Abstract
In addition to inhibiting insulin receptor and IGF1R kinase activity by directly binding to the receptors, GRB10 can also negatively regulate insulin and IGF1 signaling by mediating insulin receptor and IGF1R degradation through ubiquitination. It has been shown that GRB10 can interact with the C2 domain of the E3 ubiquitin ligase NEDD4 through its Src homology 2 (SH2) domain. Therefore, GRB10 might act as a connector, bringing NEDD4 close to IGF1R to facilitate the ubiquitination of IGF1R by NEDD4. This is the first case in which it has been found that an SH2 domain could colocalize a ubiquitin ligase and its substrate. Here we report the crystal structure of the NEDD4 C2-GRB10 SH2 complex at 2.0 Å. The structure shows that there are three interaction interfaces between NEDD4 C2 and GRB10 SH2. The main interface centers on an antiparallel β-sheet composed of the F β-strand of GRB10 SH2 and the C β-strand of NEDD4 C2. NEDD4 C2 binds at nonclassical sites on the SH2 domain surface, far from the classical phosphotyrosine-binding pocket. Hence, this interaction is phosphotyrosine-independent, and GRB10 SH2 can bind the C2 domain of NEDD4 and the kinase domain of IGF1R simultaneously. Based on these results, a model of how NEDD4 interacts with IGF1R through GRB10 has been proposed. This report provides further evidence that SH2 domains can participate in important signaling interactions beyond the classical recognition of phosphotyrosine.
Keywords: Crystal Structure, Protein-Protein Interactions, SH2 Domains, Signal Transduction, X-ray Crystallography, C2 Domain, GRB10, NEDD4
Introduction
The GRB7 (growth factor receptor-binding protein) family of adaptor proteins includes GRB7, GRB10, and GRB14. These proteins share a conserved molecular architecture: a proline-rich N-terminal region, a Ras-associating-like domain, a pleckstrin homology domain, a family-specific BPS region, and a conserved C-terminal Src homology 2 (SH2)2 domain (1, 2). Their SH2 domains have the ability to recognize phosphotyrosine-containing peptides on a variety of activated tyrosine kinase receptors. GRB7 has been shown to interact with EGF receptor, ErB2 receptor, EphB1, focal adhesion kinase, and platelet-derived growth factor receptor and to be involved in regulating cell migration (3, 4). GRB10 and GRB14 have been shown to interact with insulin receptor (IR), IGF1R (insulin-like growth factor 1 receptor), EGF receptor, Raf1 kinase, and MEK1 kinase and to be involved in cell growth regulation (5–11). The Grb10 gene is maternally imprinted in mice. When the Grb10 gene was disrupted by a gene trap insertion, the mutant mice were ∼30% greater in size than normal, with disproportionately large livers (5, 12). As adults, these mutant mice had improved glucose tolerance, increased muscle mass, and reduced adiposity (13, 14). Furthermore, Grb10 transgenic mice overexpressing GRB10 showed growth retardation and insulin resistance (15). These results indicate that GRB10 plays a negative role in cell growth, as a consequence of hypernegative regulation of the IR and IGF1R. Mice lacking the Grb14 gene were of normal size and had improved glucose tolerance and increased insulin signaling in muscle and liver (16). Therefore, GRB10 and GRB14 are tissue-specific negative regulators of insulin and IGF1 signaling.
Additional research results indicate that GRB10 and GRB14 might contribute to type 2 (non-insulin-dependent) diabetes in humans (5, 17, 18). From a genome-wide association scan in the Old Order Amish, the GRB10 gene has been identified as having the strongest association between type 2 diabetes and a single nucleotide polymorphism (SNP) (19). In subcutaneous adipose tissue, the GRB14 mRNA levels in type 2 diabetes patients were 43% higher than those in normal persons (18).
The mechanisms of the negative regulation of IR and IGF1R by GRB10 and GRB14 are not yet clear. Biochemical studies have shown that GRB10 and GRB14 can bind IR and IGF1R through their BPS and SH2 domains (20–22). The GRB14 BPS region binds as a pseudosubstrate inhibitor in the tyrosine kinase domain of IR to suppress insulin signaling (20). Suppressing endogenous GRB10 expression led to increased IR protein levels, whereas overexpression of GRB10 led to reduced IR protein levels (7). Reduced IR levels were observed in cells with prolonged insulin treatment, and this reduction was inhibited in GRB10-deficient cells (7). The insulin-induced IR reduction was largely reversed by MG-132, a proteasomal inhibitor, but not by chloroquine, a lysosomal inhibitor. IR undergoes insulin-stimulated ubiquitination in cells, and this ubiquitination was inhibited in the GRB10-suppressed cell line (7). Therefore, in addition to inhibiting IR kinase activity by directly binding to IR through its BPS and SH2 domains, GRB10 also negatively regulates insulin signaling by affecting insulin-stimulated degradation of the receptor, possibly through regulation of ubiquitination of the receptor.
GRB10 and GRB14 do not themselves have ubiquitin ligase activity. However, it has been shown that GRB10 binds the C2 domain of the E3 ubiquitin ligase NEDD4 through its SH2 domain, and formation of the complex promotes IGF1-stimulated multiubiquitination, internalization, and degradation of the IGF1R (23–25). GRB10 and NEDD4 remain associated with the IGF1R in early endosomes and caveosomes (23). Therefore, the SH2 domain of GRB10 might act as a connector between the C2 domain of NEDD4 and the kinase domain of IGF1R; proximity of NEDD4 and IGF1R would facilitate the ubiquitination of IGF1R by NEDD4. However, SH2 is typically highly specialized for the recognition of phosphotyrosine and has only one binding site for phosphotyrosine-containing peptides (the Tyr(P)-binding pocket). It was not clear how the SH2 domain of GRB10 can bind the C2 domain of NEDD4 and the kinase domain of IGF1R simultaneously, although the possibility of a second binding site was suggested by the observation that the NEDD4 C2-GRB10 SH2 interaction is phosphorylation-independent (25). Here we report the crystal structure of the NEDD4 C2-GRB10 SH2 complex, which clearly shows that the SH2 domain of GRB10 binds the C2 domain of NEDD4 through a surface region distinct from the classical Tyr(P)-binding pocket.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA encoding the mouse GRB10 SH2 domain (residues 429–536) was amplified from the plasmid pcDNA-Grb10γ (a kind gift from Dr. Junlin Guan) and inserted into the expression vector pQE80 to make a pQE80-SH2 expression construct. This construct was transferred into BL21(DE3) (Novagen) for protein expression. Protein expression was induced at 37 °C for 4 h with 0.3 mm IPTG. The cells were harvested by centrifugation and then suspended in binding buffer (500 mm NaCl, 50 mm Tris-HCl, pH 8.5, 10 mm imidazole, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride). Cell lysis was carried out by sonication. After centrifugation, the supernatant was applied to a nickel affinity column. After protein binding, the column was washed thoroughly with 100 volumes of binding buffer followed by 10 volumes of washing buffer (500 mm NaCl, 50 mm Tris-HCl, pH 8.5, 40 mm imidazole, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride). The protein was then eluted from the column with 5 volumes of elution buffer (200 mm NaCl, 300 mm imidazole-HCl, pH 7.5, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride). The protein solution was concentrated and further purified by FPLC using a Superdex 200 column (GE Healthcare) with an elution buffer containing 0.15 m NaCl and 5 mm Tris-HCl (pH 7.5).
