Abstract
A theme emerging during the past few years is that members of the small leucine-rich proteoglycan gene family affect cell growth by interacting with multiple receptor tyrosine kinases (RTKs), mostly by a physical down-regulation of the receptors, thereby depriving tumor cells of pro-survival signals. Decorin binds and down-regulates several RTKs, including Met, the receptor for hepatocyte growth factor. Here we demonstrate that decorin blocks several biological activities mediated by the Met signaling axis, including cell scatter, evasion, and migration. These effects were mediated by a profound down-regulation of noncanonical β-catenin levels. In addition, Myc, a downstream target of β-catenin, was markedly down-regulated by decorin, whereas phosphorylation of Myc at threonine 58 was markedly induced. The latter is known to destabilize Myc and target it for proteasomal degradation. We also discovered that systemic delivery of decorin using three distinct tumor xenograft models caused down-regulation of Met and a concurrent suppression of β-catenin and Myc levels. We found that decorin protein core labeled with the near infrared dye IR800 specifically targeted the tumor cells expressing Met. Even 68-h post-injection, decorin was found to reside within the tumor xenografts with little or no binding to other tissues. Collectively, our results indicate a role for a secreted proteoglycan in suppressing the expression of key oncogenic factors required for tumor progression.
Keywords: β-Catenin, Cellular Regulation, Extracellular Matrix, Myc, Receptor Tyrosine Kinase, β-Catenin, Growth Suppression, Myc, Proteoglycan, Receptor Tyrosine Kinase
Introduction
Biological activities are triggered, modulated, and maintained mostly by the complex interplay among extracellular matrix molecules, multifunctional growth factors, and their surface receptors (1). Genetic and biochemical studies have revealed that many of the matrix components are indeed signaling molecules that can affect the behavior of cells during development and disease progression (2). Decorin, a prototype member of the small leucine-rich proteoglycans (SLRPs)3 (3–8), is involved in a multitude of biological processes including collagen fibrillogenesis (9–12), modulation of various growth factors and receptors (13–22), renal diseases (23–25), angiogenesis (26, 27), wound healing (28), myocardial infarction (29), lung and tendon mechanics (30–32), infectious diseases (33, 34), bone and tooth development (35, 36), connective tissue development (37–41), and bone marrow stromal cell biology (42).
The involvement of decorin in cancer progression has been demonstrated in mutant mice. Although the decorin-null mice do not spontaneously develop malignant neoplasms, mice carrying a targeted deletion of both decorin and the tumor suppressor p53 succumb within 3–4 months to aggressive lymphomas at a rate faster than p53-null mice alone (43), suggesting that decorin is permissive for tumorigenesis. In accordance with this study, ∼30% of decorin-null animals, which were backcrossed into a different genetic background, displayed spontaneous occurrence of intestinal tumors, and both the tumor burden and frequency were enhanced by subjecting the mutant mice to a high-risk western diet (44).
Although the precise molecular mechanisms of decorin anti-oncogenic activity are not completely understood, one of the major roles of decorin is to signal through receptor tyrosine kinases (RTKs) including the epidermal growth factor receptor (EGFR) and other members of the ErbB family of RTKs (45–57) and the insulin-like growth factor receptor type I (58–60). We hypothesized that decorin might be even more promiscuous and bind to other RTKs. By screening 42 different RTKs for their Tyr phosphorylation status, we discovered that decorin interacts with Met, the receptor for hepatocyte growth factor (HGF) (61), which promotes proliferation and invasion when constitutively activated in cancer cells (62, 63). Although binding to decorin briefly activated Met, this only led to receptor rapid down-regulation via two different pathways: by intracellular degradation and by cleavage of its extracellular domain (61).
In this study we have further explored the mechanism of decorin-evoked antagonistic action toward Met and discovered that decorin evokes intracellular degradation of two key molecules, β-catenin and its downstream effector Myc. These effects occur at both transcriptional and post-translational levels via a noncanonical Wnt pathway, and cause inhibition of motility and migration, and ultimately lead to tumor growth suppression, both in vitro and in vivo. Thus, decorin could become a new therapeutic modality against various forms of solid tumors which often have increased levels of multiple growth factor receptors and can survive therapies that target only a single pathway.
EXPERIMENTAL PROCEDURES
Cells and Materials
Madin-Darby Canine Kidney (MDCK) epithelial cells, A431, MTLn3 and HeLa carcinoma cells were obtained from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's-modified Eagle's medium (DMEM) (Mediatech Inc.) supplemented with 10% fetal bovine serum (FBS) (SAFC Biosciences, Lenexa, KS) and 100 μg/ml penicillin/streptomycin (Mediatech, Manassas, VA). Primary antibodies against Met (Met-C12, Sc-10), Myc (Myc-N-262), phosphor-T58 Myc (SC-135647), and p21 (C-19) were from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit polyclonal anti-β-catenin (ab 16051) and anti-GAPDH antibodies were from Abcam Inc. (Boston, MA); mouse monoclonal anti-β-catenin, mouse anti-clathrin heavy chain, anti-caveolin-1, and MatrigelTM were from BD Biosciences (San Jose, CA); mouse monoclonal anti-Met MAB3729 antibody was from Millipore Corp. (Billerica, MA); mouse monoclonal anti-β-actin antibody, goat anti-Met blocking H9786 antibody, recombinant human HGF, and the proteasome inhibitor lactacystin were from Sigma-Aldrich. The proteasome inhibitor MG132 was purchased from Calbiochem.
Scatter, Matrigel Evasion, and In Vitro Wound Healing Assays
For cell scatter assay, MDCK cells (∼1.2 × 104/100 μl) were seeded in triplicate in gelatin-coated chamber glass slides (Nunc, Rochester, NY) within small drops. Following a 30-min incubation at 37 °C, during which the cells had attached in a small confluent layer, serum free medium was added and incubated overnight. Cells were then incubated in presence or absence of decorin protein core (500 nm) along with HGF (50 ng/ml, ∼0.71 nm) for 14 h. Cells were finally washed and fixed with 4% paraformaldehyde/phosphate-buffered saline (PFA). For the evasion assays, MDCK cells (5.8 × 106/ml) were mixed in 1:2 ratios with diluted (3.2 mg/ml) Matrigel and seeded as quadruplicate drops at corners of gelatin-coated chamber glass slides. Following a 10-min incubation at 37 °C, serum-free medium was added ± decorin (500 nm) or HGF (50 ng/ml, ∼0.71 nm) and incubated for 24 h. Images (10× magnification) were taken over time with a digital epiluminescence microscope camera (DP12; Olympus) and the number of scattered cells or cells evaded from each Matrigel drop was quantified. In vitro wound healing assays were performed as described before (61). Initial dose-response experiments using recombinant decorin (50–700 nm) showed that the most consistent data of down-regulation of Met, β-catenin, and Myc were obtained with 500 nm decorin. Therefore, all subsequent experiments in HeLa cells were carried out using decorin at 500 nm.
