Abstract
Gaucher disease is caused by the defective activity of the lysosomal hydrolase, glucosylceramidase. Although the x-ray structure of wild type glucosylceramidase has been resolved, little is known about the structural features of any of the >200 mutations. Various treatments for Gaucher disease are available, including enzyme replacement and chaperone therapies. The latter involves binding of competitive inhibitors at the active site to enable correct folding and transport of the mutant enzyme to the lysosome. We now use molecular dynamics, a set of structural analysis tools, and several statistical methods to determine the flexible behavior of the N370S Gaucher mutant at various pH values, with and without binding the chaperone, N-butyl-deoxynojirimycin. We focus on the effect of the chaperone on the whole protein, on the active site, and on three important structural loops, and we demonstrate how the chaperone modifies the behavior of N370S in such a way that it becomes more active at lysosomal pH. Our results suggest a mechanism whereby the binding of N-butyl-deoxynojirimycin helps target correctly folded glucosylceramidase to the lysosome, contributes to binding with saposin C, and explains the initiation of the substrate-enzyme complex. Such analysis provides a new framework for determination of the structure of other Gaucher disease mutants and suggests new approaches for rational drug design.
Keywords: Glycerosphingolipid, Lipid, Lysosomal Storage Disease, Molecular Dynamics, Sphingolipid, Gaucher Disease, Glucosylceramide
Introduction
Mutations in the gene encoding for glucosylceramidase (GCase),4 the lysosomal enzyme that hydrolyzes glucosylceramide (GlcCer), cause Gaucher disease (GD), the most common lysosomal storage disorder (1). GD is inherited in an autosomal recessive fashion and occurs with a particularly high frequency within the Ashkenazi Jewish population (2, 3). More than 200 mutations (4) have been reported for the GCase gene, which result in a decrease or complete loss of enzymatic activity, due either to reduced catalytic activity or to a reduced lysosomal GCase concentration. GD has been shown to increase the risk of certain types of malignancies (5–8) and of Parkinson disease (9, 10). Three common clinical subtypes of GD are known, type 1, the most common visceral form in adults, type 2, the acute neuropathic form in children, and type 3, the chronic neuropathic form.
GD is a monogenetic disease, and the current therapy of choice is enzyme replacement therapy (11–16). However, because GCase cannot pass through the blood-brain barrier, the neurological symptoms in types 2 and 3 GD do not respond to enzyme replacement therapy. Therefore, alternative treatments such as substrate reduction therapy have been developed (17). Another novel therapeutic approach involves the use of highly specific small molecules that act as chaperones, which increase the activity and stability of mutant forms of GCase as they pass through the secretory pathway (18–20). However, chaperone therapy is still in its infancy, and the rational design of highly specific, small chemical chaperones is limited by experimental and computational procedures. One chaperone that has been relatively well studied is Miglustat® (N-butyl-deoxynojirimycin (NB-DNJ)). NB-DNJ was initially used in substrate reduction therapy but later proved to be a potent chaperone because it increased the activity of wild type (WT) GCase and of the mutants S364R, N370S, V15M, and M123T (21, 22). The x-ray structure of WT GCase bound to NB-DNJ has been solved (23), and the structures of GCase bound to various other small molecules, some of which could potentially be used as chemical chaperones (21, 24–28), have been reported (21, 29–34).
Although some biochemical and cell biological information is available on the behavior of the N370S mutant (25, 31, 35–40), no structural information is available for this or for any other purified GCase mutant. The lack of such data has severely hampered efforts to understand the molecular basis of altered GCase activity in GD mutations and to rationally design chaperones that might restore their activity. To gain new mechanistic insight into the structure of the N370S mutation, we have undertaken a series of molecular dynamic (MD) simulations using several structural and statistical analysis methods. Using these tools, we are able to explain the reduced activity of the N370S mutant and the role of NB-DNJ in the partial recovery of activity; furthermore, we have explored the structural basis for the shifted pH optimum of N370S (36). Our results are in good agreement with available experimental data.
MATERIALS AND METHODS
MD Simulations
MD simulations were performed on WT, N370S, and four other common GD mutations, F213I, D409H, L444P, and R496H (41). The crystal structure of a complex of GCase with N-butyl-deoxynojirimycin (PDB code 2V3D) was used as a template for simulations of both WT GCase and of GCase mutants. Residues 28–31, which are missing in 2V3D, were modeled from another structure (PDB code 1OGS). To generate the Gaucher mutants, mutated residues were manually introduced into the crystal structure, and side chains were modeled and optimized using scwrl 3.0 (42). For both WT and N370S, 25 MD simulations were performed, with five independent runs for each group as follows: GCase, pH 7.4; NB-DNJ/GCase, pH 7.4; NB-DNJ+/GCase, pH 7.4; NB-DNJ+/GCase, pH 4.5; and GCase, pH 4.5 (Table 1). Three independent runs were performed for the other GD mutants.
TABLE 1.
GCase terminology used in the text
| Abbreviation | Description |
|---|---|
| GCase-pH 7.4 | GCase at pH 7.4 |
| NB-DNJ/GCase-pH 7.4 | GCase at pH 7.4 with unprotonated NB-DNJ bound |
| NB-DNJ+/GCase-pH 7.4 | GCase at pH 7.4 with protonated NB-DNJ bound |
| NB-DNJ+/GCase-pH 4.5 | GCase at pH 4.5 with protonated NB-DNJ bound |
| GCase-pH 4.5 | GCase at pH 4.5 |
| GCase(N370S)-pH 4.5a | GCase N370S mutant at pH 4.5 |
a Other GCase mutants are referred to appropriately in the text.
Different protonation states of titratable residues were chosen to simulate GCase and its mutants at physiological (7.4) and lysosomal (4.5) pH values. The protonation states were determined using a combination of results derived from constant pH MD simulations (43) and results produced by the prediction server H++ (44). The results, together with published data (23, 45, 46), were visually inspected to prevent wrong proton assignments in and near the active site. Because NB-DNJ can exist in either a protonated or an unprotonated form at pH 7.4, both cases were simulated. A list of protonation states for titratable residues is given in supplemental Table 1.
To prepare MD simulations, each protein was fitted into a rectangular box of water molecules; Na+ and Cl− ions were added to obtain a salt concentration of 0.2 m. The position of each protein structure in a box was optimized by Simulaid (47) to yield the smallest possible box volume, with a minimal distance of 15 Å between any protein atom and the edge of the box. Each system was energy-minimized and equilibrated for 50 ps at constant volume and constant pressure, with backbone atoms restrained to their original positions in both cases. After equilibration, each system was simulated for 10 ns at 350 K; this temperature was chosen to enhance the signal of significant observations (the high/low flexibility ratio improved on average by 3.7-fold). Intermediate structures of GCase were derived every 5 ps; these structures were used in subsequent analyses and are referred to as “snapshots.” The total of all snapshots in a MD simulation is referred to as the trajectory.
