these two possibilities.
FIGURE 4.
Load dependence of various aspects of back steps. a, load dependence of back step probability. The probability is calculated by dividing the number of tail back steps by the number of normal steps at a given external load. The back step to normal step ratio for each data point is shown in parentheses. b, force dependence of a back step followed by processive stepping (circles) versus a back step followed by either stall (another forward-backward pair) or detachment (triangles). c, force dependence of dwell time for normal steps and back steps. The curves in the figure are the fit to Equation 1. The fit yields τ0 of 430 ms for normal steps and 220 ms for back steps. Data are the mean ± S.E. (error bars).
If backward motions were a slip, then regardless of which head was labeled with a Qdot, a backward bead motion would always be linked to a backward Qdot motion of the same magnitude (Fig. 5a). For a backward step that is a reversal of a forward step, two scenarios are possible depending on whether the head that steps back is labeled with a Qdot (Fig. 5b). If the Qdot is on the leading head taking the back step, the Qdot and bead both change position coincidentally. This was the case for 45% (30 of 66) of the backward steps (Fig. 2b). If the unlabeled head takes a step back, then the bead and unlabeled head will move, but there will be no change in the position of the Qdot-labeled head, as observed in 55% (36 of 66) of the backward steps (Fig. 2c). The observation of a stationary Qdot position provides strong evidence that myoV is truly taking a back step and not slipping along the actin filament. The fact that approximately half of the backward displacements are characterized this way correlates well with an equal probability of the trailing or leading head being Qdot-labeled.
FIGURE 5.
Schematic of a slip versus a true back step. a, schematic of a slip, where one expects a step of equal magnitude for both tail and head movement. The small sphere on one of the heads represents a Qdot. b, schematic of a back step that is a reversal of a forward step. For a back step, one should observe either 36-72 nm or 36-0 nm pairs for the tail and head, respectively, depending on whether the head that is stepping back is labeled with a Qdot or not.
A Shorter Forward Step under Load Triggers a Back Step
Because the probability of a back step increases with resistive load, the forward step before a back step should provide insight into what triggers the back step. Interestingly, the forward step taken by a myoV head or tail prior to a back step (Qdot, 68 ± 11 nm; bead, 32 ± 10 nm) was significantly shorter (5 nm and 3 nm, respectively; p < 0.005) than a normal forward step during processive motion (Fig. 3a, Tables 1 and 2). A back step thus appears to be triggered by the leading head landing one actin monomer short of its preferred binding site that results in a full 73-nm step. As a result, the leading head detaches from actin and lands behind the trailing head. The histogram of back step sizes taken by the head is very broadly distributed about a mean of 49 ± 27 nm (Fig. 3c, dashed line), suggesting that the head can bind to multiple positions along the actin filament. The histogram of the back step sizes measured at the center of mass (bead) is only slightly broader (29 ± 14 nm) than a normal step or the step prior to the back step (Fig. 3b, Tables 1 and 2).
TABLE 2.
Differences between various pairs of steps
| Signal | Pairs of steps compared | Difference | pa | |
|---|---|---|---|---|
| nm | ||||
| Head | Normal step | Step before a back step | 5.0 | 0.005 |
| Tail | Normal step | Step before a back step | 2.8 | 0.00056 |
| Head | Step before a back step | Back step | 19.5 | 0.0006 |
| Tail | Step before a back step | Back step | 3.3b | 0.0016 |
a The p values are the results of Student's t test between the pairs.
b The difference between this pair of steps was also compared by a pairwise value, which is the fit to the histogram of the pairwise difference between steps. The value was 4.3 ± 5.1 nm.