Immunofluorescence and Confocal Microscopy
Immunofluorescence and confocal laser microscopy were performed as described before (64, 65). Approximately 5 × 104 HeLa cells were plated on 4-well chamber slides (BD Biosciences) and grown to full confluence in 10% FBS at 37 °C. Cells were switched to DMEM/2% FBS 2 h prior to each treatment. Slides were rinsed twice with DPBS and fixed/permeabilized with ice-cold methanol for 10 min. Subsequently slides were subjected to standard immunofluorescence protocols, and mounted with Vectashield (Vector Laboratories Inc, Burlingame, CA). Following incubation with various primary antibodies, detection was determined using goat anti-mouse IgG Alexa Fluor® 488 and goat anti-rabbit IgG Alexa Fluor® 594 (Invitrogen). Images were acquired using a LEICA DM5500B microscope with Leica Application Suite, Advanced Fluorescence 1.8 software (Leica Microsystems, Inc). Confocal analysis was performed on an Olympus IX70 microscope driven by the Laser Sharp 2000 image software. Filters were set to 488 and 564 nm for dual channel imaging. All the images were analyzed using the aforementioned software in conjunction with Adobe Photoshop CS3 (Adobe Systems, San Jose, CA). In some cases, images were captured with a SPOT-RT camera connected to an inverted fluorescence microscope (BX51; Olympus), and fluorescence quantification was done with ImageJ software.
Transient Knockdown of Met Receptor
To achieve transient knockdown of the Met receptor we utilized a mixture of three validated siRNAs specific for Met mRNA (Met siRNA sc-29397, Santa Cruz Biotechnology). Briefly, six-well plates were seeded with 2 × 105 HeLa cells, followed by incubation overnight at 37 °C until cultures were 70% confluent. Targeting siRNA duplex at a final concentration of 80 pm was added to diluted Lipofectamine 2000 (Invitrogen) in OPTI-MEM I Reduced Serum Medium (Invitrogen), according to the manufacturer's protocol. Transfection was carried out for 48 h at 37 °C and the cells were then treated with either serum-free DMEM or decorin (500 nm) for an additional 24 h. Verification of siRNA-mediated knockdown of the Met receptor was determined via immunoblotting using Met-specific antibodies.
Quantitative Real-time PCR Analysis
Gene expression analysis by quantitative real-time polymerase chain reaction (qRT-PCR) was carried out as described before (66, 67) with minor modifications. Briefly, subconfluent (∼1.5 × 106 cells) 3.5 cm2 dishes of HeLa cells were treated for 24 h with either PBS (mock) or 500 nm decorin protein core in serum-free DMEM. After incubation, cells were lysed directly in 500 μl of TRIzol reagent (Invitrogen, Grand Island, NY). Total RNA (1 μg) was annealed with oligo(dT) primers, and cDNA was synthesized utilizing the SuperScript Reverse Transcriptase II (Invitrogen) according to the manufacturer's instructions. Gene-specific primers for CTNNB1, MYC, AP4, CDKN1A, CCND1, and CDK6 were verified before use (see supplemental Table S1 for primer sequences). The target genes and endogenous housekeeping gene, ACTB, amplicons were amplified in independent reactions using the Brilliant SYBR Green Master Mix II (Agilent Technologies, Cedar Creek, TX). All samples were run in quadruplicate on an Mx3005P Real Time PCR platform (Agilent) and the cycle number (Ct) was obtained for each independent amplicon reaction. Fold change determinations were made utilizing the Comparative Ct method for gene expression analysis. Briefly, Delta Ct (ΔCt) values are representative of the normalized gene expression levels with respect to ACTB (β-actin as endogenous housekeeping control). Delta Delta Ct (ΔΔCt) values represent the experimental cDNA (samples treated with 500 nm decorin protein core) minus the corresponding gene levels of the calibrator sample (samples treated with PBS mock). Finally, the reported fold change represents an average of the fold changes as calculated using the double ΔCt method (2−ΔΔCT). All the data derive from three independent trials for each gene of interest.
Tumor Xenografts and Infrared Labeling of Decorin
All animal studies were approved by the Institutional Review Board of Thomas Jefferson University. Severe-combined immunodeficient (SCID) female mice (Charles River Lab., Malvern, PA) were injected subcutaneously with 1–2 × 106 HeLa, A431 or MTLn3 cells. The mice were randomized once tumors were established. Half the mice received a dose of 5 mg/kg decorin protein core injected intraperitoneally every 2 days. The controls received 100 μl of PBS. Six independent experiments were performed with five mice in each category. Tumor growth was measured every 2 days with a micro-caliper according to the following formula: V = a(b2/2), where a and b represent the larger and smaller diameters, respectively. On day 23, the majority of animals were sacrificed and all major organs and tumors were dissected and fixed in formalin. For infrared labeling, ∼1 mg of decorin protein core was labeled with IRDye® 800CW which contains an N-hydroxysuccinimide ester reactive group that couples to the decorin protein core and forms a stable conjugate (Li-COR Biosciences, Lincoln, NE). Fluorescent conjugates labeled with IRDye® 800CW display an absorption maximum of 774 nm and an emission maximum of 798 nm. Fluorescent proteins were separated from the free dye using a desalting spin column and aliquots of the eluates were analyzed on SDS-PAGE. The gels were scanned with an Odyssey® Infrared Imaging System to verify that the protein was successfully labeled.
Mice (n = 2–3) from each independent experiment received 100 μg of IR800-labeled decorin core via a tail vein injection. The same animals were anesthetized and scanned using a PearlTM Imager at specific time points: pre-injection, immediate post-injection, 16, 22, 24, 40, 48, and 68 h post-injection. Mice were anesthetized with isoflurane and euthanized with CO2 in accordance with institutional guidelines.
Quantification and Statistical Analysis
Immunoblots were quantified by scanning densitometry using ImageJ software or using the Odyssey software for the infrared-labeled secondary antibodies. Significance of differences was determined by unpaired Student's t test using SigmaPlot version 11.0 and SigmaStat for Windows version 3.10 (Systat Software Inc, Port Richmond, CA). Statistical significance was achieved when p values were <0.05. Fluorescence intensity and three-dimensional surface plots were quantified by measuring pixels with ImageJ software. In the scratch assay, wound closure was measured with ImageJ by measuring three linear distances between the two edges of the wound.
RESULTS AND DISCUSSION
Decorin Inhibits the HGF/Met Signaling Axis in MDCK Cells
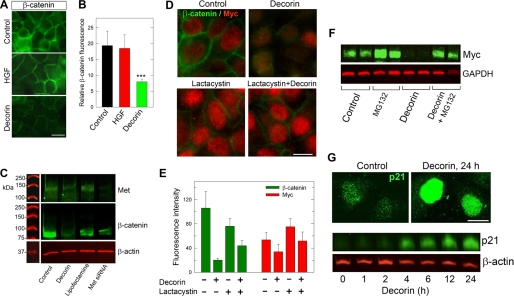
We have previously shown that decorin down-regulates Met in HeLa cells (61). To establish whether this decorin-mediated biological effect would occur also in cellular systems where HGF/Met biology was first established, we utilized MDCK cells, which have been widely used to investigate the role of HGF/scatter factor (62). First, we found that decorin had similar effects in down-regulating Met and β-catenin levels (Fig. 1A). Time course experiments showed a progressive down-regulation of Met and β-catenin levels with maximal inhibition at ∼6 h and a subsequent recovery (supplemental Fig. S1). However, even after 24 h of continuous exposure to decorin both protein levels were ∼80% of mock-treated cells.
FIGURE 1.
