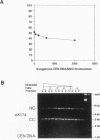
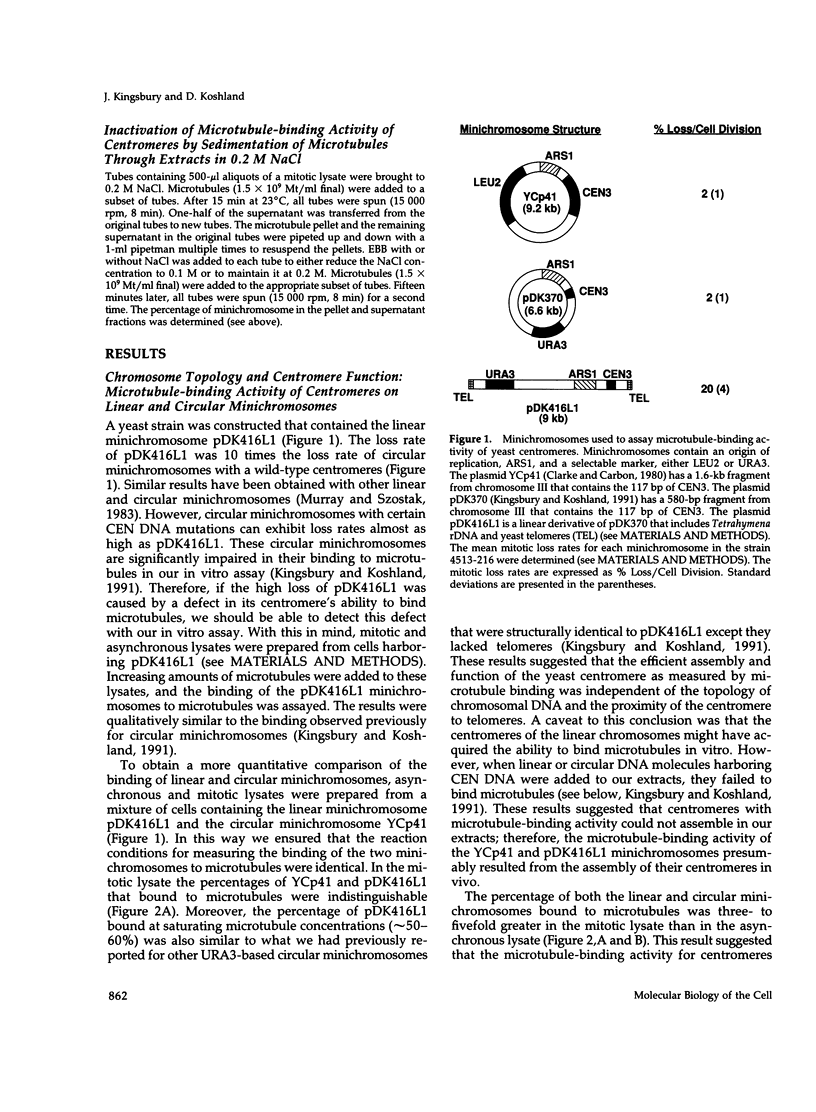
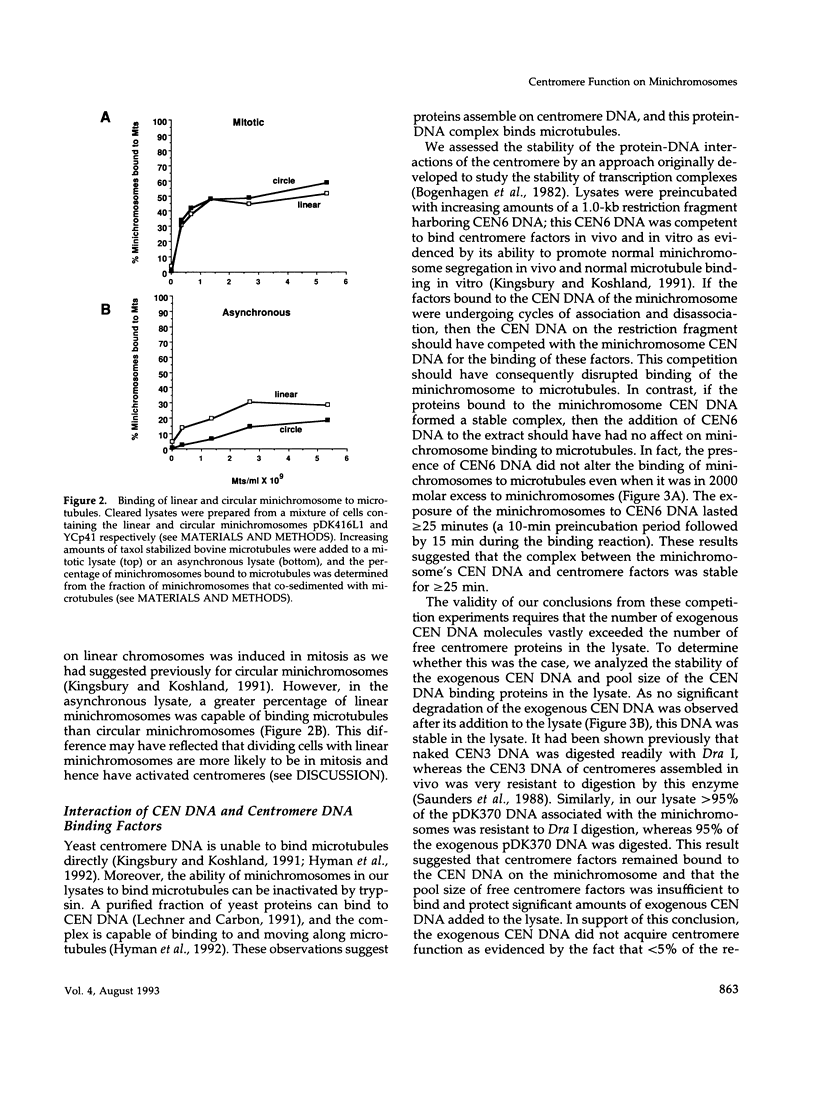
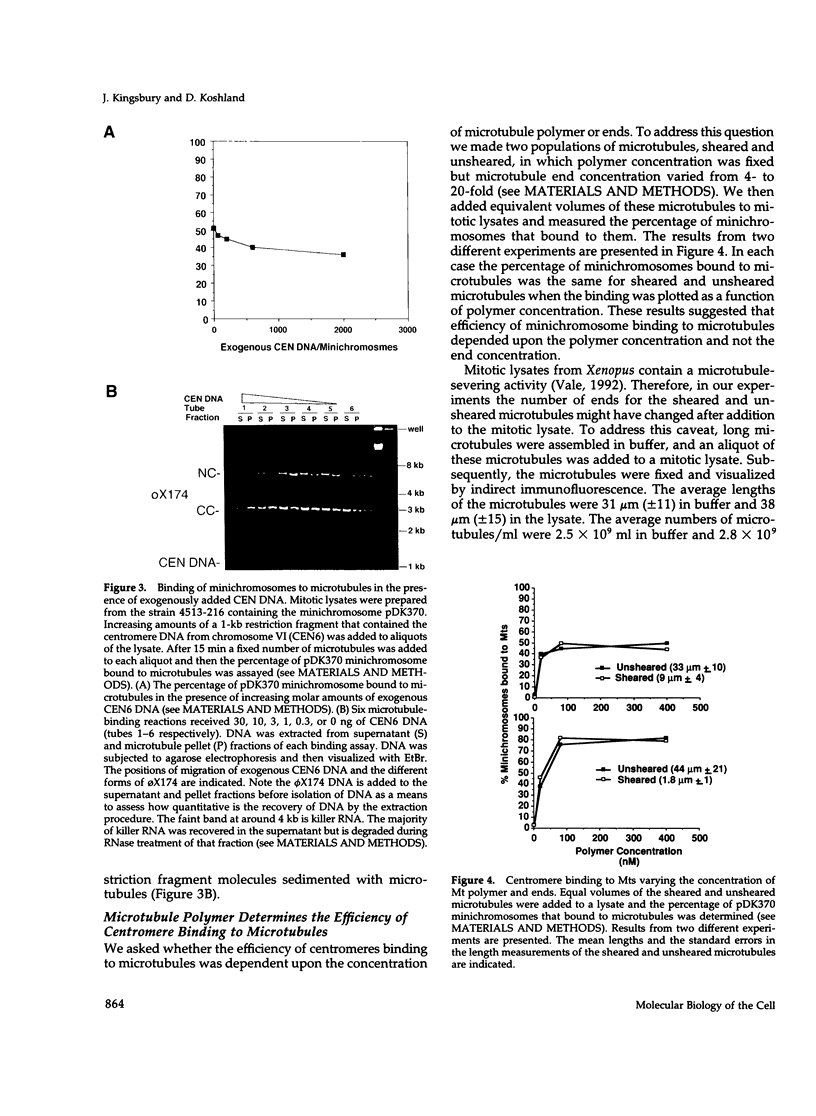
Abstract
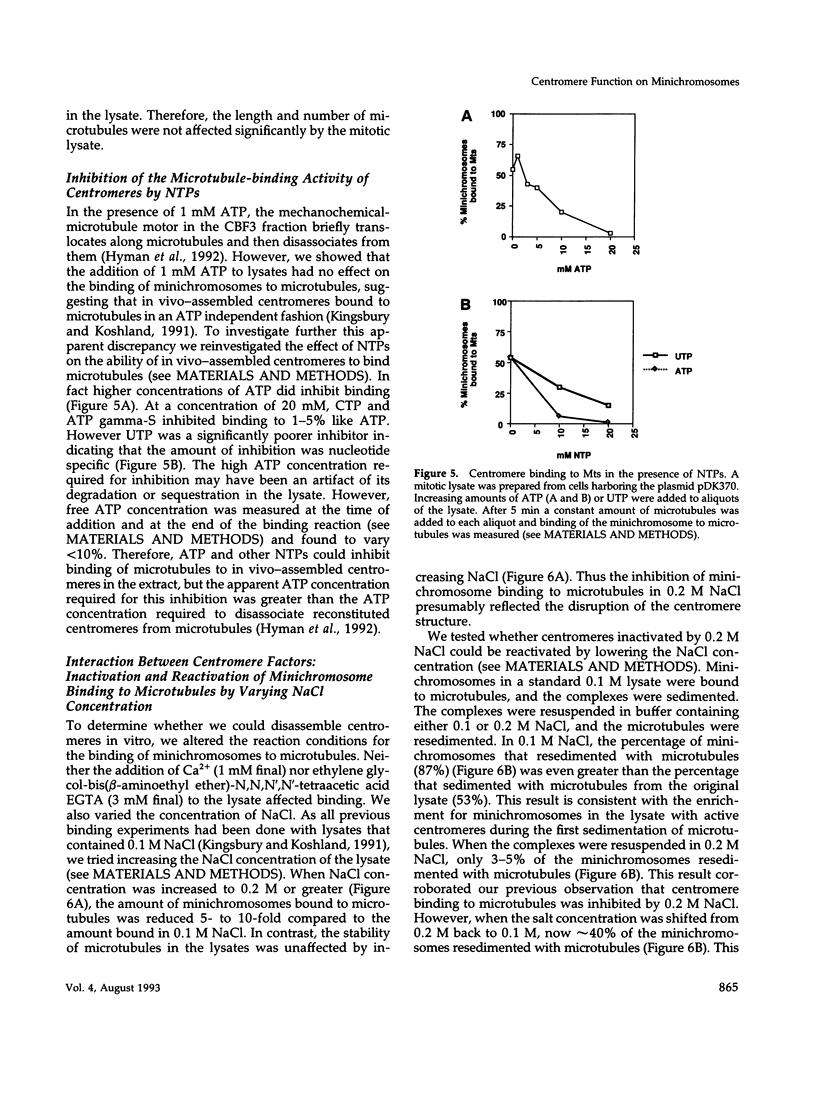
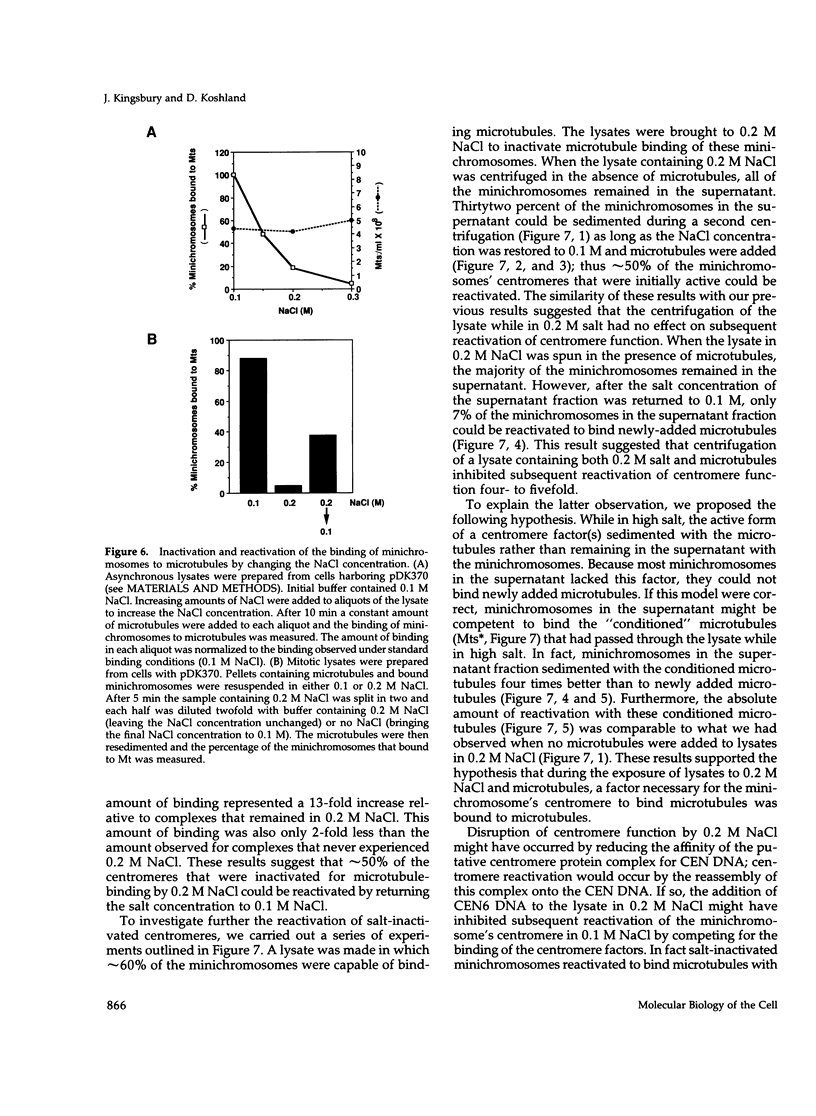
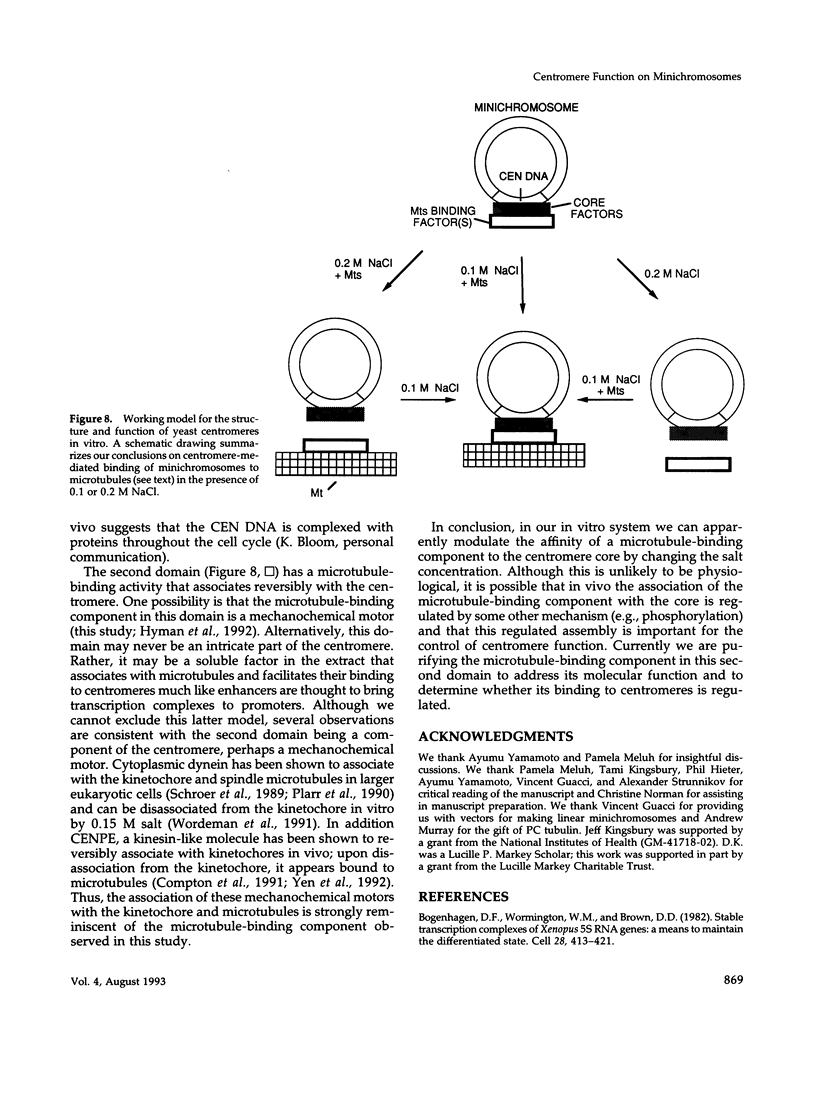
Centromeres are a complex of centromere DNA (CEN DNA) and specific factors that help mediate microtubule-dependent movement of chromosomes during mitosis. Minichromosomes can be isolated from budding yeast in a way that their centromeres retain the ability to bind microtubules in vitro. Here, we use the binding of these minichromosomes to microtubules to gain insight into the properties of centromeres assembled in vivo. Our results suggest that neither chromosomal DNA topology nor proximity of telomeres influence the cell's ability to assemble centromeres with microtubule-binding activity. The microtubule-binding activity of the minichromosome's centromere is stable in the presence of competitor CEN DNA, suggesting that the complex between the minichromosome CEN DNA and proteins directly bound to it is very stable. The efficiency of centromere binding to microtubules