The cDNA encoding the full-length mouse GRB10γ (an isoform of GRB10) was inserted into the expression vector pMAL-2C to make a pMAL-2C-Grb10γ expression construct. This construct was transferred into BL21(DE3) for protein expression. The harvested cells were lysed in the binding buffer (100 mm NaCl, 50 mm Tris-HCl, pH 8.5, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride) by sonication, and the supernatant was applied to a maltose-agarose column. The column was washed thoroughly with 100 volumes of binding buffer, and the maltose-binding protein-GRB10γ fusion protein was eluted with 3 volumes of elution buffer (100 mm NaCl, 50 mm maltose, 50 mm Tris-HCl, pH 8.0, 2 mm CaCl2). After the addition of Factor Xa, this elution solution was incubated at 4 °C for 48 h to cleave off the maltose-binding protein tag. The protein solution was then concentrated and purified with a Superdex 200 column.
The cDNA encoding the mouse NEDD4 C2 domain (residues 108–287) was amplified from the plasmid pBS-Nedd4 (a kind gift from Dr. Andrea Morrione) and inserted into the expression vector pSUMO (Invitrogen) to make a pSUMO-Nedd4 C2 expression construct. This construct was transferred into BL21(DE3) for protein expression. Protein expression was induced at 22 °C overnight with 0.3 mm IPTG. The protein was purified with a nickel column using the method described above. After the column was washed thoroughly with 100 volumes of binding buffer followed by 10 volumes of washing buffer, the protease SUMOase was added to the column and incubated at 4 °C for 24 h to cleave off the SUMO·His tag. The free protein C2 was eluted, concentrated, and further purified with a Superdex 200 column. Truncations of NEDD4 C2 (residues 108–250, 200–300, and 115–287, respectively) were cloned, expressed, and purified using the same procedure as for wild type NEDD4 C2.
To express GRB10 SH2 without any tag, the cDNA encoding the mouse GRB10 SH2 domain (residues 429–536) was inserted into the expression vector pCDFDuet-1 to make pCDFDuet-SH2. To prepare the NEDD4 C2-GRB10 SH2 complex for crystallization, the plasmids pSUMO-C2 and pCDFDuet-SH2 were co-transferred into the host cell BL21(DE3) for co-expression. In this system, the expression level of NEDD4 C2-SUMO·His was higher than that of GRB10 SH2. Protein expression was induced at 15 °C for 24 h with 0.3 mm IPTG. The cell pellet was suspended in binding buffer (200 mm NaCl, 50 mm Tris-HCl, pH 8.5, 10 mm imidazole, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride) and lysed by sonication. After centrifugation, the supernatant was applied to a nickel affinity column. After protein binding, the column was washed thoroughly with 100 volumes of binding buffer followed by 10 volumes of washing buffer (200 mm NaCl, 50 mm Tris-HCl, pH 8.5, 40 mm imidazole, 5 mm β-mercaptoethanol, and 1 mm benzamidine chloride). Then protease SUMOase was added to the column and incubated at 4 °C for 24 h to cleave off the SUMO·His tag. The free proteins (NEDD4 C2-GRB10 SH2 complex and isolated NEDD4 C2) were eluted, concentrated, and purified with a Superdex 200 column. The fractions containing the NEDD4 C2-GRB10 SH2 complex were collected and further purified using a SourceQ column (GE Healthcare) for FPLC.
Effect of Calcium on the Interaction
GRB10 SH2 (with an N-terminal His tag, GRB10 SH2·His) was mixed with NEDD4 C2 (without tag) in a 1:1 molar ratio (as judged by A280) in the incubating buffer (150 mm NaCl and 50 mm Tris-HCl, pH 8.5) containing 20 mm EGTA or 20 mm CaCl2, on ice, for 2 h. Similarly, GRB10γ (without tag) was mixed with NEDD4 C2·SUMO·His in a 1:1 molar ratio in the incubating buffer containing 20 mm EGTA or 20 mm CaCl2, on ice, for 2 h. Then EGTA or CaCl2 in the mixtures was removed by a sizing column. The fraction containing proteins from the sizing column was collected and applied onto a nickel affinity column. After protein binding, the column was washed with 100 volumes of incubating buffer and further washed with 10 volumes of washing buffer (150 mm NaCl, 50 mm Tris-HCl, and 40 mm imidazole, pH 8.5). 15 μl of gel slurry was drawn for analysis by SDS-PAGE.
Effect of Ionic Strength on the Interaction
GRB10 SH2·His and NEDD4 C2 were mixed in a 1:1 molar ratio in the incubating buffer (50 mm Tris-HCl, pH 8.5). After being incubated on ice for 2 h, the NEDD4 C2-GRB10 SH2·His complex was bound to a nickel column. Then the column was eluted with incubating buffer containing different NaCl concentrations (0, 100, 200, 300, 400, 500, and 1000 mm, sequentially). For each concentration, 2 volumes of incubating buffer were used. The eluted solutions were analyzed by SDS-PAGE.
The Specificity of the Interaction
The cDNAs encoding mouse GRB14 SH2 domain and GRB7 SH2 domain were separately inserted into the expression vector pGEX4T-1. The resulting constructs pGEX4T-GRB14 SH2 and pGEX4T-GRB7 SH2 were co-transferred, respectively, with pSUMO-NEDD4 C2 into the host cell BL21(DE3). Protein expression was induced with 0.3 mm IPTG at 15 °C for 24 h. The cells were collected and resuspended in PBS for lysis by sonication. The supernatant from the cell lysis was purified using a nickel affinity column and a glutathione-agarose column sequentially. After the column was washed thoroughly, 15 μl of gel slurry was drawn and analyzed by SDS-PAGE.