Decorin bioactivity in MDCK cells. A, immunoblot of MDCK cells treated with or without decorin (100 nm) for 6 h. Cell lysates were separated by 8% SDS-PAGE and immunoblotted for Met and β-catenin with GAPDH as loading control. Proteins were visualized with IR-Dye-labeled secondary antibodies and quantified using Odyssey Infrared Imaging system (Li-Cor). B–D, immunofluorescence images of MDCK cell untreated (control) or treated with either HGF (50 ng/ml, ∼0.71 nm) or decorin (100 nm) for 6 h. E–H, decorin attenuates HGF/SF induced MDCK cell scattering after 14-h incubation with (F and H) or without (E and G) 500 nm decorin in the presence (G and H) or absence (E and F) of HGF (0.71 nm). I, quantification of scattered cells after 14 h of HGF and decorin treatment. The values represent the mean ± S.E. (n = 40 each, ***, p < 0.001).
Immunofluorescence studies showed also a marked down-regulation of Met (not shown) and β-catenin levels (Fig. 1, B–D). Second, decorin was able to counteract the HGF-evoked scatter of MDCK cells (compare Fig. 1, H with G), and these effects were statistically significant (p < 0.001, Fig. 1I).
Next, we utilized an evasion assay where MDCK cells were first embedded in a drop of Matrigel and then exposed to decorin protein core, HGF or a combination of both in serum-free medium for 24 h. Notably, decorin blocked the HGF-evoked cell evasion (compare Fig. 2D with 2C) and these effects were highly significant (p < 0.001, Fig. 2E). In support of the scatter and evasion assays, in vitro wound healing assays showed that decorin could counteract the effects of HGF to a similar extent as the Met-blocking antibody (Fig. 3A).
FIGURE 2.
Decorin attenuates the HGF-evoked evasion of MDCK cells from Matrigel drops. Evasion of MDCK cells after 24 h incubation with (B and D) or without (A and C) decorin (500 nm) in the presence (C and D) or absence (A and B) of HGF (0.71 nm) stimulation. The dotted lines indicate the edge of the Matrigel drops. E, quantification of cells evaded from each Matrigel drop. The values represent the mean ± S.E. (n = 45 ***, p < 0.001).
FIGURE 3.
Decorin inhibits migration via the Met receptor and evokes caveolin-mediated internalization of Met. A, motility assay perfomed with a monolayer of HeLa cells scratched to cause a wound. Cells were treated for 18 h with 500 nm decorin or 0.71 nm HGF. Where indicated, 10 μg/ml H9786, a Met-blocking antibody, was added 30 min before treatment. Representative photos are shown. Bar, 60 μm. B, confocal images showing the Met receptor (green) and caveolin (red) in HeLa cells treated with or without 500 nm decorin or 0.71 nm HGF for 5 h. C, HeLa cells under the same treatment as shown in B, but stained for Met (green) and clathrin (red). All images were taken with the same exposure and gain. White arrows indicate co-localization. Bar, 10 μm.
We have previously shown that decorin induces EGFR internalization and degradation via caveolar-mediated endocytosis (64). Thus, we tested whether decorin would also induce internalization of the Met receptor via the same pathway. Using confocal microscopy, we found that decorin induced significant co-localization of Met and caveolin-1 after 5 h of treatment (Fig. 3B). In contrast, there was little or no co-localization of Met with clathrin evoked by decorin (Fig. 3C). HGF, however, induced clathrin-mediated endocytosis of the Met receptor as shown by co-localization of Met and clathrin (Fig. 3C), but no colocalization of Met and caveolin-1 (Fig. 3B), as expected of HGF-mediated signaling.
It is well established that clathrin-mediated endocytosis leads to intracellular signaling of RTKs and recycling of the receptors to the plasma membrane (68, 69). Another example is provided by the transforming growth factor-β receptor system, which utilizes both cavelae and clathrin-coated pits for internalization. Whereas the former pathway evokes receptor degradation, the latter promotes signaling (70).
Collectively, these results suggest that decorin causes a protracted degradation of the Met receptor via caveolar-mediated endocytosis which ultimately leads to transfer into caveosomes and then lysosomes, in a manner analogous to the EGFR (64).
Decorin Down-regulates β-Catenin and Myc Primarily via a Met-dependent Mechanism
Next we determined the effects of decorin on β-catenin, an established downstream effector of Met (71, 72), using a variety of approaches including confocal microscopy, validated siRNAs against the Met receptor mRNA and inhibitors of proteasomal degradation. Decorin caused a protracted degradation of β-catenin, which was statistically significant (p < 0.001) (Fig. 4, A and B). Notably, siRNA-mediated knockdown of Met showed a concurrent down-regulation of β-catenin (Fig. 4C).
FIGURE 4.
Decorin down-regulates β-catenin and Myc proteins. A, immunofluorescence images showing β-catenin epitopes in HeLa cells treated with either decorin (500 nm) for 5 h, or HGF (0.71 nm) for 30 min. B, quantification of fluorescent intensity with values representing means with their upper 95% confidence intervals (n = 30, ***, p < 0.001). C, representative immunoblot of Met and β-catenin levels in HeLa cells, which were transfected for 48 h with specific siRNAs targeting the Met receptor or treated for 24-h with decorin protein core (500 nm); β-actin was used as a loading control. D, fluorescence staining of HeLa cells for β-catenin and Myc ± decorin for 1 h. Where indicated, cells were preincubated with lactacystin (10 μm) for 30 min. E, quantification of images from D with means ± S.E. (n = 30, ***, p < 0.001). F, immunoblot of Myc levels after a 6-h treatment with decorin following a 2-h preincubation with MG132 (50 μm). G, fluorescence images of p21 induction after a 24-h decorin treatment (top panels). A representative immunoblot (bottom panel) shows time-dependent induction of p21. Decorin protein core was used at a concentration of 500 nm for all experiments shown.
Next, we investigated the potential involvement of a key downstream target of β-catenin, i.e. the Myc oncoprotein. Myc is a highly pleiotropic transcription factor that regulates the transcriptional activity of a large cohort of genes required for normal and neoplastic cell expression (73). The common mechanism of Myc oncogenic activity is due to a deregulated expression of Myc, rather than mutations, caused by upstream oncogenic signals including the Wnt/β-catenin and Met/β-catenin signaling pathways (74). Myc is a key obligatory downstream effector of the Wnt/β-catenin pathway because loss of Myc triggers abrupt growth arrest within the proliferating intestinal crypts (75, 76). Notably, Myc represses the expression of the cyclin-dependent kinase inhibitor p21WAF1/Cip1 (p21) in contrast to decorin which is known to induce its expression (77). Specifically, Myc represses p21 expression directly through complexes of Myc with Miz1 (78), indirectly through AP4, a transcriptional repressor of p21 (79), or via mitogenic signals elicited by the β-catenin/TCF pathway (80). We discovered that Myc was significantly down-regulated following a 1-h exposure to decorin protein core concurrent with β-catenin down-regulation (Fig. 4D). Notably this process was markedly inhibited by lactacystin, a proteasomal inhibitor (81), and these effects were highly significant (p < 0.001, Fig. 4E). These effects of decorin were protracted over time as shown by quantitative immunofluorescence and three-dimensional surface plot quantification (supplemental Fig. S2).