All MD simulations were performed using AMBER10 (48), with the all-atom force field ff03 (49) for the protein, and the general AMBER force field for NB-DNJ. The antechamber tool (part of the AMBER package) was used to generate topology files and to fit point charges to the electrostatic potential distribution of NB-DNJ. Geometry optimization and the calculation of the electrostatic potential distribution for NB-DNJ were performed using Gaussian03 (50). Structures for all MD simulations were in equilibrium at the beginning of the production trajectory.
Active Site Volume Calculations
To analyze the behavior of the GCase active site, CAVER (51) was used to calculate the cavity volume for each snapshot. The starting position was defined as the average of the Glu235, Glu340, and Cys342 C-β atom coordinates. The cavity volume was calculated from this position toward the active site entrance on the surface of the protein. Because the active site is situated in the middle of a β-barrel fold, it was necessary to determine whether each assigned cavity was open toward the correct side, as defined in the original crystal structure. The curvature and average diameter of the cavity were calculated using CAVER. These three geometry scores (cavity volume, curvature, and diameter) were calculated for the active sites of other GCase crystal structures (2NSX B/D and 3GXF B/D) to serve as reference values. By comparing the active site properties of each MD snapshot to the reference scores, it was possible to determine the total number of similar cavity geometries within each MD run. As some flexibility is observed for protein structures, including their active sites, a snapshot was only accepted if its values showed a maximum deviation of 20% compared with the reference values. Lower tolerance values (10 and 5%) produced lower counts; however, the ratio was preserved (data not shown).
Distance Analysis and Hydrogen Bonding
To monitor small distance perturbations in and around the active site, the shortest distances, averages, and standard deviations were calculated for several residue pairs using POPULUS (52). Based on this, distances for the following residue pairs (35) were measured: 127···179, 127···340, 127···396, 127···397, 235···340, 312···370, 315···370, 315···366, 315···349, 313···344, 313···342, and 313···340. To further investigate these interactions, hydrogen bond (HB) counts were calculated using VMD (53) for the following residue pairs: 127···179, 127···396, 312···366, 312···370, 313···340, 313···342, 315···366, and 315···370.
Root Mean Square Fluctuation (RMSF) Analysis and Statistics
To calculate the average RMSF and standard error for the whole protein or for various regions in the protein, such as loop 1 (residues 341–350), loop 2 (residues 393–396), and loop 3 (residues 312–319) (54), and of the active site residues (Glu235, Glu340, and Cys342) (55), values from all five simulations were used. The RMSF values of all residues or subsets were tested for significant differences between WT and N370S by using the ΔRMSF in each position and the p value of the t-distribution (statistical inference). Five separate simulations were conducted; a total of 25 pairwise comparisons was possible. If the p value was below 0.01 (0.05), the comparison was considered significantly different. The numbers of comparisons with a significant increase or decrease between WT and N370S RMSF values were counted. Each count of significant observations (increase or decrease) was divided by its total count (significant and nonsignificant). Finally, the absolute difference of both scores was chosen as a statistical measure, ranging from 0 (no significance) to 1 (very significant).
Loop Analysis
The affinity (interaction strength) of loops 1 and 3 was measured according to Equation 1,
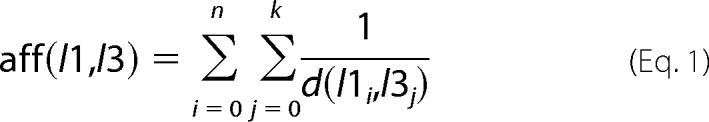 |
where l is the loops and d is the shortest distance between the side chains of the residue pairs i and j. The higher the affinity score, the stronger the spatial interaction between the loops. To compare different MD setups, the affinity scores for all snapshots were sorted in ascending order. These sorted datasets were used to derive a single comparison score between WT and N370S under each condition by calculating the root mean square difference between the paired datasets. The averages and the standard deviations of all affinity scores were calculated for WT and mutants.
The side chain torsion angles φ and ψ were measured for all residues in both loops to assign the secondary structure for each snapshot (56). The total occurrence of all observed secondary structure states was counted for each residue in the loops. These states include H/h = helix; t = turn; E = strand; C = coil. The most important residues could be identified by comparing the change in these counts.
Solvent-accessible Surface Area and Potential Docking Sites with Saposin C
The solvent-accessible surface area was measured using POPS_A (57) and correlated to the RMSF changes between WT and N370S, resulting in a list of surface patches with altered flexibility. These patches were compared with the predicted docking site of GCase and saposin C (residues 314, 317, 318, 348, 358, 362, 365, 366, 369, 370, 372, 373, 441, 443–445, 463, 464, and 487) (39), and the overlapping patches were further investigated.
RESULTS
We now report predicted structural differences between the WT GCase and the N370S mutant, based on MD simulations, and analyze the effect of NB-DNJ binding. Prior to detailed analysis, we examined the orientation of NB-DNJ within the active site and the dissociation of the ligand during MD simulations. Unprotonated NB-DNJ interacts better with N370S than with WT, whereas NB-DNJ+ at pH 7.4 binds equally well to both WT and N370S. The NB-DNJ+/GCase complex is very stable at pH 4.5 (supplemental Table 2).
RMSF Analysis
MD simulations can be used to describe the flexibility of proteins. The resulting RMSF measurements can be calculated for a whole protein, giving insights into its general flexibility, or for regions of the protein, such as loops, or for the active site. Here, we use MD to examine the flexible behavior of WT and mutant GCase under various conditions. WT and N370S were explored in the most detail. For the other GD mutants, fewer simulations were conducted; simulations of R496H, and to some extent of F213I, are of the most relevance, because D409H and L444P do not form stable proteins (32). Nevertheless, the results for the latter are included to underline the unique differences between WT and N370S GCase.
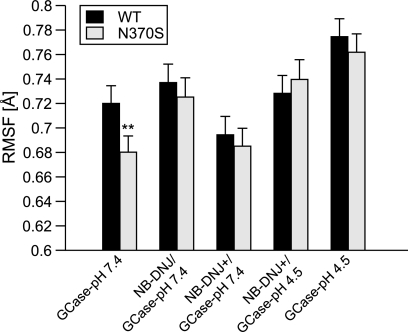
N370S is more stable than WT GCase at pH 7.4 (Fig. 1 and supplemental Table 3), consistent with observations demonstrating that the pH optimum of N370S is shifted from 4.5 to 6.4 (25). This trend is very significant, because when comparing the five WT and five N370S MD results, 21 of the total 25 comparisons have a p value <0.01, resulting in a probability score of 0.88. N370S may be more stable than WT at pH 4.5, although this does not reach statistical significance (Fig. 1) or t-distribution score (supplemental Table 3; probability score of 0.13). None of the other GD mutants display as low a flexibility as GCase(N370S)-pH 7.4 (supplemental Fig. 1). NB-DNJ+ stabilizes both WT and N370S at pH 7.4 and 4.5 (Fig. 1), whereas unprotonated NB-DNJ destabilizes both WT and N370S. NB-DNJ had no consistent effect on the other GD mutants (supplemental Fig. 1).