The cDNAs encoding mouse GRB14 and GRB7 were separately inserted into the expression vector pQE80 to express the target proteins with N-terminal His tags. Purified GRB14·His (or GRB7·His) was mixed with NEDD4 C2 (without tag) in a 1:1 molar ratio in the incubating buffer (150 mm NaCl and 50 mm Tris-HCl, pH 8.5). After being incubated on ice for 2 h, the mixture was applied to a nickel affinity column. After protein binding, the column was washed by 100 volumes of incubating buffer followed by 10 volumes of washing buffer (150 mm NaCl, 50 mm Tris-HCl, and 40 mm imidazole, pH 8.5). Then 15 μl of gel slurry was drawn for analysis by SDS-PAGE.
Crystallization and Data Collection
The purified NEDD4 C2-GRB10 SH2 complex showed two protein bands on SDS-polyacrylamide gel, corresponding to NEDD4 C2 and GRB10 SH2, respectively (Fig. 1A). The NEDD4 C2-GRB10 SH2 complex solution was desalted and concentrated to 20 mg/ml. Crystals were grown by the hanging drop vapor diffusion method at 18 °C. Typically, 2 μl of the protein stock was mixed with 2 μl of the reservoir solution consisting of 35% MPD (v/v) and 0.1 m Hepes-HCl buffer, pH 7.5. Crystals were observed after 2 days and reached a typical size of 100 × 100 × 200 μm3 1 month later. Diffraction data were collected at 100 K on beamline A1 at MacCHESS. The diffraction data were reduced using the HKL package (26), and the statistics of data collection and processing are summarized in Table 1.
FIGURE 1.
Interaction between NEDD4 C2 and GRB10 SH2. The two proteins were mixed and incubated in the incubating buffer for 2 h. After being desalted with a sizing column, the mixture was applied onto a nickel affinity column. After being washed with washing buffer, 15 μl of gel slurry was drawn for analysis by SDS-PAGE (for details, see “Experimental Procedures”). A, GRB10γ could form a stable complex with NEDD4 C2·SUMO·His in the presence of 20 mm EGTA (lane 3) or 20 mm CaCl2 (lane 4) or without any additive (lane 5); GRB10γ alone did not bind to the nickel beads (lane 2); NEDD4 C2·SUMO·His alone is shown in lane 1. GRB10 SH2·His could form a stable complex with NEDD4 C2 in the presence of 20 mm EGTA (lane 7) or 20 mm CaCl2 (lane 8) or without any additive (lane 9); NEDD4 C2 alone did not bind to the nickel beads (lane 10); GRB10 SH2·His alone is shown in lane 11. Lane 6, standards. B, wild type (lane 1) and the mutants N519A (lane 3), R533A (lane 12), R431A (lane 13), and H434A (lane 14) of GRB10 SH2·His could form a stable complex with NEDD4 C2, whereas the K505P mutant (lane 2) could not form a complex with NEDD4 C2. NEDD4 C2 alone could not bind to the nickel beads (lane 4); wild type GRB10 SH2·His alone could bind to the nickel beads (lane 10). Neither the truncated NEDD4 C2 (residues 108–250) (lane 5) nor the truncated NEDD4 C2 (residues 200–300) (lane 6) could bind to GRB10 SH2·His; the truncated NEDD4 C2 (residues 115–287) could bind to GRB10 SH2·His (lane 7). Crystals of the NEDD4 C2-GRB10 SH2 complex were picked out from the drop and analyzed by SDS-PAGE (lane 9). Lanes 8 and 11, standards. C, elution profile of the NEDD4 C2-GRB10 SH2·His complex from a Superdex 200 sizing column. The inset shows the analysis of the peak fractions by SDS-PAGE.
TABLE 1.
Data collection and structure refinement statistics
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions a, b, c (Å) | 52.31 70.30 86.47 |
| Resolution (Å) | 50-2.0 (2.03-2.00)a |
| No. of unique observations | 21,866 (1023) |
| Redundancy | 6.5 (4.4) |
| Completeness (%) | 99.4 (96.8) |
| Average I/σI | 35.7 (3.4) |
| Rmerge (%) | 5.2 (36.2) |
| Structure refinement | |
| No. of protein atoms/waters | 2011/95 |
| Resolution (Å) | 50-2.0 |
| Rwork (%) /Rfree (%) | 19.2/23.2 |
| r.m.s. deviations | |
| Bonds (Å)/Angles (degrees) | 0.006/1.057 |
| Average B factor (Å2) | 36.78 |
| Ramachandran plot | |
| Most favored (%) | 95.34 |
| Allowed (%) | 4.66 |
a Values in parentheses are for the highest resolution shell.
Structure Refinement
The structure of the NEDD4 C2-GRB10 SH2 complex was solved by the molecular replacement program Phaser (27) using the structure of human GRB10 SH2 (28) (Protein Data Bank code 1NRV) and the structure of human NEDD4 C2 (Protein Data Bank code 3B7Y) as the search models. The structure was refined using Refmac5 (29) and PHENIX (30). The refinement statistics are given in Table 1. The B factors are constant throughout the protein. For the GRB10 SH2 subunit (average B factor = 33.7 Å2), the B factors of its N terminus (residues 429–438) and C terminus (residues 526–535) are 32.1 and 37.6 Å2, respectively. For the NEDD4 C2 subunit (average B factor = 39.3 Å2), the B factors of its N terminus (residues 112–120) and C terminus (residues 281–287) are 41.1 and 40.5 Å2, respectively. The interface between GRB10 SH2 and NEDD4 C2 was analyzed using the program PISA (31) from the CCP4 suite (32). SDS-PAGE analysis of the NEDD4 C2-GRB10 SH2 crystals showed that the NEDD4 C2 and the GRB10 SH2 in the crystals had the same molecular weights as the original NEDD4 C2 and the original GRB10 SH2, respectively (Fig. 1B), suggesting that both NEDD4 C2 and GRB10 SH2 in the crystal are complete and have not been degraded.
Mutation
Site-directed mutations of GRB10 SH2 (K505P, N519A, R533A, R431A and H434A, respectively) were carried out using the Phusion mutation kit (New England Biolabs). The interaction between the GRB10 SH2 mutants and NEDD4 C2 was analyzed using the co-expression system described above.