The down-regulation of Met, β-catenin, and Myc mediated by decorin took place sequentially. The decay of Met expression level started very early with a t½ of ∼6 min (61). In contrast, down-regulation of β-catenin and Myc occurred at later time points with t½ of ∼38 and ∼68 min, respectively (supplemental Fig. S3). To further establish the role of decorin in the proteasomal degradation of Myc, we utilized an additional proteasome inhibitor, MG132, which is a specific and cell permeable inhibitor of ubiquitinated proteins (Ki = 4 nm) (81). We discovered that MG132 also blocked decorin-evoked Myc degradation. As an internal control for decorin activity we detected a marked accumulation of p21 protein in the nuclei of HeLa cells exposed to decorin for 24 h as well as a time-dependent increase in total p21 protein (Fig. 4G), thereby validating the antiproliferative effects of decorin protein core as we described before (77).
We note, however, that upon treatment with only siRNA targeting Met, no appreciable decrease in Myc was observed in three independent experiments (data not shown). This suggests that decorin might be also utilizing Met-independent pathways in suppressing Myc levels in HeLa cells. Collectively, these results report for the first time a link between a secreted proteoglycan and one of the key oncogenic signaling proteins such as the multifunctional Myc transcription factor.
Decorin Induces Phosphorylation of Myc at Threonine 58 and Its Accumulation in the Nuclei of HeLa Cells
It is well established that Myc can be regulated at the post-translational level by phosphorylation at highly conserved amino acid residues such as threonine 58 (Thr-58) and serine 62 (Ser-62) in the N-terminal transcriptional activation domain of Myc (82). Ser-62 is a target of the ERK pathway whereas Thr-58 is a target of glycogen synthase kinase-3β (83), and both sites are regulated by the Ras/Raf/Mek/Erk pathway (82, 84–86). Phosphorylation at Ser-62 increases Myc stability, whereas phosphorylation at Thr-58 stimulates ubiquitination and 26 S proteasomal degradation (87, 88). Thus, we hypothesized that decorin might evoke destabilization of Myc by inducing Thr-58 phosphorylation. The results showed a time-dependent and highly significant induction of P-T58 Myc and a concurrent down-regulation of Myc levels (Fig. 5A). Quantification of two independent experiments showed a progressive increase of P-T58 Myc evoked by decorin which reached a 7-fold increase at 24 h (Fig. 5B). Immunofluorescence microscopy using the P-T58 Myc-specific antibody showed a progressive accumulation of P-T58 Myc in the nuclei of HeLa cells (Fig. 5, C–E). Thus, decorin causes destabilization of Myc via induction of P-T58, potentially by inducing the tumor suppressor protein phosphatase 2A (89, 90), which dephosphorylates Ser-62 resulting in the unstable, singly Thr-58-phosphorylated form of Myc (91), a substrate for ubiquitination and degradation by the 26 S proteasome (82).
FIGURE 5.
Decorin induces phosphorylation of Myc at threonine 58 and its accumulation into the nuclei of HeLa cells. A, immunoblotting of HeLa cells treated for the indicated time intervals with decorin (500 nm). The blots were probed with an antibody specific for phosphothreonine 58 (P-T58) Myc, and re-probed with an anti-Myc antibody. The levels of β-actin serve as loading control. We note that to better visualize P-T58 Myc, especially at later time points, the experiments were performed in full-serum in contrast to all other experiments presented in the previous figures. Serum increases the amount of total Myc as shown by detection of Myc even after 24 h of decorin treatment. B, quantification of T58 Myc levels by scanning densitometry of two independent experiments. C–E, immunofluorescence detection of P-T58 Myc. The images were merged using differential contrast microscopy (DIC) to better visualize the cell architecture. Note the progressive accumulation of P-T58 Myc in the nuclei of HeLa cells. The white dots represent non immunoreactive endocytic vesicles and/or other cytoplasmic organelles visualized by DIC. Bar, 20 μm.
Decorin Inhibits the Transcriptional Activity of Pro-proliferative Genes
Engagement of Met by decorin induces internalization and subsequent changes in downstream signaling cascades as mediated via this receptor (61, 92). As shown above, β-catenin is a key effector of Met and proteasome-mediated degradation of this key signaling molecule is enhanced upon decorin treatment. β-Catenin is a multifunctional protein that can translocate to the nucleus thereby functioning as a transcriptional cofactor as it binds T-cell factor/lymphocyte enhancing factor (TCF/LEF) and drives transcription of pro-proliferative genes (93). Therefore, to better understand the potential role for decorin in regulating transcriptional programs through enhanced β-catenin degradation, qRT-PCR was employed to evaluate the levels of various genes known to be regulated by β-catenin. Relative mRNA levels normalized to the endogenous housekeeping gene β-actin demonstrated a significant decrease in β-catenin expression in the presence of 500 nm decorin protein core (p < 0.001 Fig. 6), implicating decorin as a negative regulator of β-catenin signaling in a noncanonical Wnt fashion. Further, reports have identified β-catenin TCF/LEF binding sites within both the promoter and downstream enhancer element for the MYC proto-oncogene (94).
FIGURE 6.

Decorin negatively regulates the transcription of critical genes. Quantitative real-time PCR showing transcriptional inhibition of key target genes represented as fold changes normalized to the housekeeping gene ACTB (β-actin). The data were obtained from HeLa cells treated with 500 nm decorin for 24 h prior to lysis and represent the average fold changes as calculated by the double ΔCt method ± S.E. (n = 12, ***, p < 0.001; NS, not significant). For additional information refer to the text.
As a possible explanation for the established role of decorin inducing cell cycle arrest through the induction of the cyclin-dependent kinase (CDK) inhibitor p21 (77), the expression of a MYC target gene, AP4, was evaluated. AP4 encodes a potent transcriptional repressor of p21 (79). We discovered a modest, but still highly significant reduction in AP4 mRNA. Subsequently, CDKN1A (p21) expression was evaluated to determine if there was a corresponding increase in mRNA levels attributable to the decrease in expression of AP4. There was, however, no significant change in CDKN1A levels (Fig. 6), suggesting p21 induction is achieved primarily via increased protein stability.
Consistent with the aforementioned role of decorin in arresting the cell cycle, an additional β-catenin target gene, Cyclin D1 was found to be significantly down-regulated. Cyclins are critical for CDK activation that ultimately drives cell cycle progression, with cyclin D1 being necessary for the early G1/S transition. Whereas the levels of other cyclin molecules (such as cyclin E, A, or B) fluctuate throughout the cell cycle, cyclin D is synthesized as long as growth factor stimulation is present (95). Therefore, it is interesting to note that the reduction in cyclin D1 levels upon decorin treatment indicates an attenuation of growth factor signaling via a reduction in β-catenin as controlled by the Met receptor. Finally the expression of CDK6, a binding partner of cyclin D1 and CDK4 to form the ternary complex responsible for partially inactivating the tumor suppressor gene pRb (95), was found to also be significantly down-regulated.
These data indicate for the first time a critical role for decorin in inhibiting downstream mitogenic responses through suppressing β-catenin transcriptional activity in a noncanonical Wnt pathway. Additionally, these data suggest the ability of decorin, an extracellular matrix molecule, to impact vast transcriptional networks, such as those mediated via MYC.