FIGURE 1.
RMSF values for WT GCase and N370S. Values are means ± S.E. n = 5. Asterisks indicate statistical significance based on the standard error and t test results (p < 0.01), which are given in supplemental Table 3.
MD results (GCase RMSF with and without bound NB-DNJ) were compared with experimentally derived measurements of the melting temperature of GCase at several pH values with and without the chemical chaperone isofagomine (IFG) (31, 58). Experimental (58) and computational results are consistent with each other and suggest that chaperones stabilize WT and N370S GCase at both pH 7.4 and 4.5 (see supplemental Fig. 2). However, when comparing the flexibility changes (RMSF) with the experimentally derived melting temperatures, the trends are only in partial agreement for GCase at pH 7.4. This is likely due to the difference in chemical structure between the two chaperones, viz. IFG and NB-DNJ. Nevertheless, the simulations suggest that binding of NB-DNJ+ to GCase at pH 7.4 results in stabilization, in agreement with the experimental melting temperature data.
Comparative RMSF analysis of the three loops and the active site residues for WT and N370S revealed several trends (supplemental Fig. 3). Loop 1 (supplemental Table 3), including Cys342, and loop 3 are less flexible in GCase(N370S)-pH 7.4 (probability score of 0.69 and 0.67, respectively). Loop 2, Glu235, and Glu340 show no significant changes between WT and N370S at pH 7.4 and pH 4.5. When NB-DNJ+ is bound, loops 2 and 3 and Glu235 are stabilized at pH 7.4 and 4.5 for WT and N370S (supplemental Fig. 3).
In summary, RMSF analysis demonstrates that N370S is more stable than WT at pH 7.4, particularly in loops 1 and 3, including Cys342, a residue important for catalytic activity (32). The stability of loop 1 may contribute to the shifted pH optimum observed for N370S. The results also suggest that NB-DNJ destabilizes GCase at pH 7.4 and that NB-DNJ+ forms a stable complex with GCase.
Interaction of GCase with Saposin C
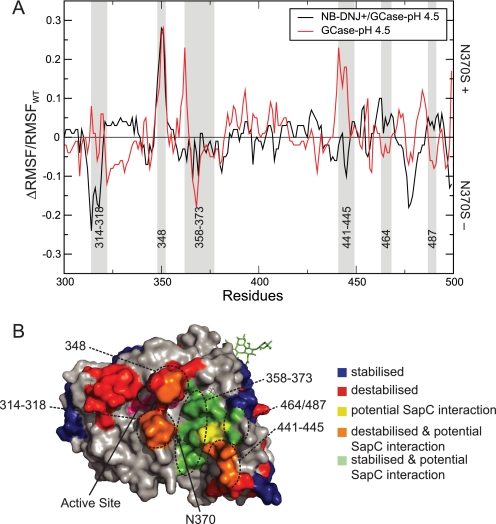
Saposin C (SapC) activates GCase and enhances its binding to anionic phospholipids in the lysosome (59); binding is compromised in N370S (38) and can be restored by IFG (25). A direct interaction between GCase and SapC has been proposed (60), and a potential mode of interaction has been reported (39). As the interaction is based on a computational docking model, the following conclusions are dependent on the validity of this previously published data. However, the predicted changes correlate with the predicted SapC binding interface. MD simulations (supplemental Fig. 4) demonstrate changes in the putative SapC/GCase interaction interface (39) in N370S. More stabilized patches were observed for N370S compared with WT at pH 7.4 (75%), whereas destabilized patches are observed with GCase(N370S)-pH 4.5 (45%). Loop 1 of N370S is stable at pH 7.4 but not at pH 4.5. NB-DNJ+ has a significant effect on the predicted SapC/GCase interaction interface (Fig. 2) and stabilizes residues 314–318, 441–445, and loop 1, but it has little effect on residues 358–373, 464, and 487, which were stable in the absence of NB-DNJ. The stabilization of a protein-protein interface of this scale (ΔRMSF/RMSFWT) can enhance the binding affinity within a protein complex (61); in contrast, a destabilization can decrease the ability to interact. These results imply that NB-DNJ+ can enhance the interaction between GCase and SapC at pH 4.5 by reducing the flexibility of residues in the protein-binding interface.
FIGURE 2.
Effect of NB-DNJ on stability of the GCase/SapC interface. A, ΔRMSF/RMSFWT between WT and N370S at pH 4.5 with (black) and without (red) NB-DNJ+. Predicted interaction sites with SapC are highlighted in gray (39). Positive values indicate increased flexibility for N370S; negative values decreased flexibility. B, GCase surface color-coded for RMSF changes between WT and N370S. Blue patches, decreased flexibility for N370S; red patches, increased flexibility for N370S; orange patches, residues with increased flexibility for N370S that are part of the GCase/SapC interface; green patches, residues with decreased flexibility for N370S that are part of the GCase/SapC interface; yellow patches, residues that are part of the GCase/SapC interface and show no change in flexibility for N370S. The active site is colored pink. Residue numbers for each altered surface patch are given.
Hydrogen Bonding Network and Distance Analysis of the Active Site
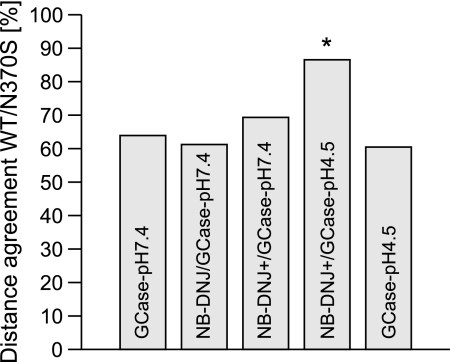
Distances between important residues in and around the active site (supplemental Fig. 5), as well as HB interactions, were determined for WT and for N370S (Table 2). As expected, the altered side chain of N370S results in longer distances to any other interacting amino acid, but differences are also observed for other residue pairs, including the active-site residues Glu235, Glu340, and Cys342. For GCase-pH 7.4 and 4.5, the agreement between all measured distances in WT and N370S is ∼60% (Fig. 3). Binding of unprotonated and protonated NB-DNJ at pH 7.4 does not significantly improve this agreement. At pH 4.5, NB-DNJ+ significantly improves the agreement between the measured distances of WT and N370S to 87%. The most dramatic and statistically significant changes between WT and N370S in HBs were observed for the amino acid pairs 312···370, 313···340, 313···342, and 315···366 (Table 2), which are likely to affect the integrity of the active site, causing the reduced catalytic activity of GCase(N370S)-pH 4.5.