RESULTS
The NEDD4 C2-GRB10 SH2 Interaction Is Ca2+-independent and Phosphorylation-independent
As shown by the yeast two-hybrid method, GRB10 can interact with the C2 domain of NEDD4 through its SH2 domain (25). The C2 domain is a conserved module of about 120 residues, which was first found in protein kinase C (33). In most proteins, the C2 domain binds phospholipids in a Ca2+-dependent manner (34, 35); in some proteins, it mediates protein-protein interactions (36). The C2 domain of NEDD4 has been shown to target NEDD4 to the plasma membrane in response to Ca2+ (37). However, the interaction between NEDD4 and GRB10 has been demonstrated to be Ca2+-independent by the co-immunoprecipitation method (25). We performed a series of in vitro interaction experiments using purified C2 domain and purified SH2 domain (or full-length GRB10). As shown in Fig. 1A, NEDD4 C2 could form a complex with GRB10 SH2 in the presence of either 20 mm EGTA or 20 mm CaCl2, suggesting that the interaction between NEDD4 C2 and GRB10 SH2 was Ca2+-independent. NEDD4 C2 could also form a complex with full-length GRB10γ (an isoform of GRB10), and this interaction was also Ca2+-independent (Fig. 1A).
The SH2 domain is a conserved module of about 100 residues, which is highly specialized for the recognition of phosphotyrosine, with only a few exceptions to date (38). However, GRB10 has been suggested to preferentially associate with unphosphorylated NEDD4 (based on co-immunoprecipitation experiments) (25). In our work, NEDD4 C2 was expressed in Escherichia coli, in which expressed proteins cannot be phosphorylated. The NEDD4 C2 domain produced in this system could form a complex with GRB10 SH2 or GRB10γ, indicating that binding of GRB10 SH2 to NEDD4 C2 is phosphorylation-independent.
Both the NEDD4 C2-GRB10 SH2 complex and the NEDD4 C2-GRB10γ complex were stable in low ionic strength solution (<200 mm NaCl). The complexes dissociated in a solution containing more than 500 mm NaCl, in the presence or absence of 20 mm EGTA (or 20 mm CaCl2) (data not shown). These results suggested that the main interaction between NEDD4 C2 and GRB10 SH2 was not hydrophobic and was unaffected by Ca2+.
The molecular mass of GRB10 SH2 is about 12 kDa. The molecular mass of NEDD4 C2 is about 19 kDa. As determined by a Superdex200 sizing column, the molecular mass of the NEDD4 C2-GRB10 SH2 complex was about 30 kDa, suggesting that this complex is a heterodimer in solution (Fig. 1C).
The Specificity of the Interaction
All members of the GRB7 adaptor protein family (GRB7, GRB10, and GRB14) contain a Ras-associating-like domain, a pleckstrin homology domain, a family-specific BPS region, and a C-terminal SH2 domain (1, 2). Because the SH2 domains are highly conserved among this family (Fig. 2), we investigated whether NEDD4 C2 could also bind the SH2 domains of GRB7 and GRB14, using the co-expression method. In the E. coli co-expression system, NEDD4 C2 was expressed with a SUMO·His tag, which could bind to a nickel-agarose column, whereas GRB7 SH2 and GRB14 SH2 were expressed with a GST tag, which could bind to a glutathione-agarose column but not a nickel column. If the SH2 domain can form a complex with C2·SUMO·His, it should be co-purified with C2·SUMO·His by a nickel column. As shown by SDS-PAGE analysis, the protein purified by a nickel column contained only NEDD4 C2·SUMO·His, whereas the protein purified by a glutathione-agarose column contained only GRB7 SH2·GST or GRB14 SH2·GST. Although both NEDD4 C2·SUMO·His and GRB7 SH2·GST (or GRB14 SH2·GST) were co-expressed in the same cell at high levels, neither of the SH2 domains could be co-purified with NEDD4 C2·SUMO·His by a nickel column, suggesting that neither GRB7 SH2 nor GRB14 SH2 could form a complex with NEDD4 C2. As a control, we showed that GRB10 SH2·GST could form a complex with NEDD4 C2·SUMO·His, suggesting that neither the GST tag nor the SUMO tag could inhibit the interaction between NEDD4 C2 and the SH2 domain.
FIGURE 2.
Alignment of the amino acid sequences of the SH2 domains of GRB7, GRB10, and GRB14. The α-helices and β-strands of GRB10 SH2 are highlighted with red lines and blue arrows, respectively. The underlined peptide of GRB10 is the F β-strand, which forms an antiparallel β-sheet with the C β-strand of NEDD4 C2 in the NEDD4 C2-GRB10 SH2 complex. The residues buried in the interface with NEDD4 C2 are shown with red letters. The two residues in interface I (Lys505 and Asn519) chosen for mutation are highlighted as red boldface letters.
In vitro interactions between NEDD4 C2 and full-length GRB7 (or GRB14) were also investigated using purified proteins. As shown by SDS-PAGE analysis, NEDD4 C2 could not form a complex with GRB7 or GRB14.
Crystal Structure of the NEDD4 C2-GRB10 SH2 Complex
To gain further insights into the interaction between NEDD4 C2 and GRB10 SH2, we co-expressed NEDD4 C2 and GRB10 SH2 in E. coli and determined the crystal structure of the NEDD4 C2-GRB10 SH2 complex at 2.0 Å resolution (Protein Data Bank code 3M7F). Data collection and refinement statistics are given in Table 1.
The NEDD4 C2-GRB10 SH2 complex exists as a heterodimer in the crystal. The most interesting features of the NEDD4 C2-GRB10 SH2 complex structure are the interaction interfaces between the two proteins (Fig. 3A). Interface I is the major interface, with a buried area of 530 Å2. Central to this interface is a small antiparallel β-sheet formed from the F β-strand (residues 502–508) of GRB10 SH2 and the C β-strand (residues 153–160) of NEDD4 C2 (Fig. 3B). It has been shown that a Pro residue can disrupt a β-strand in proteins (39). Therefore, the Lys505 residue of GRB10 SH2 was mutated to Pro to disrupt the F β-strand. The resultant K505P mutant of GRB10 SH2 could not form a complex with NEDD4 C2 in the E. coli co-expression system as wild type GRB10 SH2 did (Fig. 1B). This result suggests that this antiparallel β-sheet plays an important role in stabilizing the NEDD4 C2-GRB10 SH2 complex. Residues in β-strand E and helix B of GRB10 SH2 and in β-strands A, B, and D of NEDD4 C2 also contribute to interface I. There are a total of eight hydrogen bonds between SH2 and C2 in this interface, four main chain-main chain in the small β-sheet and four side chain-main chain elsewhere. Two of the latter are from the side chain of Asn519 in helix B of SH2 to main chain nitrogen and oxygen atoms of Leu177 in the D β-strand of C2 (Fig. 3C). The N519A mutant of GRB10 SH2 could form a complex with NEDD4 C2 (Fig. 1B), suggesting that this pair of hydrogen bonds is not critical to the formation of the GRB10 SH2-NEDD4 C2 complex. There is a possible salt bridge between the acid residue Asp514 of GRB10 SH2 and the alkaline residue Arg179 of NEDD4 C2 (distance ∼4 Å). In this interface, there are also hydrophobic interactions between the two proteins, involving residues Phe496, Phe506, Leu512, Phe515, Tyr516, and Leu518 of GRB10 SH2 with Leu148, Ile155, Leu156, Val159, Ile176, Leu177, and Phe178 of NEDD4 C2 (Fig. 3C).