Fate of Motor after a Back Step Is Load-dependent
Of 71 back steps, 5 resulted in the motor detaching from actin (Fig. 2d), whereas the remaining 66 were followed by a forward step. The step size for this subsequent forward step (recovery step) was 61 ± 22 nm as measured from the Qdot and 31 ± 10 nm as measured from the bead (Table 1). The fate of the motor following the 71 back steps fell into two categories. In 32 instances the motor continued processive forward stepping (at least two forward steps following the back step). In the remaining 39 instances the motor either stalled by taking alternating backward and forward steps, or detached from actin prior to or after taking its forward recovery step. Because the probability of a back step occurring increases with resistive load (Fig. 4a), we asked whether the fate of the motor following a back step was also load-dependent. The frequency of the motor continuing to processively step, versus stalling or detaching, was plotted as a function of the resistive load that triggered the back step. These relationships were fit by Gaussians with a mean resistive load of 0.7 ± 0.005 pN for continuing to step, in contrast to a higher mean load of 0.9 ± 0.03 pN when the motor subsequently stalled or detached (Fig. 4b).
Force Dependence of the Dwell Time for Both Forward and Backward Steps
Because twice as many steps were observed from the bead signal, we analyzed the bead steps to determine the effect of resistive load on the dwell time for both forward (excluding steps prior to and after a backstep) and backward steps (Fig. 4c). These data were fitted to a modified Arrhenius/Eyring formula of the form,
 |
in which τ0 is the dwell time under no external load, F is the resistive load, and δ is the distance over which the load acts. The fit to the forward steps yielded a τ0 of 430 ± 76 ms and δ of 3.3 ± 0.5 nm, while the fit to the back steps gave a τ0 of 220 ± 32 ms and δ of 4.5 ± 0.5 nm.
DISCUSSION
The movement of both the head and center of mass (neck/tail junction) of myoV during processive motion was simultaneously observed for the first time. Head motion was monitored by a Qdot attached to only one of the two motor domains, whereas the center of mass was monitored by the position of a bead attached to the tail. This experimental setup allowed us to show directly that the center of mass of the molecule moves forward by half the distance of each of its heads and that the heads take turns stepping, in agreement with the commonly accepted hand-over-hand model for processive movement (15). When resistive loads are experienced, this process is reversed, allowing the motor to maintain contact with the actin filament by stepping backward rather than slipping along the filament.
Missing the Optimum Spot on Actin Increases Likelihood of a Back Step
Under unloaded conditions, myoV takes at least 25 consecutive forward steps on average along the actin filament before terminating its run (18). Under the resistive load of the laser trap, the number of steps is dramatically reduced to ∼7 steps under the conditions of this study. The probability of the motor taking a back step and/or terminating its run increased with load (Fig. 4a). For normal forward steps each head takes an ∼73-nm step onto the 13th actin monomer, matching the ∼36-nm pseudo-repeat of the actin filament. Under resistive load, the forward step is reduced in length by the distance equal to one actin monomer (Tables 1 and 2), suggesting that the motor steps one monomer short along the same actin strand and in doing so increases the probability of a backward step (Fig. 6a).
FIGURE 6.
Schematic diagram summarizing the types of steps myoV takes. a, difference between a normal step (leading head bound to the 13th actin subunit) and the shorter step myoV takes before a back step (leading head bound to the 11th actin subunit). When viewed down the actin axis, the two lever arms are in the same plane for a normal step, whereas for a step before a back step, there is an azimuthal angle of ∼28° between the two lever arms. b, schematic of a back step. The lever arms in green show the possible binding positions of the new trailing head. During a back step, the tail position changes by 29 ± 14 nm, whereas that of the heads is 49 ± 27.
Why does a short forward step trigger a backstep only under load? Under unloaded conditions, shorter steps occur within the normal distribution of forward steps (Fig. 3a), but they do not lead to a back step. Under unloaded conditions, myoV carrying a bead along a suspended actin filament will spiral around the filament. This suggests that the motor will step occasionally on the 11th actin monomer because it would not spiral if it stepped faithfully on each 13th monomer (19). Based on a lever arm model of myoV, it was estimated that nearly four times more energy is required for the leading head to bind onto the 11th actin monomer compared with the 13th, which is limited only by thermal energy (i.e. 1.1 kT, where k is the Boltzmann constant, and T is temperature in Kelvin) when the leading head is in the pre-powerstroke state and the trailing head is in the post-powerstroke state (20). When myoV takes a shorter forward step, an off-axis component to the resistive load would be experienced by the two lever arms (Fig. 6a), which in turn may weaken the actomyosin bond of the leading head. This was recently reported by Ishiwata and co-workers (21), who applied off-axis loads to single headed myoV S1 constructs. In a similar study, Rief and co-workers (22) show that as little as 1.7 pN was sufficient to detach a 6IQ myoV S1 construct in the absence of ATP. Our back step data show directly for the first time that the leading head (Qdot signal) detaches due to externally applied resistive loads in the piconewton range and reattaches behind the trailing head to then become the new trailing head.