Systemic Delivery of Decorin Inhibits Met and β-Catenin Expression in Two Orthotopic Tumor Xenografts
To determine whether the effects of decorin on Met and β-catenin could also occur in vivo, we established tumor xenografts utilizing two mouse models of orthotopic squamous cell carcinoma (human A431 cells) and orthotopic mammary carcinoma (rat MTLn3 cells). In both cases, systemic decorin protein core treatment (5 mg/kg with injections every other day) caused a significant reduction of tumor growth after ∼3 weeks of treatment (Fig. 7, A and C, p < 0.01). Both treated tumors displayed a significant down-regulation of Met expression compared with controls treated with vehicle as detected by immunoblot (Fig. 7, B and D) and immunofluorescence with 3D surface plot imaging (Fig. 7, E–J). The effects of systemic delivery of recombinant human decorin on the tumor Met levels are also supported by the observation that Met levels were elevated in the decorin-null liver tissues (supplemental Fig. S4), further reinforcing an antagonism between decorin and Met.
FIGURE 7.
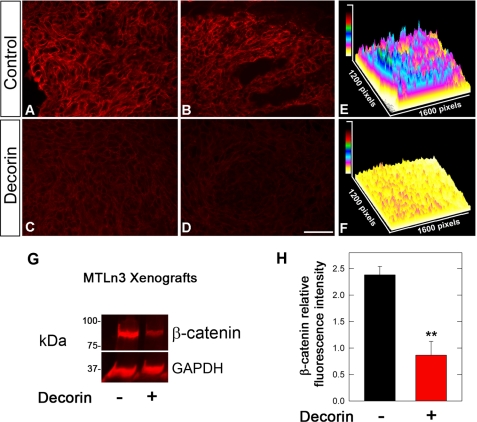
Systemic delivery of decorin protein core inhibits Met expression in two tumor xenografts. A, bar graph showing inhibition of A431 squamous tumor growth in SCID mice at day 23 of decorin treatment (5 mg/kg). Data represent the mean ± S.E. (n = 8, **, p < 0.01). B, representative immunoblot of A431 tumor lysates showing down-regulation of Met expression following decorin treatment. Tubulin was used as loading control. C and D, same as in A and B using MTLn3 mammary carcinoma xenografts. E-I, detection of Met (green) using immunofluorescence of frozen sections from A431 tumor xenografts of vehicle (control) or decorin-treated mice. All the micrographs were taken using the same exposure and gain. Mice bearing A431 tumor xenografts were treated with intraperitoneal injections of decorin (5 mg/kg) every other day for 26 days. Bar, 250 μm. G and J, three-dimensional surface plots generated with ImageJ software depicting levels of Met expression corresponding to the staining above. The scale bars for signal intensity are included in the top left corner.
To further prove an in vivo link between decorin and Met/β-catenin we determined the expression of β-catenin in the A431 and MTLn3 tumor xenografts with or without decorin treatment. We discovered that in A431 cells the levels of β-catenin were even more suppressed than those of Met (Fig. 8, A–F). Similar results were obtained with MTLn3 tumor xenografts (Fig. 8, G and H). These data correlate well with a recent study utilizing decorin-null animals, which has shown that lack of decorin is associated with increased β-catenin levels in vivo, in spontaneous intestinal tumors (44). Thus, these in vivo data further strengthen our rationale and provide a proof of principle that decorin affects this important pathway.
FIGURE 8.
Decorin treatment leads to down-regulation of β-catenin in A431 and MTLn3 tumor xenografts. A–D, immunofluorescence images of two control and two decorin-treated A431 tumor xenografts, reacted with anti-β-catenin antibodies. A431 tumors were treated with decorin every other day for 23 days (5 mg/kg) as in Fig. 7. All the micrographs were taken using the same exposure and gain. Bar, 250 μm. E and F, three-dimensional surface plot analyses of immunofluorescence images obtained by staining for β-catenin in control and decorin-treated tumors, respectively. The scale bars for signal intensity are included in the top left corners. G, immunoblot for β-catenin and GAPDH of tumor lysates from MTLn3 mammary carcinoma xenografts grown in SCID mice. The tumors were treated on alternate days after the tumors became palpable with either vehicle (control) or 5 mg/kg human recombinant decorin protein core until the end point as in Fig. 7. The proteins were visualized with near-infrared labeled secondary antibody and measured with the Odyssey software (Li-COR). H, quantification of immunoblots of β-catenin expression normalized to GAPDH, n = 3. Values represent mean ± S.E. (**, p < 0.01).
Near Infrared-labeled Decorin Protein Core Targets Human Tumor Xenografts
To determine whether decorin specifically targeted tumor xenografts, we generated decorin protein core visible in the infrared spectrum by covalently attaching IRDye® 800CW to the primary amines of the protein core. SCID mice bearing HeLa xenografts and not previously treated with decorin were injected with the IR800-tagged decorin and the distribution of the proteins in the animals following injection was visualized using the PearlTM infrared Imager and software (Li-COR). IR800-labeled decorin targeted the tumor xenograft (Fig. 9B) and this uptake lasted for up to 68 h (Fig. 9, C and D). Initially, decorin was taken up by the liver (Fig. 9E) and then by the kidneys (Fig. 9F). Scanning of isolated organs after 68-h post-injection showed very little or no uptake by brain, heart, lungs, and spleen whereas liver and kidneys were still labeled (Fig. 9G). At later time points (72 h), the hepatic uptake of decorin decreased while the kidneys still contained high levels of decorin (not shown). This suggests that decorin is taken up primarily by the liver, at least partially via the Met receptor, which is highly expressed in this organ, and then slowly cleared via the urinary tract. Notably, IR800-decorin was taken up by the tumor xenografts and even after 68-h post-injection there was prominent labeling of the HeLa xenografts (Fig. 9H).
FIGURE 9.
IR800-labeled decorin protein core targets specifically the human tumor xenografts. A, preinjection scan (white channel), B–D, lateral side scans using pseudocolor intensity map of the same tumor-bearing mouse at various times post-injection, as indicated. The tumor is circled in white. About 100 μg of IR800-labeled decorin protein core was injected via the tail vein. E, ventral view showing liver at 68 h post-injection. F, dorsal view showing kidneys and tumor as indicated at 22 h post-injection. G, isolated organs after 68 h. Bar, 10 mm. H, an example of an isolated tumor xenograft after 68 h of IR800-decorin injection. I, detection of IR800-labeled decorin in kidney and tumor xenograft lysates. Lane 1, Coomassie Blue-prestained protein standards (note that using the infrared detection system the Coomassie Blue-prestained proteins appear as red); lanes 2–6, increasing concentrations of purified IR800-decorin protein core (5, 10, 20, 30, and 40 ng, respectively); lane 7, kidney lysate at 68 h (calculated to correspond to ∼1.4 ng/mg wet weight); lane 8, tumor lysate at 68 h (calculated to correspond to ∼0.64 ng/mg wet weight). SCID mice were subcutaneously injected with 2 × 106 HeLa cells into the upper left dorsal areas, and tumor xenografts were allowed to establish for ∼3 weeks.
Next, we extracted various tissues and the treated tumor xenografts and analyzed them by SDS-PAGE. Notably, we could identify intact IR800-labeled decorin protein core in the kidney and tumor lysates (Fig. 9I, lanes 7 and 8, respectively). Based on a standard curve with known concentrations of IR800-decorin (Fig. 9I, lanes 2–6) we estimated that after 68 h of injection with 100 μg of labeled decorin core there was ∼1.4 ng and ∼0.64 ng/mg wet weight in the kidney and tumor cell lysates, respectively. Currently there is no microscope available that can detect infrared-tagged proteins within a tissue section at the cellular and subcellular level. However, in a previous study from our laboratory we have shown that decorin targets the cell surface of carcinoma cells (52).