FIGURE 3.
Crystal structure of the NEDD4 C2-GRB10 SH2 complex. A, ribbon diagram of the entire complex. There are three interaction interfaces between NEDD4 C2 and GRB10 SH2. Interface I includes the F β-strand and the B helix of GRB10 SH2 and (primarily) the C and D β-strands of NEDD4 C2. Interface II includes the N termini of both NEDD4 C2 and GRB10 SH2. Interface III includes the N- and C-terminal regions of GRB10 SH2 and the C terminus of NEDD4 C2. Both the Tyr(P)-binding pocket on GRB10 SH2 and the Ca2+-binding site on NEDD4 C2 are far from these interfaces. The peptides without electron density are shown as dashed lines. B, the antiparallel β-sheet formed between the C β-strand of NEDD4 C2 and the F β-strand of GRB10 SH2 in interface I, containing four hydrogen bonds (shown as red lines with lengths in angstroms). Only the main chains are shown. C, the hydrophobic interactions between NEDD4 C2 and GRB10 SH2 in interface I. The two hydrogen bonds between Asn519 of GRB10 SH2 and Leu177 of NEDD4 C2 are shown as red lines with lengths in angstroms. Only the side chains are shown (except for residue Leu177 of NEDD4 C2, whose main chain and side chain are shown).
Interface II is the smallest interface, with an area of 90 Å2, comprising the N terminus (residues 429–434) and part of the B helix (residue Ile510) of GRB10 SH2 and the N terminus (residues 112–114) of NEDD4 C2 (Fig. 3A). There is one hydrogen bond between the side chain of residue Gln433 of GRB10 SH2 and the carbonyl group of residue Glu112 of NEDD4 C2 (Fig. 4B). A truncated NEDD4 C2 (residues 115–287) lacking the N terminus could form a stable complex with GRB10 SH2 (Fig. 1B), suggesting that interface II does not play an important role in stabilizing the NEDD4 C2-GRB10 SH2 complex.
FIGURE 4.
Superposition of free GRB10 SH2 (blue; Protein Data Bank code 1NRV), GRB7 SH2 (yellow; Protein Data Bank 2QMS), and GRB14 SH2 (red; Protein Data Bank 2AUG) with the GRB10 SH2 subunit (cyan) of the NEDD4 C2-GRB10 SH2 complex. The NEDD4 C2 subunit of the NEDD4 C2-GRB10 SH2 complex is shown in magenta. A, the structure of GRB10 SH2 is similar to those of GRB14 SH2 and GRB7 SH2, with significant conformational differences located at the BC loop, the DE loop, the N-terminal region, and the C-terminal region (marked with arrows). B, detailed structure of the N-terminal region around residue Gln433. The side chain of residue Gln433 of GRB10 SH2 can form a hydrogen bond with the carbonyl group of residue Glu112 of NEDD4 C2 (shown with a red line), whereas the side chain of residue Gln437 of GRB14 SH2 is too close to the carbonyl group of residue Glu112 of NEDD4 C2 (shown with a blue arrow, 1.49 Å). The N-terminal part of GRB7 SH2 (side chains of His426 and Arg427) clashes with the N-terminal part of NEDD4 C2 (residues Leu113 and His114). C, detailed structure of the C-terminal region around residue Arg533. The side chain of residue Arg533 of GRB10 SH2 can form a hydrogen bond with the carbonyl group of residue Pro284 of NEDD4 C2 (shown with a red line), whereas the side chain of residue Arg537 of GRB14 SH2 is too far away from the carbonyl group of residue Pro284 of NEDD4 C2 (shown with a blue arrow, 3.91 Å) to form a hydrogen bond. Moreover, the side chain of residue Arg537 of GRB14 SH2 is too close to residue Pro286 of NEDD4 C2 (<1.5 Å). D, detailed structure of the N-terminal region around residue Arg431. There is a salt bridge interaction between the side chain of residue Arg431 of GRB10 SH2 and the carboxyl group of residue Pro287 of NEDD4 C2 (shown with a red line, 3.22 Å), whereas the side chain of residue Arg435 of GRB14 SH2 is too far away from the carboxyl group of residue Pro287 of NEDD4 C2 (shown with a blue arrow, 8.01 Å) to form a salt bridge interaction. Moreover, the side chain of residue Arg537 of GRB14 SH2 is too close to residue Pro286 of NEDD4 C2 (<1.5Å). The side chain of residue His442 of GRB14 SH2 clashes with residue Pro286 of NEDD4 C2, whereas the corresponding residues in GRB10 SH2 and GRB7 SH2 are glycines that have no side chain.
Interface III is a medium-sized interface, with an area of 312 Å2. The proline-rich C terminus (residues 283–287) of NEDD4 C2 rests against a surface made up of an N-terminal part (residues 431–440) and a C-terminal part (residues 532–535) of GRB10 SH2 (Fig. 3A). There is a hydrogen bond between the side chain of residue Arg533 of GRB10 SH2 and the carbonyl group of residue Pro284 of NEDD4 C2 (Fig. 4C) and a salt bridge between the side chain of Arg431 in SH2 and the C-terminal carboxyl group of C2 (residue Pro287) (Fig. 4D). The particular importance of this interface is suggested by the observation that a 28-residue stretch preceding the C-terminal Pro-rich cluster of C2 is disordered (and therefore invisible) in the crystal, but the cluster itself is well ordered and clearly defined in electron density maps. A truncated NEDD4 C2 (residues 108–250), lacking the C-terminal 37 residues, could not form a stable complex with GRB10 SH2 (Fig. 1B), suggesting that interface III plays an important role in stabilizing the NEDD4 C2-GRB10 SH2 complex. A single site mutation of SH2 at this interface (R533A, R431A, or H434A) could not disrupt the NEDD4 C2-GRB10 SH2 complex (Fig. 1B), indicating that interface III involves a number of interacting residues and is not easily disrupted. NEDD4 C2 truncated to residues 200–300, however, could not form a stable complex with GRB10 SH2 (Fig. 1B), suggesting that interface III by itself is not sufficient for the formation of the NEDD4 C2-GRB10 SH2 complex.