A Back Step Is a Reversal of Normal Forward Stepping
Because a back step appears to be a reversal of the hand-over-hand stepping pattern, we also assume that the strongly bound trailing head that remains bound to actin during the back step undergoes a reversal of its powerstroke. Powerstroke reversals from a 6IQ myoV S1 construct were observed under resistive loads in the 2–5 pN range, which were assumed to be a post- to pre-powerstroke transition with ADP in the active site (23). Given that our studies were performed at low ATP concentrations (5 μm), the powerstroke reversal of the trailing head seen here presumably occurs while the head is in the rigor state. This is based on the observed dwell time for back steps of 220 ms under unloaded conditions, which agrees with that estimated from a second order ATP-binding rate (kATP) of ∼ 1 μm−1 s−1 (2) and 5 μm ATP. The similarity of the load-dependence of the back step and forward step dwell times seen here, together with additional data under load from Kad et al. (6), suggests that the kinetic state of the motor following a back step is probably rate limited by the same process that limits a forward step. This suggests that a back step returns the motor to the same or a comparable “waiting” state that the motor adopts during normal processive stepping. The back step must therefore involve a reversal of the trailing head's post-powerstroke state to a pre-powerstroke-like conformation, but without ADP in the active site. If the powerstroke is not fully reversed to a normally occupied pre-powerstroke state with ADP and Pi in the active site, it may account for the smaller than expected 29-nm back step of the center of mass of the motor compared with the 32-nm forward step that preceded the back step (Table 1). In addition, detachment of the leading head, which is presumably in the pre-powerstroke conformation with ADP, or ADP and Pi in the active state, is then followed by the head reattaching in the post-powerstroke ADP state behind the trailing head. This is supported by a similar distance parameter for the back step and forward step dwell time (Fig. 4c), both of which are consistent with the distance parameter of 4.3 nm reported for the kinetics of the ADP release step following the powerstroke (24).
Position of Detached Head during and after a Back Step
During a back step, the leading head detaches and reattaches behind the stationary actin-bound head. Given the broad distribution of back step sizes taken by the Qdot-labeled head, the detached head has the capacity to reattach to one of multiple binding sites behind the stationary head (Fig. 6b). The probability of the head binding to any given actin monomer will be determined by geometric constraints and the energy required to overcome the intramolecular strain needed to bend the lever arms as the motor adopts its new conformation (20).
After the back step, the motor then dwells for a period that is limited by the rate of ATP binding to the new trailing head. At this point the motor can resume forward stepping given that the resistive loads used here are well below the 5-pN superstall loads required to force the motor to step backward continuously (7). The next forward recovery step effectively returns the motor to its position on actin that initially triggered the back step, when considering all of the data (Table 1). After the recovery step the motor either continues stepping processively or stalls and detaches, with the fate of motor determined by the resistive load (Fig. 4b). Presumably, at loads <1 pN, the forward recovery step returns the head to the preferred 13th actin monomer, thus allowing the motor to resume normal processive stepping. For higher loads, the motor most likely takes a short forward recovery step once again, resulting in a higher probability of another back step (i.e. stall) or run termination.
CONCLUSIONS
Our results suggest that myoV is designed to manage effectively the complexities of the intracellular environment, having the capacity to alter its stepping pattern as necessary when presented with resistive loads. MyoV takes a back step to compensate for a less than optimal forward step, in essence attempting to correct its “mistake.” If the motor were to encounter an obstacle such as another motor or an actin-associated protein while transporting cargo, then the motor could effectively back up and try stepping forward again or dissociate if a barrier on the actin filament precludes its ability to continue. Either scenario would help prevent myoV from getting stuck on an actin track when cargo delivery is required.