TABLE 2.
HB occupancies at the active site
HB occupancies were calculated using VMD (53). Occupancies of >100 are obtained if multiple HBs are formed between two residues. Values with an asterisk are significant according to nonoverlapping standard deviations.
| Pairs | 7.4 |
7.4 NB-DNJ |
7.4 NB-DNJ+ |
4.5 NB-DNJ+ |
4.5 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | N370S | WT | N370S | WT | N370S | WT | N370S | WT | N370S | |
| 127···179 | 9.3 | 55.5* | 3.1 | 12.2* | 0.1 | 0 | 0 | 0 | 11.6 | 24.3 |
| 127···396 | 42.3* | 21.9 | 44.0* | 15.4 | 43.6 | 42.2 | 44.5 | 47.1 | 39.1 | 23.2 |
| 312···366 | 12.1 | 16.5 | 9.8 | 9.8 | 14.2* | 5.0 | 16.0* | 10.1 | 10.3 | 8.2 |
| 312···370a | 8.8* | 0.1 | 8.9* | 0.0 | 6.1* | 0.0 | 9.9* | 0.7 | 17.9* | 0.0 |
| 315···366a | 0.1* | 0.1 | 0.1 | 0.0 | 0.1 | 0. | 7.9* | 2.7 | 0.3 | 17.7* |
| 313···340a | 100.8 | 114.9 | 97.5 | 113.0* | 126.6* | 115.0 | 115.6 | 114.8 | 77.4 | 119.9* |
| 313···342a | 27.6 | 41.7* | 38.6* | 31.5 | 55.5* | 48.1 | 41.1* | 36.2 | 15.2 | 21.6* |
a HB pairs with the most significant changes for the active site are shown.
FIGURE 3.
Comparison of distances of pairs of important amino acids in and around the active site in WT and N370S. The total distance agreement for pairs between WT and N370S for all simulation settings is given as a percentage score. An asterisk indicates statistical significance.
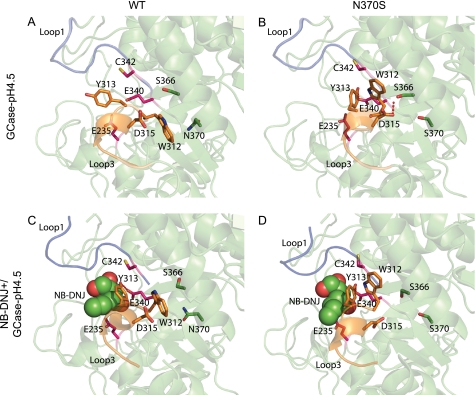
According to MD, Trp312 in WT GCase forms a π/H interaction (62) with Asn370 at pH 4.5 (Fig. 4A). This interaction has not been observed in any GCase crystal structure; rather, an HB was observed in some crystal structures (32, 35, 58, 63) (difference in the frequency of Trp312···Asn370 HB between WT and N370S, p < 0.03). Upon interaction of Trp312 with Asn370, formation of an HB between residues Asp315 and Ser366 is blocked. Because the HB between Trp312 and Asn370 is missing in GCase(N370S)-pH 4.5, a new HB between residues Asp315 and Ser366 can be formed (Fig. 4B). Consequently, because Asp315 is part of loop 3, which itself interacts directly with loop 1, a change in the behavior of the loops occurs. For NB-DNJ+/GCase-pH 4.5, the active site is stabilized, and the Trp312···Asn370 and Asp315···Ser366 HBs show a similar occupancy without affecting loop 3 (Fig. 4C). For NB-DNJ+/GCase(N370S)-pH 4.5, a 6-fold decrease in the occupancy of the HB between Asp315 and Ser366 is observed (Fig. 4D). These changes in WT and N370S at pH 4.5 upon binding of NB-DNJ+ cause both to show similar trends for the Trp312···Asn370 (no significant difference, p > 0.08) and Asp315···Ser366 (p > 0.1) HBs. Thus, NB-DNJ+ induces a conformational change in GCase(N370S), which prevents the abnormal behavior of HBs and loops 3 and 1 at pH 4.5.
FIGURE 4.
Active site of WT and N370S. A, for GCase-pH 4.5, a π/H interaction is observed between Trp312 and Asn370, which stabilizes the conformation of loop 3 and of interacting loop 1, maintaining a correct active site configuration. B, for GCase(N370S)-pH 4.5, residues 313 and 370 cannot interact, and Trp313 cannot shield the loops and the helix and is therefore not able to prevent a HB between Asp315 and Ser366, and loop 1 and 3 are rearranged. C, NB-DNJ+/GCase-pH 4.5. D, NB-DNJ+/GCase(N370S)-pH 4.5 behave similarly, and no bond is observed between Asp315 and Ser366 for N370S. Loop 1 is in blue; loop 3 is orange, the active site residues are pink, and residues from the interacting helix to the right of the active site are in green.
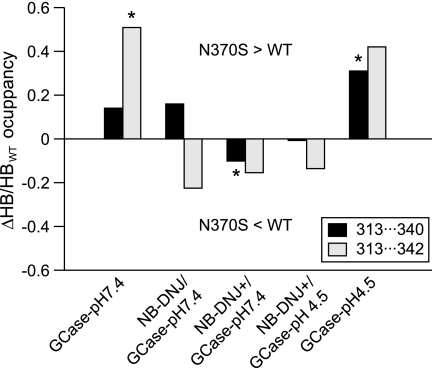
The interaction between Tyr313, which is part of loop 3, and Glu340/Cys342 is important for the stability of the active site residues (46). The difference in HB occupancy was calculated between WT and N370S (Fig. 5). Because the conformation of loop 3 is changed in GCase(N370S)-pH 4.5, Trp312 shows an altered behavior, allowing formation of an HB between Tyr313 and Cys342 at a significantly higher frequency compared with WT (p < 0.03). For GCase(N370S)-pH 4.5, a Tyr313···Glu340 HB (p < 0.02) is observed more often than in the WT. The increase in these HB occupancies results in a decreased distance (measured in Å) between Glu235 and Glu340 (acid catalyst and base) (supplemental Fig. 6), which should be ∼5.5 Å for optimal enzymic activity of glycosyl hydrolases (64). Once NB-DNJ+ is bound to GCase, the effect is reversed, with a small increase in HB occupancies for 313···340 and 313···342 in WT (Tyr313···Glu340, p > 0.4; Tyr313···Cys342, p > 0.15). The smallest differences with respect to HB occupancies between WT and N370S GCase are found for NB-DNJ+/GCase-pH 4.5, where the optimal distance of ∼5.5 Å between Glu235 and Glu340 is restored for GCase(N370S).
FIGURE 5.