In another set of experiments, we used IR800-labeled EGF and compared its distribution to that of IR800-decorin. We note that labeling HGF is cumbersome insofar the commercially available HGF is not carrier free and needs to be proteolytically processed to be active. Immediately post-injection both decorin and EGF showed a wide distribution, with initial (4–24 h) labeling in the liver and subsequent (48–68 h) clearance from the liver and accumulation in the kidneys (supplemental Fig. S5). Notably, IR800-decorin was taken up by the tumor xenografts and even 68-h post-injection there was prominent labeling of the HeLa xenografts at a level even higher than EGF as better visualized by the heat map of isolated organs (supplemental Fig. S5). Essentially, decorin was found to be long-lived (up to 3 days post-injection) in areas within the tumor xenografts and showed slower clearance than EGF.
Finally, we determined whether systemic delivery of decorin could down-regulate the in vivo expression of β-catenin and Myc in HeLa tumor xenografts. We found that, in agreement with the data presented above, HeLa xenografts responded to systemic delivery of decorin (supplemental Fig. S6, A and B). Moreover, both β-catenin and Myc were down-regulated in the HeLa tumor xenografts systemically treated with recombinant decorin protein core vis-à-vis the vehicle-treated animals (supplemental Fig. S6C). Similar inhibition was also detected at the mRNA level (supplemental Fig. S6D). Collectively, these studies show that decorin targets specifically the tumor xenografts where it resides for protracted periods of time as an intact protein, and indicate that decorin down-regulates key transcription factors controlling tumor cell survival and growth.
CONCLUSIONS
The multifaceted ability of decorin to retard in vivo tumor growth and metastatic spreading has a mechanistic explanation in the ability of decorin to down-regulate multiple signaling pathways (96). Our results indicate that decorin is a pan-RTK inhibitor insofar as it directly interacts with and down-regulates the Met receptor as well as the EGFR. Because decorin is highly expressed in the tumor stroma (97, 98), it is possible for secreted matrix proteases to liberate physiological concentrations of decorin during the course of tumorigenesis. Indeed, both the EGFR and Met activate the Ras-MAPK and PI3 kinase-Akt/PKB pathways thereby controlling cell growth, motility, and survival. The unique activity of decorin as a Met antagonist is manifested by a rapid induction of both Met receptor shedding and internalization with consequent downstream degradation of β-catenin, which is required for cell survival. The novel part of this report is the discovery that Myc, a key oncogene involved in the pathogenesis of multiple types of cancers, is also markedly down-regulated by decorin treatment both at the transcriptional and post-translational levels. Decorin-evoked phosphorylation of Myc at threonine 58 could explain its rapid and protracted down-regulation because phosphorylation at this site targets Myc for proteasomal degradation. Further transcriptional down-regulation of β-catenin and Myc target genes involved in the control of cell division and proliferation could contribute to the cytostatic effects of decorin.
It is well established that coactivation of RTKs affects the response of tumor cells to targeted therapies (99) and amplification of the Met-encoding gene promotes drug resistance in ErbB-driven cancers (100). Whereas in the past, main efforts were aimed at developing highly specific inhibitors acting on single RTKs, now there is general consensus that molecules interfering simultaneously with multiple RTKs might be more effective than single target agents (101, 102). In this perspective, the activity of decorin, and perhaps of other molecules sharing leucine-rich repeats (56), might represent a novel therapeutic modality against metastatic cancer.
Supplementary Material
Acknowledgments
We thank D. Wasserman for invaluable help with the Li-Cor imaging and for providing the IR800-labeled EGF, and S. Goldoni and A. Nÿstrom for contributions in the initial stages of this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA39481, R01 CA47282, and R01 CA120975 (to R. V. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
- SLRPs
- small leucine-rich proteoglycans
- EGFR
- epidermal growth factor receptor
- HGF
- hepatocyte growth factor
- RTK
- receptor tyrosine kinase
- MDCK
- Madine-Darby canine kidney
- p21
- the cyclin-dependent kinase inhibitor p21WAF1/Cip1
- SCID
- severe combined immunodeficiency
- qRT-PCR
- quantitative real-time polymerase chain reaction.
REFERENCES
- 1.Ramirez F., Rifkin D. B. (2003) Matrix Biol. 22, 101–107 [DOI] [PubMed] [Google Scholar]
- 2.Bateman J. F., Boot-Handford R. P., Lamandé S. R. (2009) Nat. Rev. Genet. 10, 173–183 [DOI] [PubMed] [Google Scholar]
- 3.Iozzo R. V., Murdoch A. D. (1996) FASEB J. 10, 598–614 [PubMed] [Google Scholar]
- 4.Ameye L., Young M. F. (2002) Glycobiology 12, 107R–116R [DOI] [PubMed] [Google Scholar]
- 5.Iozzo R. V. (1999) J. Biol. Chem. 274, 18843–18846 [DOI] [PubMed] [Google Scholar]
- 6.Schaefer L., Iozzo R. V. (2008) J. Biol. Chem. 283, 21035–21039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer L., Schaefer R. M. (2010) Cell Tissue Res. 339, 237–246 [DOI] [PubMed] [Google Scholar]
- 8.Kalamajski S., Oldberg A. (2010) Matrix Biol. 29, 248–253 [DOI] [PubMed] [Google Scholar]
- 9.Santra M., Skorski T., Calabretta B., Lattime E. C., Iozzo R. V. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7016–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danielson K. G., Baribault H., Holmes D. F., Graham H., Kadler K. E., Iozzo R. V. (1997) J. Cell Biol. 136, 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed C. C., Iozzo R. V. (2002) Glycoconj. J. 19, 249–255 [DOI] [PubMed] [Google Scholar]
- 12.Rühland C., Schönherr E., Robenek H., Hansen U., Iozzo R. V., Bruckner P., Seidler D. G. (2007) FEBS J. 274, 4246–4255 [DOI] [PubMed] [Google Scholar]
- 13.Schmidt G., Robenek H., Harrach B., Glössl J., Nolte V., Hörmann H., Richter H., Kresse H. (1987) J. Cell Biol. 104, 1683–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumdieck R., Höök M., Rosenberg L. C., Volanakis J. E. (1992) J. Immunol. 149, 3695–3701 [PubMed] [Google Scholar]