Acknowledgments
We thank Carol Bookwalter and Elena Krementsova for cloning of constructs and providing the proteins that made this study possible.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078097 (to K. M. T.) and HL059408 and GM094229 (to D. M. W.).
- myoV
- myosin Va
- pN
- piconewton
- Qdot
- quantum dot
- TIRF
- total internal reflectance fluorescence
- TRITC
- tetramethylrhodamine isothiocyanate
- HMM
- heavy meromyosin
- IQ motif
- motif with consensus sequence IQxxxRGxxxR.
REFERENCES
- 1.Mehta A. D., Rock R. S., Rief M., Spudich J. A., Mooseker M. S., Cheney R. E. (1999) Nature 400, 590–593 [DOI] [PubMed] [Google Scholar]
- 2.De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rief M., Rock R. S., Mehta A. D., Mooseker M. S., Cheney R. E., Spudich J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9482–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veigel C., Wang F., Bartoo M. L., Sellers J. R., Molloy J. E. (2002) Nat. Cell Biol. 4, 59–65 [DOI] [PubMed] [Google Scholar]
- 5.Trybus K. M. (2008) Cell. Mol. Life Sci. 65, 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kad N. M., Trybus K. M., Warshaw D. M. (2008) J. Biol. Chem. 283, 17477–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemen A. E., Vilfan M., Jaud J., Zhang J., Bärmann M., Rief M. (2005) Biophys. J. 88, 4402–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., Krementsova E. B., Trybus K. M. (2006) J. Biol. Chem. 281, 31987–31994 [DOI] [PubMed] [Google Scholar]
- 9.Rovner A. S., Fagnant P. M., Trybus K. M. (2006) Biochemistry 45, 5280–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krementsov D. N., Krementsova E. B., Trybus K. M. (2004) J. Cell Biol. 164, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodges A. R., Bookwalter C. S., Krementsova E. B., Trybus K. M. (2009) Curr. Biol. 19, 2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilford W. H., Dupuis D. E., Kennedy G., Wu J., Patlak J. B., Warshaw D. M. (1997) Biophys. J. 72, 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kad N. M., Rovner A. S., Fagnant P. M., Joel P. B., Kennedy G. G., Patlak J. B., Warshaw D. M., Trybus K. M. (2003) J. Cell Biol. 162, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali M. Y., Lu H., Bookwalter C. S., Warshaw D. M., Trybus K. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warshaw D. M., Kennedy G. G., Work S. S., Krementsova E. B., Beck S., Trybus K. M. (2005) Biophys. J. 88, L30–L32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 17.Dupuis D. E., Guilford W. H., Wu J., Warshaw D. M. (1997) J. Muscle Res. Cell Motil. 18, 17–30 [DOI] [PubMed] [Google Scholar]
- 18.Baker J. E., Krementsova E. B., Kennedy G. G., Armstrong A., Trybus K. M., Warshaw D. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5542–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali M. Y., Uemura S., Adachi K., Itoh H., Kinosita K., Ishiwata S. I. (2002) Nat. Struct. Mol. Biol. 9, 464–467 [DOI] [PubMed] [Google Scholar]
- 20.Vilfan A. (2005) Biophys. J. 88, 3792–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oguchi Y., Mikhailenko S. V., Ohki T., Olivares A. O., De La Cruz E. M., Ishiwata S. (2010) Nat. Chem. Biol. 6, 300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebhardt J. C., Okten Z., Rief M. (2010) Biophys. J. 98, 277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sellers J. R., Veigel C. (2010) Nat. Struct. Mol. Biol. 17, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veigel C., Schmitz S., Wang F., Sellers J. R. (2005) Nat. Cell Biol. 7, 861–869 [DOI] [PubMed] [Google Scholar]