HB occupancy for 313···340 and 313···342. The Δratio (Δoccupancy/occupancyWT) between WT and N370S HB occupancy is shown for the residue pairs 313···340 and 313···342. A value >0 indicates a higher occupancy for N370S and <0 a higher occupancy for WT GCase. An asterisk indicates statistical significance.
In summary, the interaction between loops 1 and 3 is altered in N370S due to formation of a different HB network. This causes a suboptimal configuration of the active site residues Glu235 and Glu340, which can be reversed upon NB-DNJ+ binding. Binding of unprotonated NB-DNJ is less effective in the WT compared with N370S, also due to changes in the HB network.
Interactions of Loops 1 and 3
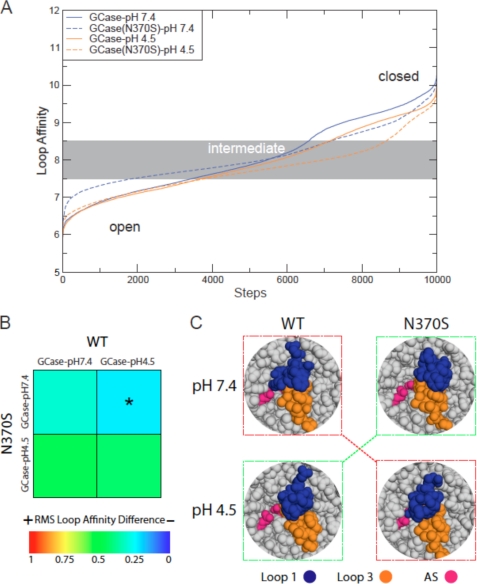
Of the three flexible loops located at the entrance to the active site (23), loops 1 and 3 display the most significant RMSF differences between WT and N370S. The transition from closed to open conformations (Fig. 6A) demonstrates that GCase(N370S)-pH 7.4 shows the highest similarity to GCase-pH 4.5 (Fig. 6B; root mean square loop affinity <0.25). GCase-pH 7.4 shows an interaction between loops 1 and 3, which allows loop 1 to cover most of the active site (Fig. 6C). In contrast, the stabilization observed for loop 1 in GCase(N370S)-pH 7.4 results in a closed conformation of loops 1 and 3, which does not cover all of the active site; this behavior is reversed at pH 4.5, probably due to changes in loop 3, as exemplified by a secondary structure change of a helical turn within this loop (supplemental Fig. 7). Such a secondary structure change had already been suggested based on crystal structures (35), but the MD simulations show the transition of this helical turn into a helix and finally into an unfolded coil structure. Addition of NB-DNJ+ to GCase-pH 7.4/4.5 reduces the differences between WT and N370S (supplemental Fig. 8, root mean square loop affinity <0.25). Comparison of the average affinity scores of WT and N370S with the other four GD mutations reveals that only R496H shows some similarity at pH 4.5 (supplemental Fig. 9). In summary, we have shown that changes in the conformations of loops 1 and 3 are critical for the activity of GCase, because they cause an altered loop interaction and changes in the active site topology in N370S at pH 4.5 and in WT at pH 7.4.
FIGURE 6.
Interaction of loops 1 and 3. A, interaction strength between loops 1 and 3 is plotted for all snapshots, starting at an open configuration, via a medium and, subsequently, a closed loop configuration. B, overall difference for the loop 1 and 3 interaction is presented as a heat plot. Different behavior is colored red (score = 1) and similar behavior is dark-blue (score = 0). The asterisk indicates statistical significance. C, snapshot of a medium and strong interaction for loop 1 (blue) and 3 (orange). Active site residues are in pink. For GCase-pH 4.5 and GCase(N370S)-pH 7.4 (green boxes), the majority of the active site residues are visible for a medium and strong loop interaction. In contrast, for GCase-pH 7.4 and GCase(N370S)-pH 4.5 (red boxes), the majority of the active site is inaccessible.
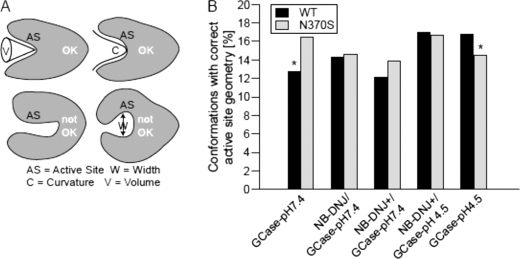
Active Site Geometry
Finally, the impact on active site properties, such as cavity volume, size, and curvature were investigated (Fig. 7A). Snapshots for each simulation were analyzed for all three scores and assigned as “correct” if they did not deviate significantly (>20% for each score) from a set of selected GCase reference crystal structures. At pH 7.4, N370S has a significantly higher percentage of correct cavity geometry (30%) than GCase WT (Fig. 7B). This scenario changes at pH 4.5 where WT GCase shows a significantly higher percentage of structures with correct cavity geometry versus N370S (16%). At pH 4.5, NB-DNJ+ eliminates the differences in cavity geometry between WT and N370S. Analysis of the other Gaucher mutants (supplemental Fig. 10) shows no consistent behavior; F213I, which shows improved activity when binding some small chaperones (65), displays improved cavity geometry with unprotonated NB-DNJ at pH 7.4. As observed for loop affinity, R496H behaves similarly to N370S and WT at pH 4.5. In summary, N370S displays more conformations with nonoptimal cavity geometry at pH 4.5, but not at pH 7.4, compared with WT.
FIGURE 7.
Active site properties. A, depiction of acceptable (accessible, top) and incorrect (inaccessible, bottom) active site cavities. B, percentage of snapshots in the MD simulations for WT and N370S with a correct active site geometry. An asterisk indicates statistical significance.
DISCUSSION
In the current MD study, we have analyzed the structural changes responsible for the reduced activity of the N370S GCase mutation and have examined the effect of the chaperone, NB-DNJ. The major findings are summarized in Fig. 8 and in the supplemental movies 1–4, in which the importance of loops 1 and 3 can be clearly distinguished. At pH 7.4, a destabilization of loop 1 is observed for the WT relative to N370S, which causes restricted access to the active site. At pH 4.5, loop 1 is stabilized in the WT; however, due to changes in the HB network in N370S, specifically the bond between Asp315 and Ser366, loop 3 is displaced, causing a dramatic change in the active site, in the interacting loops, and at the SapC/GCase interface. Binding of NB-DNJ+ can compensate for the majority of these differences. It should be emphasized that all of the MD simulations reported in this study need to be experimentally verified. For instance, changes in the loop structure in N370S could be experimentally verified by amide hydrogen/deuterium exchange mass spectroscopy, a method previously used for GCase (31).
FIGURE 8.