- 15.Winnemöller M., Schön P., Vischer P., Kresse H. (1992) Eur.J. Cell Biol. 59, 47–55 [PubMed] [Google Scholar]
- 16.Hildebrand A., Romaris M., Rasmussen L. M., Heinegård D., Twardzik D. R., Border W. A., Ruoslahti E. (1994) Biochem.J. 302, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santra M., Mann D. M., Mercer E. W., Skorski T., Calabretta B., Iozzo R. V. (1997) J. Clin. Invest. 100, 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iozzo R. V. (1997) Crit. Rev. Biochem. Mol. Biol. 32, 141–174 [DOI] [PubMed] [Google Scholar]
- 19.Ferdous Z., Wei V. M., Iozzo R., Höök M., Grande-Allen K. J. (2007) J. Biol. Chem. 282, 35887–35898 [DOI] [PubMed] [Google Scholar]
- 20.Iozzo R. V., Moscatello D. K., McQuillan D. J., Eichstetter I. (1999) J. Biol. Chem. 274, 4489–4492 [DOI] [PubMed] [Google Scholar]
- 21.Santra M., Reed C. C., Iozzo R. V. (2002) J. Biol. Chem. 277, 35671–35681 [DOI] [PubMed] [Google Scholar]
- 22.Ferdous Z., Peterson S. B., Tseng H., Anderson D. K., Iozzo R. V., Grande-Allen K. J. (2010) J. Biomed. Mater. Res. 93, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer L., Macakova K., Raslik I., Micegova M., Gröne H.-J., Schönherr E., Robenek H., Echtermeyer F. G., Grässel S., Bruckner P., Schaefer R. M., Iozzo R. V., Kresse H. (2002) Am. J. Pathol. 160, 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer L., Mihalik D., Babelova A., Krzyzankova M., Gröne H. J., Iozzo R. V., Young M. F., Seidler D. G., Lin G., Reinhardt D. P., Schaefer R. M. (2004) Am. J. Pathol. 165, 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams K. J., Qiu G., Usui H. K., Dunn S. R., McCue P., Bottinger E., Iozzo R. V., Sharma K. (2007) Am. J. Pathol. 171, 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant D. S., Yenisey C., Rose R. W., Tootell M., Santra M., Iozzo R. V. (2002) Oncogene 21, 4765–4777 [DOI] [PubMed] [Google Scholar]
- 27.Schönherr E., Sunderkötter C., Schaefer L., Thanos S., Grässel S., Oldberg A., Iozzo R. V., Young M. F., Kresse H. (2004) J. Vasc. Res. 41, 499–508 [DOI] [PubMed] [Google Scholar]
- 28.Järveläinen H., Puolakkainen P., Pakkanen S., Brown E. L., Höök M., Iozzo R. V., Sage H., Wight T. N. (2006) Wound Rep. Reg. 14, 443–452 [DOI] [PubMed] [Google Scholar]
- 29.Weis S. M., Zimmerman S. D., Shah M., Covell J. W., Omens J. H., Ross J., Jr., Dalton N., Jones Y., Reed C. C., Iozzo R. V., McCulloch A. D. (2005) Matrix Biol. 24, 313–324 [DOI] [PubMed] [Google Scholar]
- 30.Fust A., LeBellego F., Iozzo R. V., Roughley P. J., Ludwig M. S. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L159–L166 [DOI] [PubMed] [Google Scholar]
- 31.Robinson P. S., Lin T. W., Jawad A. F., Iozzo R. V., Soslowsky L. J. (2004) Ann. Biomed. Eng. 32, 924–931 [DOI] [PubMed] [Google Scholar]
- 32.Robinson P. S., Huang T. F., Kazam E., Iozzo R. V., Birk D. E., Soslowsky L. J. (2005) J. Biomechanical Eng. 127, 181–185 [DOI] [PubMed] [Google Scholar]
- 33.Brown E. L., Wooten R. M., Johnson B. J. B., Iozzo R. V., Smith A., Dolan M. C., Guo B. P., Weis J. J., Höök M. (2001) J. Clin. Inv. 107, 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang F. T., Brown E. L., Wang T., Iozzo R. V., Fikrig E. (2004) Am. J. Pathol. 165, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corsi A., Xu T., Chen X. D., Boyde A., Liang J., Mankani M., Sommer B., Iozzo R. V., Eichstetter I., Robey P. G., Bianco P., Young M. F. (2002) J. Bone Miner. Res. 17, 1180–1189 [DOI] [PubMed] [Google Scholar]
- 36.Häkkinen L., Strassburger S., Kähäri V. M., Scott P. G., Eichstetter I., Iozzo R. V., Larjava H. (2000) Lab. Invest. 80, 1869–1880 [DOI] [PubMed] [Google Scholar]
- 37.Haruyama N., Sreenath T. L., Suzuki S., Yao X., Wang Z., Wang Y., Honeycutt C., Iozzo R. V., Young M. F., Kulkarni A. B. (2009) Matrix Biol. 28, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldberg M., Septier D., Rapoport O., Iozzo R. V., Young M. F., Ameye L. G. (2005) Calcif. Tissue. Int. 77, 297–310 [DOI] [PubMed] [Google Scholar]
- 39.Zhang G., Ezura Y., Chervoneva I., Robinson P. S., Beason D. P., Carine E. T., Soslowsky L. J., Iozzo R. V., Birk D. E. (2006) J. Cell. Biochem. 98, 1436–1449 [DOI] [PubMed] [Google Scholar]
- 40.Zhang G., Chen S., Goldoni S., Calder B. W., Simpson H. C., Owens R. T., McQuillan D. J., Young M. F., Iozzo R. V., Birk D. E. (2009) J. Biol. Chem. 284, 8888–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanches J. C., Jones C. J., Aplin J. D., Iozzo R. V., Zorn T. M., Oliveira S. F. (2010) J. Anat. 216, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bi Y., Stueltens C. H., Kilts T., Wadhwa S., Iozzo R. V., Robey P. G., Chen X. D., Young M. F. (2005) J. Biol. Chem. 280, 30481–30489 [DOI] [PubMed] [Google Scholar]
- 43.Iozzo R. V., Chakrani F., Perrotti D., McQuillan D. J., Skorski T., Calabretta B., Eichstetter I. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3092–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi X., Tong C., Dockendorff A., Bancroft L., Gallagher L., Guzman G., Iozzo R. V., Augenlicht L. H., Yang W. (2008) Carcinogenesis 29, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moscatello D. K., Santra M., Mann D. M., McQuillan D. J., Wong A. J., Iozzo R. V. (1998) J. Clin. Investig. 101, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel S., Santra M., McQuillan D. J., Iozzo R. V., Thomas A. P. (1998) J. Biol. Chem. 273, 3121–3124 [DOI] [PubMed] [Google Scholar]
- 47.Csordás G., Santra M., Reed C. C., Eichstetter I., McQuillan D. J., Gross D., Nugent M. A., Hajnóczky G., Iozzo R. V. (2000) J. Biol. Chem. 275, 32879–32887 [DOI] [PubMed] [Google Scholar]
- 48.Santra M., Eichstetter I., Iozzo R. V. (2000) J. Biol. Chem. 275, 35153–35161 [DOI] [PubMed] [Google Scholar]
- 49.Reed C. C., Gauldie J., Iozzo R. V. (2002) Oncogene 21, 3688–3695 [DOI] [PubMed] [Google Scholar]
- 50.Tralhão J. G., Schaefer L., Micegova M., Evaristo C., Schönherr E., Kayal S., Veiga-Fernandes H., Danel C., Iozzo R. V., Kresse H., Lemarchand P. (2003) FASEB J. 17, 464–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed C. C., Waterhouse A., Kirby S., Kay P., Owens R. T., McQuillan D. J., Iozzo R. V. (2005) Oncogene 24, 1104–1110 [DOI] [PubMed] [Google Scholar]
- 52.Seidler D. G., Goldoni S., Agnew C., Cardi C., Thakur M. L., Owens R. T., McQuillan D. J., Iozzo R. V. (2006) J. Biol. Chem. 281, 26408–26418 [DOI] [PubMed] [Google Scholar]
- 53.Zafiropoulos A., Nikitovic D., Katonis P., Tsatsakis A., Karamanos N. K., Tzanakakis G. N. (2008) Mol. Cancer Res. 6, 785–794 [DOI] [PubMed] [Google Scholar]