Because the main interactions between NEDD4 C2 and GRB10 SH2 are hydrogen bonds and salt bridges, the complex is expected to be unstable in high salt solution. In fact, as shown above, the complex dissociates in a solution containing more than 500 mm NaCl.
Phosphotyrosine-containing peptides/proteins bind to the Tyr(P)-binding pocket on the surface of an SH2 domain (the classical SH2 binding site) (Fig. 3A). On the surface of GRB10 SH2, this pocket is more than 15 Å away from the three NEDD4 C2-GRB10 SH2 interfaces and has no overlap with them (Fig. 3A); NEDD4 C2 binds to a nonclassical binding site on GRB10 SH2, which explains why the interaction between NEDD4 C2 and GRB10 SH2 is phosphorylation-independent.
Usually, a C2 domain binds Ca2+ through its AB and EF loops (Fig. 3A). This Ca2+-binding site is more than 12 Å away from the three NEDD4 C2-GRB10 SH2 interaction interfaces, and no Ca2+ was found in the interfaces. Hence, the NEDD4 C2-GRB10 SH2 interaction is expected to be Ca2+-independent, as observed in binding studies (Fig. 1A).
Structure Comparison
Superposition of the GRB10 SH2 subunit of the NEDD4 C2-GRB10 SH2 complex with the free GRB10 SH2 domain shows that binding of NEDD4 C2 has not resulted in significant conformational change of GRB10 SH2 (r.m.s. deviation = 0.37 Å) (Fig. 4A), except that the side chain of residue Gln511 has moved from a position that would clash with the main chain of residue Leu177 of NEDD4 C2 to a location well clear of Leu177.
The structure of GRB10 SH2 is similar to those of GRB14 SH2 (r.m.s. deviation 0.92 Å) and GRB7 SH2 (r.m.s. deviation 0.95 Å). Superposition of GRB14 SH2 and GRB7 SH2 onto the GRB10 SH2 subunit of the NEDD4 C2-GRB10 SH2 complex shows that the main differences among these three structures are located at the BC loop, the DE loop, and the N and the C termini (Fig. 4A).
Almost all of the residues involved in interface I between SH2 and NEDD4 C2 are identical or very similar among the three species. One point of difference is the contact between residues 494–496 (QTF) of GRB10 SH2 and residues 153–155 (SGI) of NEDD4 C2. The sequence of the corresponding SH2 residues is EMF in GRB14 and RLY in GRB7 (i.e. a charged rather than merely polar residue is introduced in the first position, and, in the case of GRB7, the large Tyr replaces Phe in the third position). Additionally, the position of the main chain is slightly shifted, relative to the central β-sheet contact, in the other two proteins versus GRB10.
In the other two interfaces, significant differences are apparent among the superposed structures. In interface II, the conformation of the N terminus of GRB7 SH2 is quite different from that of GRB10 and would clash with NEDD4 C2 (Fig. 4B). GRB14 SH2 is less discrepant but does show a positional shift relative to GRB10 as well as a sequence difference in one position, which could affect binding.
In interface III, both GRB7 and GRB14 exhibit significant differences in their SH2 C-terminal conformations from GRB10, such that the fit of the NEDD4 C2 Pro-rich cluster against SH2 becomes very poor (Fig. 4, C and D). The possibility exists that the GRB7 and/or GRB14 SH2 conformation could be modified in the presence of NEDD4 C2 to create more favorable interactions with C2, although in the case of GRB14, a sequence change from Gly to His at position 438 (GRB10 numbering) would create a clash with Pro286 in NEDD4 C2 even if the main chain conformation were the same as in GRB10 (Fig. 4D). In any case, the binding studies described above argue strongly against such a conformational change.
NEDD4 C2 contains eight β-strands and no α-helices. Comparison of the NEDD4 C2 subunit of the NEDD4 C2-GRB10 SH2 complex with the free NEDD4 C2 domain (superposition r.m.s. deviation 0.53 Å) shows that the main differences between the two structures are located at the BC loop and the N-terminal part, regions that interact with the GRB10 SH2 subunit.
DISCUSSION
SH2 domains are well known as binding modules for phosphotyrosine-containing peptides. The classical SH2/Tyr(P)-containing peptide interactions play important roles in numerous cell signaling pathways (38, 40). A typical SH2 domain comprises a seven-stranded β-sheet core flanked by two α-helices (41). SH2 domains are highly specialized for the recognition of Tyr(P) residues. On the classical substrate-binding site of an SH2 domain, the target Tyr(P)-containing peptide is usually bound in an extended conformation. The highly conserved Tyr(P)-binding site contains an invariant arginine and a second positively charged residue coordinating the phosphate moiety (42). Residues in the target peptide that are located downstream of the Tyr(P) residue confer specificity on the interaction (43).
It has been shown that the SH2 domains of GRB10 and GRB14 can bind the activation loop of the kinase domains of IR and IGF1R through classical Tyr(P)-SH2 interactions (44). On the other hand, there was evidence showing that GRB10 could form a complex with the E3 ubiquitin ligase NEDD4 through the GRB10 SH2-NEDD4 C2 interaction. This interaction is phosphotyrosine-independent and Ca2+-independent (25). Furthermore, there was also evidence suggesting the existence of a GRB10-NEDD4-IGF1R complex (23). The crystal structure of the GRB10 SH2-NEDD4 C2 complex, reported here, provides a structural basis for how the SH2 domain of GRB10 can bind the C2 domain of NEDD4 and the kinase domain of IGF1R simultaneously. All of the three NEDD4 C2 recognition sites on GRB10 SH2 are far away (more than 15 Å) from, and do not overlap with, the classical Tyr(P)-containing peptide binding pocket (Fig. 3A); binding of the kinase domain of IGF1R at the Tyr(P)-binding pocket (the classical site) does not interfere with binding of the C2 domain of NEDD4. In the NEDD4-GRB10-IGF1R complex, GRB10 serves as a connector to form a bridge between NEDD4 and IGF1R.