Significant changes between WT and N370S in and around the active site. WT (blue) and N370S (red) are superimposed for GCase-pH 7.4/4.5 and NB-DNJ+/GCase-pH 7.4/4.5. Only loop 1 (top), loop 3 (bottom left with cylinder), and the helix (bottom right) where Asn370 is situated are shown. Important side chains are shown as sticks. The distance between Asp315 on loop 3 and Ser366 on the helix is shown as a dashed line (indicated by #). Residue N370(S) is shown at the bottom end of the helix on the left side; the active site residues are depicted above loop 3. NB-DNJ+ is shown as sticks. The orange circle highlights destabilized loop 1 in the WT compared with the more stable N370S loop 1. For GCase, pH 4.5, loop 3 is dislocated for N370S due to a bond between Asp315 and Ser366. NB-DNJ+ can stabilize both loops 1 and 3 for WT and N370S; at pH 4.5 NB-DNJ+ is less flexible in the active site compared with pH 7.4. The supplemental movies 1–4 illustrate the conformational changes.
In most cases, excellent agreement was found between published experimental data and our MD analyses (Table 3). Moreover, to ensure the integrity and statistical significance of our data, we ran five independent simulations per setting for WT and N370S, and three simulations for the other mutations, producing 110 simulations of 10 ns each. This extremely large computational analysis provides a comprehensive framework to investigate mutations occurring in GCase. However, ultimately it will be only possible to confirm the MD predictions by using additional experimental techniques such as NMR or x-ray crystallography.
TABLE 3.
Comparison of literature observations and MD
| Observation | Reference | Experimental dataa | MD simulations in the current study |
|---|---|---|---|
| N370S pH optimum of 6.2 | 25 | + | N370S less flexible, loop 1 stabilized, better loop 1 and 3 interaction, active site geometry more often correct |
| Loop 3b destabilized at pH 4.5 | 35 | − | Destabilization |
| NB-DNJ stabilizes GCase WT and N370S | 58 | + | Stabilization |
| In crystal structure with chaperones, loops 1 and 3 interact less well | 35 | + | Less interaction for bound NB-DNJ+ |
| Secondary structural change in loop 3 | 35 | + | For N370S, changes are observed at pH 4.5 |
| The activity of N370S at higher pH is similar to the activity of WT at lysosomal pH | 40 | + | Similar trend in the active site configuration and the arrangement of loops 1 and 3 |
| Loop 3 interacts less with the N370S helix, which might favor interaction between residues 315 and 366 | 35 | − | Confirmed in MD simulations |
| 3 orders of magnitude more IFG needed to increase activity of WT compared to N370S | 25 | + | The affinity of unprotonated NB-DNJ at pH 7.4 is lower for the WT than for N370S |
| N370S reduces the interaction of GCase and SapC, which can be reversed by IFG | 25, 38 | + | Interaction with SapC is likely to be disrupted but can be restored by binding of NB-DNJ+ |
a The availability of experimental data is indicated by a plus sign, and lack of experimental data by a minus sign; the information is based on analysis of GCase crystal structures.
b In Ref. 35, loop 3 is referred to as loop 1.
The main differences between WT and N370S at pH 4.5 originate in the altered HB network within the active site. WT GCase can form a stabilizing π/H bond for Trp312···Asn370, particularly at pH 4.5. This bond stabilizes loop 3 and, as a consequence, interacting loop 1. Furthermore, it balances the interaction between Tyr313 and Glu235/Glu340, ensuring the correct distance between Glu235 and Glu340 (64).
For N370S a completely different scenario emerges, with loop 3 destabilized to adopt an altered secondary structure (35). Because Trp312 cannot interact with Asn370, the interaction between Asp315 and Ser366 is not blocked. This new HB causes loop 3 to be pulled toward the helix in which N370S is located, reducing its interaction with loop 1. This rearrangement causes a stronger interaction of Tyr313 with the two active site residues, Glu235 and Glu340, resulting in a reduced distance between them of ∼4.5 Å. In extreme cases, loop 1 can cover the majority of the active site, as reflected in its inferior cavity geometry. Finally, the changes in the loops cause increased flexibility for nearby surface residues. Most of these surface patches are part of the predicted binding interface of GCase with SapC (39). Thus, N370S affects both the activity of GCase at pH 4.5, but may also interfere with the activation of GCase and its ability to bind anionic phospholipids (25, 38). These results demonstrate the importance of loop 1, which is thought to be responsible for membrane association (54).
Our MD analyses also reveal that upon binding of NB-DNJ+, Asp315 and Ser366 interact less in N370S; however, an optimal distance of 5.5 Å is obtained between Glu235 and Glu340 in both N370S and WT. Loops 1 and 3 interact less upon NB-DNJ+ binding (35). Thus, once NB-DNJ+ has stabilized the GCase structure, it enhances the interaction with SapC. Subsequently, NB-DNJ+ is released and replaced by GlcCer, similarly to IFG (25).
NB-DNJ occurs in both an unprotonated and a protonated state at pH 7.4, consistent with its pKa value of 7.1 (23). Thus, a fraction of NB-DNJ (unprotonated) is less soluble at pH 7.4, allowing its binding to the GCase active site. At pH 4.5, protonated NB-DNJ is more soluble and therefore competes less with GlcCer. Our data also suggest that the binding of unprotonated NB-DNJ is less strong, which may destabilize the GCase/NB-DNJ complex at pH 7.4. For the WT, unprotonated NB-DNJ may even dissociate from the active site, which may explain why IFG only increases WT GCase activity at concentrations 3 orders of magnitude higher than for N370S (25). However, as the majority of NB-DNJ at pH 4.5 is protonated, it forms a stable complex with GCase until GlcCer replaces it.
Some of the results of this study also help explain the shifted pH optimum of N370S from pH 5.2 to 6.4 (25). A lower RMSF is observed for N370S, which mainly originates from loop 1 (which is highly flexible in the WT at pH 7.4). This stabilization also includes Cys342, a residue important for GCase activity (32). Together, these changes favorably affect the interaction between loops 1 and 3 in N370S.
Analysis of the results of the other GD mutations (F213I, D409H, L444P, and R496H), demonstrates that only N370S displays a significantly more stable RMSF at pH 7.4. Furthermore, even with bound chaperone, the other mutants do not show any consistent trends, with only F213I significantly stabilized by NB-DNJ. These findings underlie the unique character and behavior of N370S. The novel analytical framework presented here could also be used to investigate the impact of known chaperones on the less frequent GD mutations, and may help for in silico screening of potential new chaperones that could be used in rational drug design.
In summary, we have shown the structural mechanism responsible for the reduced activity of the N370S GCase mutation. Our data suggest that NB-DNJ, and possibly other chaperones, not only ensures the correct initial folding and targeting of GCase to the lysosome, but also mediates the formation of the enzyme-substrate complex in the lysosome and the interaction of GCase with SapC.
Supplementary Material
Acknowledgments
MD calculations were performed at the Academic Computer Centre Cyfronet of Stanislaw Staszic University of Mining and Metallurgy in Krakow, Poland.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3, Figs. 1–10, and Movies 1–4.