is dependent upon the concentration of microtubule polymer and is inhibited by ATP. These properties are similar to those exhibited by mechanochemical motors. The binding of minichromosomes to microtubules can be inactivated by the presence of 0.2 M NaCl and then reactivated by restoring NaCl to 0.1 M. In 0.2 M NaCl, some centromere factor(s) bind to microtubules, whereas other(s) apparently remain bound to the minichromosome's CEN DNA. Therefore, the yeast centromere appears to consist of two domains: the first consists of a stable core containing CEN DNA and CEN DNA-binding proteins; the second contains a microtubule-binding component(s). The molecular functions of this second domain are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Yen T. J., Cleveland D. W. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J Cell Biol. 1991 Mar;112(6):1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S., Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987 Oct;117(2):203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993 May 21;73(4):761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Buhler J. M., Cooper T. G., Carbon J. Isolation and subcloning analysis of functional centromere DNA (CEN11) from Saccharomyces cerevisiae chromosome XI. Mol Cell Biol. 1982 Jan;2(1):82–87. doi: 10.1128/mcb.2.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Gaudet A., Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993 May;121(3):503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Hyman A. A., Middleton K., Centola M., Mitchison T. J., Carbon J. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature. 1992 Oct 8;359(6395):533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- Hyman A. A., Mitchison T. J. Two different microtubule-based motor activities with opposite polarities in kinetochores. Nature. 1991 May 16;351(6323):206–211. doi: 10.1038/351206a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J., Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991 Aug 9;66(3):483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Mitchison T. J., Kirschner M. W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988 Feb 11;331(6156):499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991 Feb 22;64(4):717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Kirschner M. W. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985 Sep;101(3):766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Coordinating cell cycle events. Cold Spring Harb Symp Quant Biol. 1991;56:399–408. doi: 10.1101/sqb.1991.056.01.047. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Schultes N. P., Szostak J. W. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986 May 23;45(4):529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction of artificial chromosomes in yeast. Nature. 1983 Sep 15;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Koval M., Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989 Dec;109(6 Pt 2):3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr C. M., Coue M., Grissom P. M., Hays T. S., Porter M. E., McIntosh J. R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990 May 17;345(6272):263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Alexander S. P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990 Jan;110(1):81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M., Fitzgerald-Hayes M., Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci U S A. 1988 Jan;85(1):175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T. A., Steuer E. R., Sheetz M. P. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989 Mar 24;56(6):937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Furusawa K., Ohashi S., Toyoshima Y. Y., Okuno M., Malik F., Vale R. D. Nucleotide specificity of the enzymatic and motile activities of dynein, kinesin, and heavy meromyosin. J Cell Biol. 1991 Mar;112(6):1189–1197. doi: 10.1083/jcb.112.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer E. R., Wordeman L., Schroer T. A., Sheetz M. P. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990 May 17;345(6272):266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Vale R. D. Microtubule motors: many new models off the assembly line. Trends Biochem Sci. 1992 Aug;17(8):300–304. doi: 10.1016/0968-0004(92)90440-k. [DOI] [PubMed] [Google Scholar]
- Wordeman L., Steuer E. R., Sheetz M. P., Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol. 1991 Jul;114(2):285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. J., Li G., Schaar B. T., Szilak I., Cleveland D. W. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992 Oct 8;359(6395):536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]