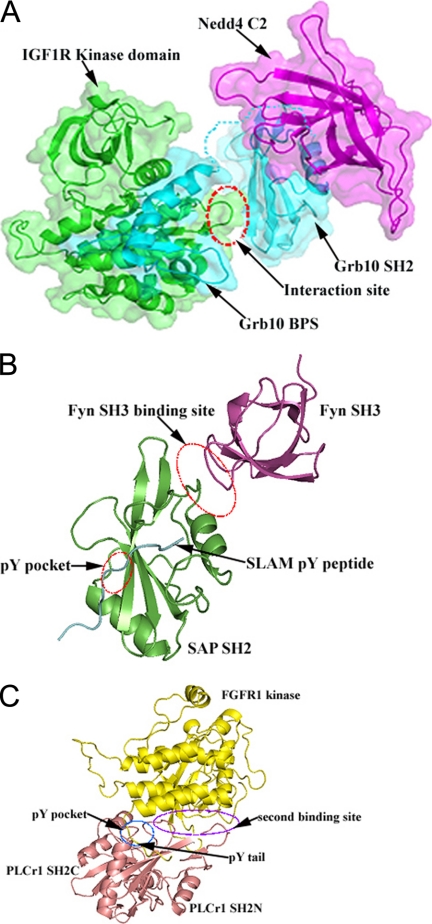
Although GRB10 can form a complex with the E3 ubiquitin ligase NEDD4, GRB10 is not ubiquitinated by NEDD4 inside the cell. However, IGF1R is ubiquitinated by NEDD4 inside the cell, and binding of GRB10 to NEDD4 is critical for this ubiquitination (25). This is explained by the predicted structure of the NEDD4-GRB10-IGF1R complex, in which GRB10 acts as an adaptor to bring NEDD4 close enough to IGF1R to facilitate ubiquitination of IGF1R by NEDD4, through the C2-SH2-kinase domain interaction (Fig. 5A).
FIGURE 5.
Model for the interaction of NEDD4 with IGF1R through GRB10 and examples of SH2 domains that use binding sites different from and not overlapping with the classical Tyr(P)-binding pocket to colocalize a kinase and substrate. A, the interaction between GRB10 BPS (cyan) and the IGF1R kinase domain (green) is modeled based on the crystal structure of the GRB14 BPS-IR kinase complex (25). The interaction between GRB10 SH2 (cyan) and the IGF1R kinase domain is modeled based on the crystal structure of the APS SH2-IR kinase complex (48). The dashed red oval highlights the interaction between GRB10 SH2 and the activation loop of the kinase domain. The missing linker between the BPS and the SH2 domain of GRB10 is shown as a dashed cyan line. B, structure of the SAP SH2-Fyn SH3-SLAM complex (45). The two dashed red ovals highlight the Tyr(P)-binding pocket and the interaction interface between SAP SH2 and Fyn SH3, respectively. C, structure of the PLCγ1 SH2N-SH2C-FGFR1 kinase domain complex (46). The dashed blue oval highlights the Tyr(P)-binding pocket. The dashed purple oval highlights the secondary interaction interface between PLCγ1 SH2N and the FGFR1 kinase domain.
There are a few other cases in which an SH2 domain binds proteins using binding sites other than the classical Tyr(P)-binding pocket (45, 46). For example, the Itk kinase domain docking site on the PLCγ1 SH2C domain surface, which includes residues Glu709, Arg748, Met750, Lys751, and Arg753, is far from and does not overlap with the classical Tyr(P)-binding pocket (47).
Some instances have been reported in which SH2 domains use binding sites different from and not overlapping with the classical Tyr(P)-binding pocket to colocalize a kinase and substrate (45, 46). As shown in Fig. 5B, the SAP SH2 domain binds simultaneously to the SH3 domain of Fyn kinase and to the Tyr(P)-containing peptide of SLAM. This interaction results in the colocalization of Fyn with its SLAM substrate to facilitate the phosphorylation of SLAM by Fyn (45). In the complex, SLAM binds to the classical Tyr(P)-binding pocket of SAP SH2, whereas Fyn SH3 binds SAP SH2 in a phosphotyrosine-independent manner, at a binding site outside of the classical Tyr(P)-binding pocket, involving the DE loop and part of the B helix (45). Another example is the interaction between the fibroblast growth factor receptor kinase (FGFR1) and the N-terminal SH2 domain (SH2N) of PLCγ1 (Fig. 5C) (46). In this complex, there are two interaction sites between PLCγ1 SH2N and the kinase domain of FGFR1; the Tyr(P)-containing tail of the kinase domain binds the classical Tyr(P)-binding pocket on SH2N, whereas a second interaction site involves the BC and DE loops of SH2N.
Unlike the above cases in which the SH2 domain colocalizes a kinase and its substrate, in the GRB10-NEDD4-IGF1R complex, GRB10 SH2 colocalizes a ubiquitin ligase (NEDD4) and its substrate (IGF1R). This case provides further evidence that SH2 domains have a diverse set of other interaction surfaces besides the classical Tyr(P)-binding pocket and that SH2 domains can colocalize an enzyme and its substrate to facilitate the reaction between them. Our work also supports the conclusion that SH2 domains can participate in important signaling interactions beyond the recognition of phosphotyrosine.
This work was supported, in whole or in part, by National Institutes of Health, NCRR, Grant RR-01646. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under Award DMR 0225180.
The atomic coordinates and structure factors (code 3M7F) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SH2
- Src homology 2
- IR
- insulin receptor
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- r.m.s.
- root mean square.