- GCase
- glucosylceramidase
- GD
- Gaucher disease
- GlcCer
- glucosylceramide
- HB
- hydrogen bond
- IFG
- isofagomine
- MD
- molecular dynamics
- NB-DNJ
- N-butyl-deoxynojirimycin
- RMSF
- root mean square fluctuation
- SapC
- Saposin C
- SRT
- substrate reduction therapy.
REFERENCES
- 1.Futerman A. H., Zimran A. (2006) Gaucher Disease, Taylor and Francis Group, Boca Raton, FL [Google Scholar]
- 2.Grabowski G. A. (1993) Adv. Hum. Genet. 21, 377–441 [PubMed] [Google Scholar]
- 3.Lee R. E. (1982) Prog. Clin. Biol. Res. 95, 177–217 [PubMed] [Google Scholar]
- 4.Stenson P. D., Mort M., Ball E. V., Howells K., Phillips A. D., Thomas N. S., Cooper D. N. (2009) Genome Med. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimran A., Liphshitz I., Barchana M., Abrahamov A., Elstein D. (2005) Blood Cells Mol. Dis. 34, 197–200 [DOI] [PubMed] [Google Scholar]
- 6.Shiran A., Brenner B., Laor A., Tatarsky I. (1993) Cancer 72, 219–224 [DOI] [PubMed] [Google Scholar]
- 7.Rosenbloom B. E., Weinreb N. J., Zimran A., Kacena K. A., Charrow J., Ward E. (2005) Blood 105, 4569–4572 [DOI] [PubMed] [Google Scholar]
- 8.de Fost M., Vom Dahl S., Weverling G. J., Brill N., Brett S., Häussinger D., Hollak C. E. (2006) Blood Cells Mol. Dis. 36, 53–58 [DOI] [PubMed] [Google Scholar]
- 9.Sidransky E., Nalls M. A., Aasly J. O., Aharon-Peretz J., Annesi G., Barbosa E. R., Bar-Shira A., Berg D., Bras J., Brice A., Chen C. M., Clark L. N., Condroyer C., De Marco E. V., Dürr A., Eblan M. J., Fahn S., Farrer M. J., Fung H. C., Gan-Or Z., Gasser T., Gershoni-Baruch R., Giladi N., Griffith A., Gurevich T., Januario C., Kropp P., Lang A. E., Lee-Chen G. J., Lesage S., Marder K., Mata I. F., Mirelman A., Mitsui J., Mizuta I., Nicoletti G., Oliveira C., Ottman R., Orr-Urtreger A., Pereira L. V., Quattrone A., Rogaeva E., Rolfs A., Rosenbaum H., Rozenberg R., Samii A., Samaddar T., Schulte C., Sharma M., Singleton A., Spitz M., Tan E. K., Tayebi N., Toda T., Troiano A. R., Tsuji S., Wittstock M., Wolfsberg T. G., Wu Y. R., Zabetian C. P., Zhao Y., Ziegler S. G. (2009) N. Engl. J. Med. 361, 1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsui J., Mizuta I., Toyoda A., Ashida R., Takahashi Y., Goto J., Fukuda Y., Date H., Iwata A., Yamamoto M., Hattori N., Murata M., Toda T., Tsuji S. (2009) Arch. Neurol. 66, 571–576 [DOI] [PubMed] [Google Scholar]
- 11.Weinreb N. J., Charrow J., Andersson H. C., Kaplan P., Kolodny E. H., Mistry P., Pastores G., Rosenbloom B. E., Scott C. R., Wappner R. S., Zimran A. (2002) Am. J. Med. 113, 112–119 [DOI] [PubMed] [Google Scholar]
- 12.Pastores G. M., Sibille A. R., Grabowski G. A. (1993) Blood 82, 408–416 [PubMed] [Google Scholar]
- 13.Grabowski G. A., Hopkin R. J. (2003) Annu. Rev. Genomics Hum. Genet. 4, 403–436 [DOI] [PubMed] [Google Scholar]
- 14.Grabowski G. A., Barton N. W., Pastores G., Dambrosia J. M., Banerjee T. K., McKee M. A., Parker C., Schiffmann R., Hill S. C., Brady R. O. (1995) Ann. Intern. Med. 122, 33–39 [DOI] [PubMed] [Google Scholar]
- 15.Beutler E. (2004) PLoS Med. 1, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton N. W., Brady R. O., Dambrosia J. M., Di Bisceglie A. M., Doppelt S. H., Hill S. C., Mankin H. J., Murray G. J., Parker R. I., Argoff C. E., et al. (1991) N. Engl. J. Med. 324, 1464–1470 [DOI] [PubMed] [Google Scholar]
- 17.Platt F. M., Jeyakumar M., Andersson U., Dwek R. A., Butters T. D. (2005) Adv. Exp. Med. Biol. 564, 117–126 [DOI] [PubMed] [Google Scholar]
- 18.Beck M. (2007) Hum. Genet. 121, 1–22 [DOI] [PubMed] [Google Scholar]
- 19.Pastores G. M., Barnett N. L. (2003) Expert Opin. Investig. Drugs 12, 273–281 [DOI] [PubMed] [Google Scholar]
- 20.Fan J. Q. (2003) Trends Pharmacol. Sci. 24, 355–360 [DOI] [PubMed] [Google Scholar]
- 21.Alfonso P., Pampín S., Estrada J., Rodríguez-Rey J. C., Giraldo P., Sancho J., Pocoví M. (2005) Blood Cells Mol. Dis. 35, 268–276 [DOI] [PubMed] [Google Scholar]
- 22.Sawkar A. R., Cheng W. C., Beutler E., Wong C. H., Balch W. E., Kelly J. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15428–15433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brumshtein B., Greenblatt H. M., Butters T. D., Shaaltiel Y., Aviezer D., Silman I., Futerman A. H., Sussman J. L. (2007) J. Biol. Chem. 282, 29052–29058 [DOI] [PubMed] [Google Scholar]
- 24.Luan Z., Higaki K., Aguilar-Moncayo M., Ninomiya H., Ohno K., García-Moreno M. I., Ortiz Mellet C., García Fernández J. M., Suzuki Y. (2009) ChemBioChem 10, 2780–2792 [DOI] [PubMed] [Google Scholar]
- 25.Steet R. A., Chung S., Wustman B., Powe A., Do H., Kornfeld S. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G. N., Reinkensmeier G., Zhang S. W., Zhou J., Zhang L. R., Zhang L. H., Butters T. D., Ye X. S. (2009) J. Med. Chem. 52, 3146–3149 [DOI] [PubMed] [Google Scholar]
- 27.Yu Z., Sawkar A. R., Whalen L. J., Wong C. H., Kelly J. W. (2007) J. Med. Chem. 50, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng W., Padia J., Urban D. J., Jadhav A., Goker-Alpan O., Simeonov A., Goldin E., Auld D., LaMarca M. E., Inglese J., Austin C. P., Sidransky E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfonso P., Rodríguez-Rey J. C., Gañán A., Pérez-Calvo J. I., Giralt M., Giraldo P., Pocoví M. (2004) Blood Cells Mol. Dis. 32, 218–225 [DOI] [PubMed] [Google Scholar]
- 30.Hodanová K., Melková Z., Horowitz M., Hrebícek M. (2003) Eur. J. Hum. Genet. 11, 369–374 [DOI] [PubMed] [Google Scholar]