- 54.Goldoni S., Seidler D. G., Heath J., Fassan M., Baffa R., Thakur M. L., Owens R. T., McQuillan D. J., Iozzo R. V. (2008) Am. J. Pathol. 173, 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Pennisi A., Yaccoby S. (2008) Blood 112, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldoni S., Iozzo R. V. (2008) Int. J. Cancer 123, 2473–2479 [DOI] [PubMed] [Google Scholar]
- 57.Hu Y., Sun H., Owens R. T., Wu J., Chen Y. Q., Berquin I. M., Perry D., O'Flaherty J. T., Edwards I. J. (2009) Neoplasia 11, 1042–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schönherr E., Sunderkötter C., Iozzo R. V., Schaefer L. (2005) J. Biol. Chem. 280, 15767–15772 [DOI] [PubMed] [Google Scholar]
- 59.Schaefer L., Tsalastra W., Babelova A., Baliova M., Minnerup J., Sorokin L., Gröne H. J., Reinhardt D. P., Pfeilschifter J., Iozzo R. V., Schaefer R. M. (2007) Am. J. Pathol. 170, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merline R., Lazaroski S., Babelova A., Tsalastra-Greul W., Pfeilschifter J., Schluter K. D., Gunther A., Iozzo R. V., Schaefer R. M., Schaefer L. (2009) J. Physiol. Pharmacol. 60, Suppl. 4, 5–13 [PMC free article] [PubMed] [Google Scholar]
- 61.Goldoni S., Humphries A., Nyström A., Sattar S., Owens R. T., McQuillan D. J., Ireton K., Iozzo R. V. (2009) J. Cell Biol. 185, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. (2003) Nat. Rev. Mol. Cell. Biol. 4, 915–925 [DOI] [PubMed] [Google Scholar]
- 63.Lai A. Z., Abella J. V., Park M. (2009) Trends Cell Biol. 19, 542–551 [DOI] [PubMed] [Google Scholar]
- 64.Zhu J. X., Goldoni S., Bix G., Owens R. T., McQuillan D. J., Reed C. C., Iozzo R. V. (2005) J. Biol. Chem. 280, 32468–32479 [DOI] [PubMed] [Google Scholar]
- 65.Zoeller J. J., McQuillan A., Whitelock J., Ho S. Y., Iozzo R. V. (2008) J. Cell Biol. 181, 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoeller J. J., Pimtong W., Corby H., Goldoni S., Iozzo A. E., Owens R. T., Ho S. Y., Iozzo R. V. (2009) J. Biol. Chem. 284, 11728–11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sims K., Jr., Eble D. M., Iovine M. K. (2009) Dev. Biol. 327, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. (2008) Dev. Cell 15, 209–219 [DOI] [PubMed] [Google Scholar]
- 70.Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. (2003) Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 71.Danilkovitch-Miagkova A., Miagkov A., Skeel A., Nakaigawa N., Zbar B., Leonard E. J. (2001) Mol. Cell. Biol. 21, 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasola A., Fassetta M., De Bacco F., D'Alessandro L., Gramaglia D., Di Renzo M. F., Comoglio P. M. (2007) Oncogene 26, 1078–1087 [DOI] [PubMed] [Google Scholar]
- 73.Adhikary S., Eilers M. (2005) Nat. Rev. Mol. Cell. Biol. 6, 635–645 [DOI] [PubMed] [Google Scholar]
- 74.van der Flier L. G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., van Gijn M. E., Suijkerbuijk S., van de Wetering M., Marra G., Clevers H. (2007) Gastroenterology 132, 628–632 [DOI] [PubMed] [Google Scholar]
- 75.Muncan V., Sansom O. J., Tertoolen L., Phesse T. J., Begthel H., Sancho E., Cole A. M., Gregorieff A., de Alboran I. M., Clevers H., Clarke A. R. (2006) Mol. Cell. Biol. 26, 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., Evan G. I. (2008) Nature 455, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Luca A., Santra M., Baldi A., Giordano A., Iozzo R. V. (1996) J. Biol. Chem. 271, 18961–18965 [DOI] [PubMed] [Google Scholar]
- 78.Seoane J., Le H. V., Massagué J. (2002) Nature 419, 729–734 [DOI] [PubMed] [Google Scholar]
- 79.Jung P., Menssen A., Mayr D., Hermeking H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15046–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamei J., Toyofuku T., Hori M. (2003) Biochem. Biophys. Res. Commun. 312, 380–387 [DOI] [PubMed] [Google Scholar]
- 81.Foveau B., Ancot F., Leroy C., Petrelli A., Reiss K., Vingtdeux V., Giordano S., Fafeur V., Tulasne D. (2009) Mol. Biol. Cell 20, 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Albihn A., Johnsen J. I., Henriksson M. A. (2010) Adv. Cancer Res. 107, 163–224 [DOI] [PubMed] [Google Scholar]
- 83.Gregory M. A., Qi Y., Hann S. R. (2003) J. Biol. Chem. 278, 51606–51612 [DOI] [PubMed] [Google Scholar]
- 84.Lutterbach B., Hann S. R. (1994) Mol. Cell. Biol. 14, 5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J. R. (2000) Genes Dev. 14, 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sears R. C. (2004) Cell Cycle 3, 1133–1137 [PubMed] [Google Scholar]
- 87.Malempati S., Tibbits D., Cunningham M., Akkari Y., Olson S., Fan G., Sears R. C. (2006) Leukemia 20, 1572–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salghetti S. E., Kim S. Y., Tansey W. P. (1999) EMBO J. 18, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold H. K., Sears R. C. (2008) Cancer Metastasis Rev. 27, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perrotti D., Neviani P. (2008) Cancer Metastasis Rev. 27, 159–168 [DOI] [PubMed] [Google Scholar]
- 91.Arnold H. K., Sears R. C. (2006) Mol. Cell. Biol. 26, 2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iozzo R. V., Schaefer L. (2010) FEBS J. 277, 3864–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 94.Dang C. V., O'Donnell K. A., Zeller K. I., Nguyen T., Osthus R. C., Li F. (2006) Semin. Cancer Biol. 16, 253–264 [DOI] [PubMed] [Google Scholar]
- 95.Malumbres M., Barbacid M. (2009) Nat. Rev.Cancer 9, 153–166 [DOI] [PubMed] [Google Scholar]
- 96.Iozzo R. V. (1998) Annu. Rev. Biochem. 67, 609–652 [DOI] [PubMed] [Google Scholar]
- 97.Adany R., Heimer R., Caterson B., Sorrell J. M., Iozzo R. V. (1990) J. Biol. Chem. 265, 11389–11396 [PubMed] [Google Scholar]
- 98.Adany R., Iozzo R. V. (1991) Biochem. J. 276, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stommel J. M., Kimmelman A. C., Ying H., Nabioullin R., Ponugoti A. H., Wiedemeyer R., Stegh A. H., Bradner J. E., Ligon K. L., Brennan C., Chin L., DePinho R. A. (2007) Science 318, 287–290 [DOI] [PubMed] [Google Scholar]
- 100.Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., Jänne P. A. (2007) Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 101.Comoglio P. M., Giordano S., Trusolino L. (2008) Nature Rev. Drug Discov. 7, 504–516 [DOI] [PubMed] [Google Scholar]
- 102.Migliore C., Giordano S. (2008) Eur. J. Cancer 44, 641–651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.