REFERENCES
- 1.Han D. C., Shen T. L., Guan J. L. (2001) Oncogene 20, 6315–6321 [DOI] [PubMed] [Google Scholar]
- 2.Holt L. J., Siddle K. (2005) Biochem. J. 388, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen T. L., Guan J. L. (2004) Front. Biosci. 9, 192–200 [DOI] [PubMed] [Google Scholar]
- 4.Han D. C., Shen T. L., Miao H., Wang B., Guan J. L. (2002) J. Biol. Chem. 277, 45655–45661 [DOI] [PubMed] [Google Scholar]
- 5.Holt L. J., Lyons R. J., Ryan A. S., Beale S. M., Ward A., Cooney G. J., Daly R. J. (2009) Mol. Endocrinol. 23, 1406–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tezuka N., Brown A. M., Yanagawa S. (2007) Biochem. Biophys. Res. Commun. 356, 648–654 [DOI] [PubMed] [Google Scholar]
- 7.Ramos F. J., Langlais P. R., Hu D., Dong L. Q., Liu F. (2006) Am. J. Physiol. Endocrinol Metab. 290, E1262–E1266 [DOI] [PubMed] [Google Scholar]
- 8.Dufresne A. M., Smith R. J. (2005) Endocrinology 146, 4399–4409 [DOI] [PubMed] [Google Scholar]
- 9.Murdaca J., Treins C., Monthouël-Kartmann M. N., Pontier-Bres R., Kumar S., Van Obberghen E., Giorgetti-Peraldi S. (2004) J. Biol. Chem. 279, 26754–26761 [DOI] [PubMed] [Google Scholar]
- 10.Morrione A. (2003) J. Cell. Physiol. 197, 307–311 [DOI] [PubMed] [Google Scholar]
- 11.Giorgetti-Peraldi S., Murdaca J., Mas J. C., Van Obberghen E. (2001) Oncogene 20, 3959–3968 [DOI] [PubMed] [Google Scholar]
- 12.Charalambous M., Smith F. M., Bennett W. R., Crew T. E., Mackenzie F., Ward A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith F. M., Holt L. J., Garfield A. S., Charalambous M., Koumanov F., Perry M., Bazzani R., Sheardown S. A., Hegarty B. D., Lyons R. J., Cooney G. J., Daly R. J., Ward A. (2007) Mol. Cell. Biol. 27, 5871–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Balas B., Christ-Roberts C. Y., Kim R. Y., Ramos F. J., Kikani C. K., Li C., Deng C., Reyna S., Musi N., Dong L. Q., DeFronzo R. A., Liu F. (2007) Mol. Cell. Biol. 27, 6497–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiura H., Miyoshi N., Konishi A., Wakisaka-Saito N., Suzuki R., Muguruma K., Kohda T., Wakana S., Yokoyama M., Ishino F., Kaneko-Ishino T. (2005) Biochem. Biophys. Res. Commun. 329, 909–916 [DOI] [PubMed] [Google Scholar]
- 16.Cooney G. J., Lyons R. J., Crew A. J., Jensen T. E., Molero J. C., Mitchell C. J., Biden T. J., Ormandy C. J., James D. E., Daly R. J. (2004) EMBO J. 23, 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Paola R., Wojcik J., Succurro E., Marucci A., Chandalia M., Padovano L., Powers C., Merla G., Abate N., Sesti G., Doria A., Trischitta V. (2010) J. Intern. Med. 267, 132–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cariou B., Capitaine N., Le Marcis V., Vega N., Béréziat V., Kergoat M., Laville M., Girard J., Vidal H., Burnol A. F. (2004) FASEB J. 18, 965–967 [DOI] [PubMed] [Google Scholar]
- 19.Rampersaud E., Damcott C. M., Fu M., Shen H., McArdle P., Shi X., Shelton J., Yin J., Chang Y. P., Ott S. H., Zhang L., Zhao Y., Mitchell B. D., O'Connell J., Shuldiner A. R. (2007) Diabetes 56, 3053–3062 [DOI] [PubMed] [Google Scholar]
- 20.Depetris R. S., Hu J., Gimpelevich I., Holt L. J., Daly R. J., Hubbard S. R. (2005) Mol. Cell. 20, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein E. G., Gustafson T. A., Hubbard S. R. (2001) FEBS Lett. 493, 106–111 [DOI] [PubMed] [Google Scholar]
- 22.He W., Rose D. W., Olefsky J. M., Gustafson T. A. (1998) J. Biol. Chem. 273, 6860–6867 [DOI] [PubMed] [Google Scholar]
- 23.Monami G., Emiliozzi V., Morrione A. (2008) J. Cell. Physiol. 216, 426–437 [DOI] [PubMed] [Google Scholar]
- 24.Vecchione A., Marchese A., Henry P., Rotin D., Morrione A. (2003) Mol. Cell. Biol. 23, 3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrione A., Plant P., Valentinis B., Staub O., Kumar S., Rotin D., Baserga R. (1999) J. Biol. Chem. 274, 24094–24099 [DOI] [PubMed] [Google Scholar]
- 26.Otwinoski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein E. G., Ghirlando R., Hubbard S. R. (2003) J. Biol. Chem. 278, 13257–13264 [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G., Vagin A., Dodson E. (1997) Acta Cryst. D53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 33.Newton A. C., Johnson J. E. (1998) Biochim. Biophys. Acta 1376, 155–172 [DOI] [PubMed] [Google Scholar]
- 34.Corbalán-García S., Gómez-Fernández J. C. (2010) Biofactors 36, 1–7 [DOI] [PubMed] [Google Scholar]
- 35.Bai J., Chapman E. R. (2004) Trends Biochem. Sci. 29, 143–151 [DOI] [PubMed] [Google Scholar]
- 36.Benes C. H., Wu N., Elia A. E., Dharia T., Cantley L. C., Soltoff S. P. (2005) Cell 121, 271–280 [DOI] [PubMed] [Google Scholar]
- 37.Plant P. J., Yeger H., Staub O., Howard P., Rotin D. (1997) J. Biol. Chem. 272, 32329–32336 [DOI] [PubMed] [Google Scholar]
- 38.Pawson T., Gish G. D., Nash P. (2001) Trends Cell Biol. 11, 504–511 [DOI] [PubMed] [Google Scholar]
- 39.Li S. C., Goto N. K., Williams K. A., Deber C. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filippakopoulos P., Müller S., Knapp S. (2009) Curr. Opin. Struct. Biol. 19, 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawyer T. K. (1998) Biopolymers 47, 243–261 [DOI] [PubMed] [Google Scholar]
- 42.Waksman G., Kumaran S., Lubman O. (2004) Expert Rev. Mol. Med. 6, 1–18 [DOI] [PubMed] [Google Scholar]
- 43.Sondermann H., Kuriyan J. (2005) Cell 121, 158–160 [DOI] [PubMed] [Google Scholar]
- 44.Ceccarelli D. F., Sicheri F. (2009) Nat. Struct. Mol. Biol. 16, 803–804 [DOI] [PubMed] [Google Scholar]
- 45.Chan B., Lanyi A., Song H. K., Griesbach J., Simarro-Grande M., Poy F., Howie D., Sumegi J., Terhorst C., Eck M. J. (2003) Nat. Cell Biol. 5, 155–160 [DOI] [PubMed] [Google Scholar]
- 46.Bae J. H., Lew E. D., Yuzawa S., Tomé F., Lax I., Schlessinger J. (2009) Cell 138, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min L., Joseph R. E., Fulton D. B., Andreotti A. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21143–21148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J., Liu J., Ghirlando R., Saltiel A. R., Hubbard S. R. (2003) Mol. Cell. 12, 1379–1389 [DOI] [PubMed] [Google Scholar]