- 31.Kornhaber G. J., Tropak M. B., Maegawa G. H., Tuske S. J., Coales S. J., Mahuran D. J., Hamuro Y. (2008) ChemBioChem 9, 2643–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liou B., Kazimierczuk A., Zhang M., Scott C. R., Hegde R. S., Grabowski G. A. (2006) J. Biol. Chem. 281, 4242–4253 [DOI] [PubMed] [Google Scholar]
- 33.Montfort M., Chabás A., Vilageliu L., Grinberg D. (2004) Hum. Mutat. 23, 567–575 [DOI] [PubMed] [Google Scholar]
- 34.Kacher Y., Brumshtein B., Boldin-Adamsky S., Toker L., Shainskaya A., Silman I., Sussman J. L., Futerman A. H. (2008) Biol. Chem. 389, 1361–1369 [DOI] [PubMed] [Google Scholar]
- 35.Lieberman R. L., Wustman B. A., Huertas P., Powe A. C., Jr., Pine C. W., Khanna R., Schlossmacher M. G., Ringe D., Petsko G. A. (2007) Nat. Chem. Biol. 3, 101–107 [DOI] [PubMed] [Google Scholar]
- 36.Steet R., Chung S., Lee W. S., Pine C. W., Do H., Kornfeld S. (2007) Biochem. Pharmacol. 73, 1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H. H., Asano N., Ishii S., Ichikawa Y., Fan J. Q. (2006) FEBS J. 273, 4082–4092 [DOI] [PubMed] [Google Scholar]
- 38.Salvioli R., Tatti M., Scarpa S., Moavero S. M., Ciaffoni F., Felicetti F., Kaneski C. R., Brady R. O., Vaccaro A. M. (2005) Biochem. J. 390, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atrian S., López-Viñas E., Gómez-Puertas P., Chabás A., Vilageliu L., Grinberg D. (2008) Proteins 70, 882–891 [DOI] [PubMed] [Google Scholar]
- 40.Liou B., Grabowski G. A. (2009) Mol. Genet. Metab. 97, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawkar A. R., D'Haeze W., Kelly J. W. (2006) Cell. Mol. Life Sci. 63, 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canutescu A. A., Shelenkov A. A., Dunbrack R. L., Jr. (2003) Protein Sci. 12, 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mongan J., Case D. A., McCammon J. A. (2004) J. Comput. Chem. 25, 2038–2048 [DOI] [PubMed] [Google Scholar]
- 44.Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., Onufriev A. (2005) Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dvir H., Harel M., McCarthy A. A., Toker L., Silman I., Futerman A. H., Sussman J. L. (2003) EMBO Rep. 4, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Premkumar L., Sawkar A. R., Boldin-Adamsky S., Toker L., Silman I., Kelly J. W., Futerman A. H., Sussman J. L. (2005) J. Biol. Chem. 280, 23815–23819 [DOI] [PubMed] [Google Scholar]
- 47.Mezei M. (1997) J. Comp. Chem. 18, 812–815 [Google Scholar]
- 48.Case D. A., Darden T., Cheatham T. E., 3rd, Wang J., Duke R., Luo R., Merz K., Pearlman D., Crowley M., Walker R., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. A. (2006) AMBER, version 9, University of California, San Francisco [Google Scholar]
- 49.Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 50.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J. E., Hratchian H. P., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Ayala P. Y., Morokuma K., Voth G. A., Salvador P., Dannenberg J. J., Zakrzewski V. G., Dapprich S., Daniels A. D., Strain M. C., Farkas O., Malick D. K., Rabuck A. D., Raghavachari K., Foresman J. B., Ortiz J. V., Cui Q., Baboul A. G., Clifford S., Cioslowski J., Stefanov B. B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R. L., Fox D. J., Keith T., Al-Laham M. A., Peng C. Y., Nanayakkara A., Challacombe M., Gill P. M. W., Johnson B., Chen W., Wong M. W., Gonzalez C., Pople J. A. (2004) Gaussian 03. Revision C.02 Ed., Gaussian, Inc., Wallingford, CT [Google Scholar]
- 51.Damborský J., Petrek M., Banás P., Otyepka M. (2007) Biotechnol. J. 2, 62–67 [DOI] [PubMed] [Google Scholar]
- 52.Offman M. N., Fitzjohn P. W., Bates P. A. (2006) Bioinformatics 22, 1838–1845 [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 54.Brumshtein B., Wormald M. R., Silman I., Futerman A. H., Sussman J. L. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1458–1465 [DOI] [PubMed] [Google Scholar]
- 55.Porter C. T., Bartlett G. J., Thornton J. M. (2004) Nucleic Acids Res. 32, D129–D133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivasan R., Rose G. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14258–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavallo L., Kleinjung J., Fraternali F. (2003) Nucleic Acids Res. 31, 3364–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieberman R. L., D'aquino J. A., Ringe D., Petsko G. A. (2009) Biochemistry 48, 4816–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaccaro A. M., Tatti M., Ciaffoni F., Salvioli R., Barca A., Scerch C. (1997) J. Biol. Chem. 272, 16862–16867 [DOI] [PubMed] [Google Scholar]
- 60.Wilkening G., Linke T., Sandhoff K. (1998) J. Biol. Chem. 273, 30271–30278 [DOI] [PubMed] [Google Scholar]
- 61.Benedix A., Becker C. M., de Groot B. L., Caflisch A., Böckmann R. A. (2009) Nat. Methods 6, 3–4 [DOI] [PubMed] [Google Scholar]
- 62.Burley S. K., Petsko G. A. (1986) FEBS Lett. 203, 139–143 [DOI] [PubMed] [Google Scholar]
- 63.Brumshtein B., Salinas P., Peterson B., Chan V., Silman I., Sussman J. L., Savickas P. J., Robinson G. S., Futerman A. H. (2010) Glycobiology 20, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies G., Henrissat B. (1995) Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 65.Lin H., Sugimoto Y., Ohsaki Y., Ninomiya H., Oka A., Taniguchi M., Ida H., Eto Y., Ogawa S., Matsuzaki Y., Sawa M., Inoue T., Higaki K., Nanba E., Ohno K., Suzuki Y. (2004) Biochim. Biophys. Acta 1689, 219–228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.